Short-Chain Fatty-Acid-Producing Micro-Organisms Regulate the Pancreatic FFA2-Akt/PI3K Signaling Pathway in a Diabetic Rat Model Affected by Pumpkin Oligosaccharides
Abstract
:1. Introduction
2. Methods and Materials
2.1. Oligosaccharide Preparation
2.2. Animal Experimental Design
2.3. Sample Collection
2.4. Gut Microbiota Detected Using Illumina MiSeq Pyrosequencing Platform
2.5. Metabolic Profile Quantified via Gas Chromatography–Mass Spectrometry (GC–MS)
2.6. Quantification of the SCFA Profiles
2.7. Gene Expression Detection
2.8. Statistical Analysis
3. Results and Discussion
3.1. Physiological and Biochemical Properties
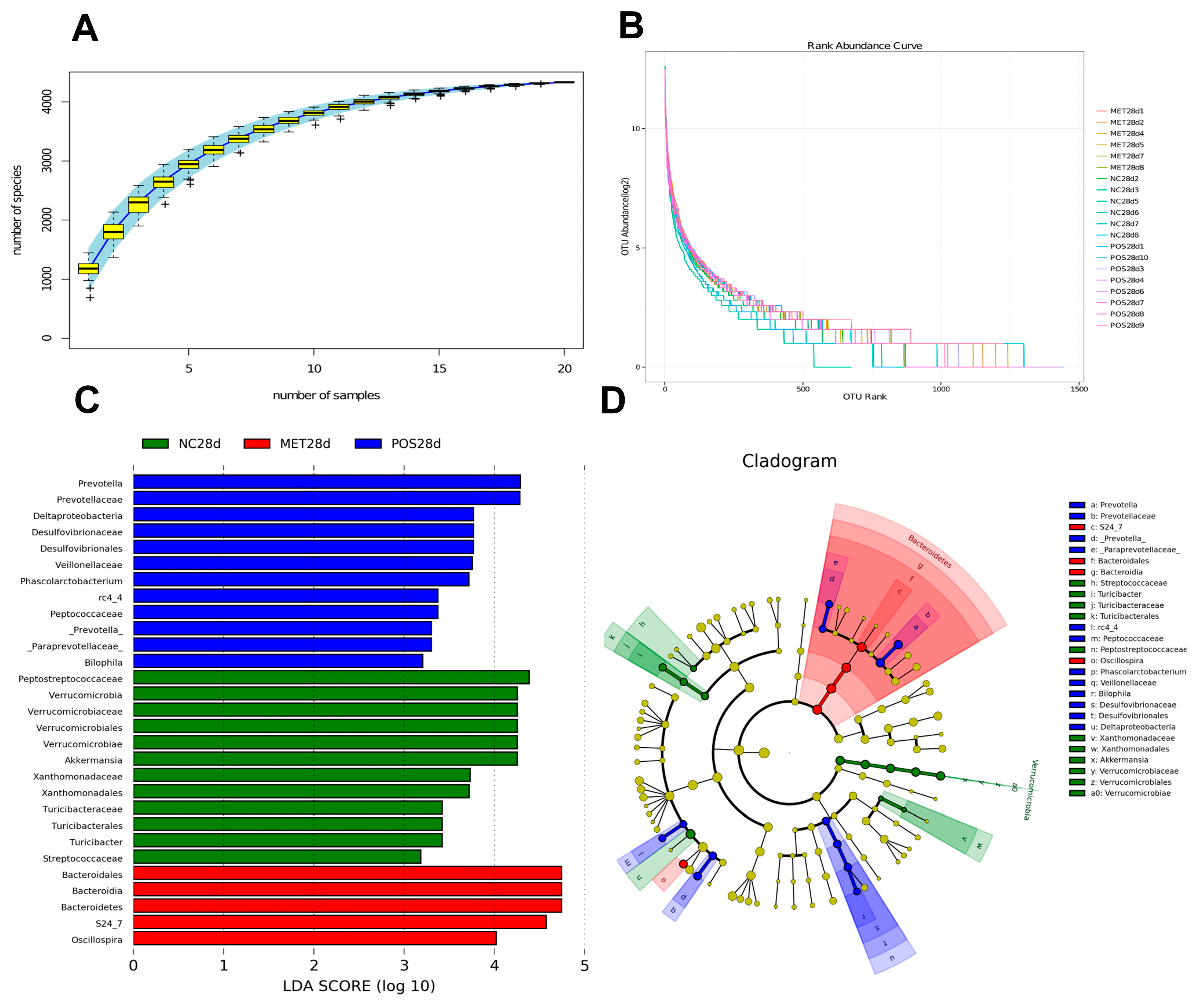
3.2. Gut Microbiota Shifts and Profile
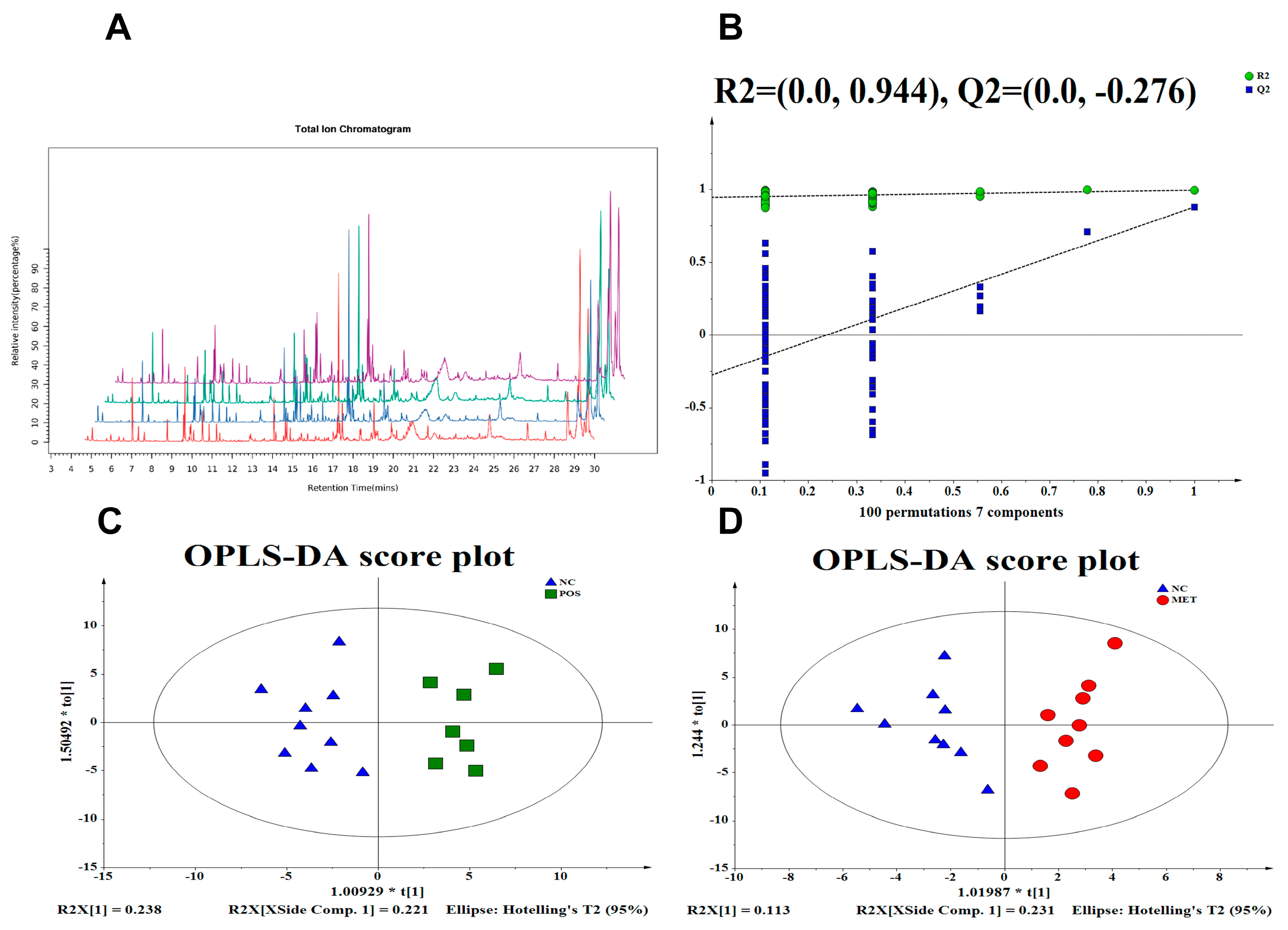
3.3. Gut Metabolic Profiling via GC–MS
3.4. SCFA Concentration in the Gut
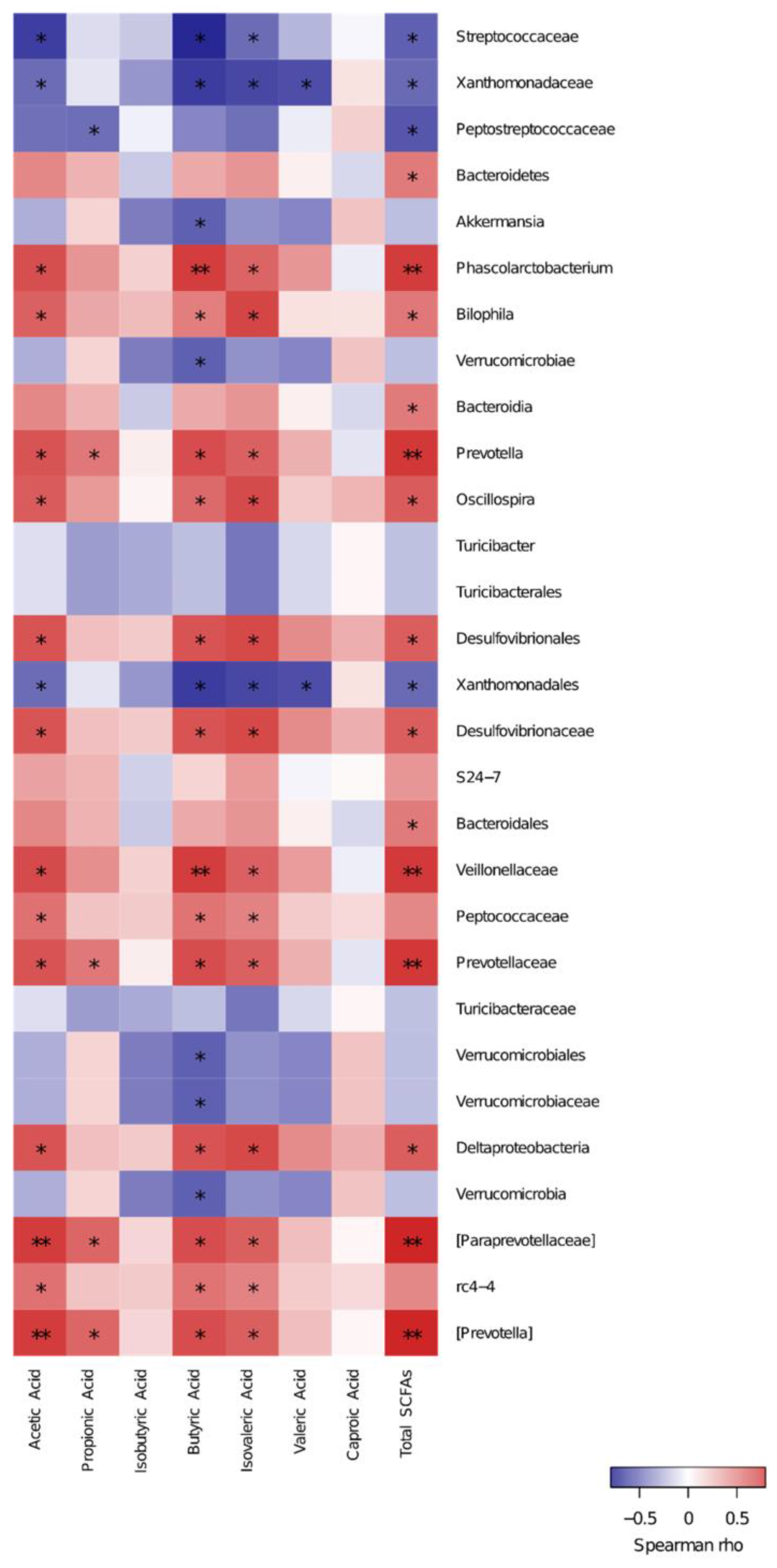
3.5. Spearman Correlation of Key Species and SCFAs
3.6. SCFA-Related Gene Expression in Pancreatic Tissues
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, G.; Liang, L.; Yu, G.; Li, Q. Pumpkin polysaccharide modifies the gut microbiota during alleviation of type 2 diabetes in rats. Int. J. Biol. Macromol. 2018, 115, 711–717. [Google Scholar] [CrossRef]
- Song, Y.; Ni, Y.; Hu, X.; Li, Q. Effect of phosphorylation on antioxidant activities of pumpkin (Cucurbita pepo, Lady godiva) polysaccharide. Int. J. Biol. Macromol. 2015, 81, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, G.; Yu, G.; Song, Y.; Li, Q. Simultaneous decoloration and purification of crude oligosaccharides from pumpkin (Cucurbita moschata Duch) by macroporous adsorbent resin. Food Chem. 2019, 277, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Courtois, J. Oligosaccharides from land plants and algae: Production and applications in therapeutics and biotechnology. Curr. Opin. Microbiol. 2009, 12, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Cloete, L. Diabetes mellitus: An overview of the types, symptoms, complications and management. Nurs. Stand. 2022, 37, 61–66. [Google Scholar] [CrossRef]
- Das, U.N. Is there a Role for Bioactive Lipids in the Pathobiology of Diabetes Mellitus? Front. Endocrinol. 2017, 8, 182. [Google Scholar] [CrossRef]
- Senthilkumar, G.P.; Anithalekshmi, M.S.; Yasir, M.; Parameswaran, S.; Packirisamy, R.M.; Bobby, Z. Role of omentin 1 and IL-6 in type 2 diabetes mellitus patients with diabetic nephropathy. Diabetes Metab. Syndr. 2018, 12, 23–26. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef]
- Napolitano, A.; Miller, S.; Nicholls, A.W.; Baker, D.; Van Horn, S.; Thomas, E.; Rajpal, D.; Spivak, A.; Brown, J.R.; Nunez, D.J. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS ONE 2014, 9, e100778. [Google Scholar] [CrossRef]
- Bielka, W.; Przezak, A.; Pawlik, A. The Role of the Gut Microbiota in the Pathogenesis of Diabetes. Int. J. Mol. Sci. 2022, 23, 480. [Google Scholar] [CrossRef] [PubMed]
- Iatcu, C.O.; Steen, A.; Covasa, M. Gut Microbiota and Complications of Type-2 Diabetes. Nutrients 2021, 14, 166. [Google Scholar] [CrossRef]
- Nie, Y.; Lin, Q.; Luo, F. Effects of Non-Starch Polysaccharides on Inflammatory Bowel Disease. Int. J. Mol. Sci. 2017, 18, 1372. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, Z.; Li, L.; Liang, X.; Wu, Y.; Wang, X.; Ma, H.; Cheng, J.; Zhang, A.; Tang, P.; et al. Gut microbiota induces DNA methylation via SCFAs predisposing obesity-prone individuals to diabetes. Pharmacol. Res. 2022, 182, 106355. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, Y.T.; Yang, Y.X.; Shou, D.; Li, C.Y. Urinary Metabolomic Profiling in Zucker Diabetic Fatty Rats with Type 2 Diabetes Mellitus Treated with Glimepiride, Metformin, and Their Combination. Molecules 2016, 21, 1446. [Google Scholar] [CrossRef]
- Marti, A.; Morell-Azanza, L.; Rendo-Urteaga, T.; Garcia-Calzon, S.; Ojeda-Rodriguez, A.; Martin-Calvo, N.; Moreno-Aliaga, M.J.; Martinez, J.A.; Azcona-San Julian, M.C. Serum and gene expression levels of CT-1, IL-6, and TNF-alpha after a lifestyle intervention in obese children. Pediatr. Diabetes 2018, 19, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, H.; Yu, X.; Wu, Y.; Sui, D. Metformin ameliorates diabetic nephropathy in a rat model of low-dose streptozotocin-induced diabetes. Exp. Ther. Med. 2017, 14, 383–390. [Google Scholar] [CrossRef]
- Miller, D.N.; Bryant, J.E.; Madsen, E.L.; Ghiorse, W.C. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar] [CrossRef]
- Niu, Q.; Li, P.; Hao, S.; Zhang, Y.; Kim, S.W.; Li, H.; Ma, X.; Gao, S.; He, L.; Wu, W.; et al. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015, 5, 9938. [Google Scholar] [CrossRef]
- Liu, G.M.; Hanigan, M.D.; Lin, X.Y.; Zhao, K.; Jiang, F.G.; White, R.R.; Wang, Y.; Hu, Z.Y.; Wang, Z.H. Methionine, leucine, isoleucine, or threonine effects on mammary cell signaling and pup growth in lactating mice. J. Dairy. Sci. 2017, 100, 4038–4050. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; Huang, Q.; Ren, Q.; Chen, S.; Zhang, A.; Zhao, L.; Zhen, Q.; Peng, Y. Impaired beta cell function in Chinese newly diagnosed type 2 diabetes mellitus with hyperlipidemia. J. Diabetes Res. 2014, 2014, 493039. [Google Scholar] [CrossRef] [PubMed]
- Nasri, H.; Rafieian-Kopaei, M. Metformin: Current knowledge. J. Res. Med. Sci. 2014, 19, 658–664. [Google Scholar] [PubMed]
- Choudhary, N.; Ahlawat, R.S. Interleukin-6 and C-reactive protein in pathogenesis of diabetic nephropathy: New evidence linking inflammation, glycemic control, and microalbuminuria. Iran. J. Kidney Dis. 2008, 2, 72–79. [Google Scholar]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Hylckama Vlieg, J.E. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Gibson, G.R. Commentary on: Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2022, 127, 554–555. [Google Scholar] [CrossRef]
- Salgaco, M.K.; Oliveira, L.G.S.; Costa, G.N.; Bianchi, F.; Sivieri, K. Relationship between gut microbiota, probiotics, and type 2 diabetes mellitus. Appl. Microbiol. Biotechnol. 2019, 103, 9229–9238. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Sakamoto, M.; Ohkuma, M. Reclassification of Xylanibacter oryzae Ueki et al. 2006 as Prevotella oryzae comb. nov., with an emended description of the genus Prevotella. Int. J. Syst. Evol. Microbiol. 2012, 62, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Nyman, M.; Fak, F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol. Nutr. Food Res. 2015, 59, 2066–2076. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. A J. Chemom. Soc. 2010, 16, 119–128. [Google Scholar] [CrossRef]
- Sanchez, J.I.; Marzorati, M.; Grootaert, C.; Baran, M.; Van Craeyveld, V.; Courtin, C.M.; Broekaert, W.F.; Delcour, J.A.; Verstraete, W.; Van de Wiele, T. Arabinoxylan-oligosaccharides (AXOS) affect the protein/carbohydrate fermentation balance and microbial population dynamics of the Simulator of Human Intestinal Microbial Ecosystem. Microb. Biotechnol. 2009, 2, 101–113. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl. Environ. Microbiol. 2007, 73, 2009–2012. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Sonnenburg, J.L.; Bäckhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019, 59, 3227–3236. [Google Scholar] [CrossRef] [PubMed]
- Zagato, E.; Pozzi, C.; Bertocchi, A.; Schioppa, T.; Saccheri, F.; Guglietta, S.; Fosso, B.; Melocchi, L.; Nizzoli, G.; Troisi, J.; et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 2020, 5, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; van Esch, B.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharmacol. 2018, 831, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshini, M.; Kotlo, K.U.; Dudeja, P.K.; Layden, B.T. Role of Short Chain Fatty Acid Receptors in Intestinal Physiology and Pathophysiology. Compr. Physiol. 2018, 8, 1091. [Google Scholar]
- Vieira, A.T.; Macia, L.; Galvao, I.; Martins, F.S.; Canesso, M.C.; Amaral, F.A.; Garcia, C.C.; Maslowski, K.M.; De Leon, E.; Shim, D.; et al. A Role for Gut Microbiota and the Metabolite-Sensing Receptor GPR43 in a Murine Model of Gout. Arthritis Rheumatol. 2015, 67, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Tang, C.; Offermanns, S. FFA2 and FFA3 in Metabolic Regulation. Free. Fat. Acid. Recept. 2016, 236, 205–220. [Google Scholar]
- Zhao, L.; Sun, T.; Wang, L. Chitosan oligosaccharide improves the therapeutic efficacy of sitagliptin for the therapy of Chinese elderly patients with type 2 diabetes mellitus. Ther. Clin. Risk Manag. 2017, 13, 739–750. [Google Scholar] [CrossRef]
- Ryuta, M.; Makoto, S.; Yasushi, F.; Naotaka, O.; Mari, M.; Akihito, I.; Shinya, I.; Yoshihiro, K.; Yoshitsugi, I.; Yasuhiro, T. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm. Bowel Dis. 2013, 19, 2848–2856. [Google Scholar]
- Koyasu, S. The role of PI3K in immune cells. Nat. Immunol. 2003, 4, 313–319. [Google Scholar] [CrossRef] [PubMed]
| NC | POS | MET | p Value | |
|---|---|---|---|---|
| Glu (mmol/L) | 26.66 ± 5.24 a | 17.62 ± 3.92 b | 21.68 ± 3.50 b | 0.03 |
| TC (mmol/L) | 5.23 ± 0.35 a | 3.13 ± 0.40 b | 3.03 ± 0.33 b | 0.03 |
| TG (mmol/L) | 1.78 ± 0.21 a | 1.00 ± 0.09 b | 1.05 ± 0.23 b | 0.01 |
| LDL-C (mmol/L) | 2.59 ± 0.32 a | 1.31 ± 0.28 b | 2.59 ± 0.56 a | 0.06 |
| HDL-C (mmol/L) | 1.23 ± 0.14 | 1.47 ± 0.09 | 1.09 ± 0.10 | 0.69 |
| Metabolites | VIP | p Value | Log2fc_ | |||
|---|---|---|---|---|---|---|
| POS | MET | POS | MET | POS | MET | |
| 4-hydroxy-proline | 1.10 | 0.82 | 0.00 | 0.38 | −2.61 | −0.73 |
| 9-(Z)-hexadecenoic acid | 1.73 | 0.60 | 0.00 | 0.48 | −1.21 | −0.17 |
| 9-(Z)-octadecenoic acid | 1.44 | 0.52 | 0.01 | 0.33 | −1.08 | −0.21 |
| Alanine | 1.49 | 0.70 | 0.01 | 0.09 | −1.00 | −0.31 |
| Aspartic acid | 1.51 | 1.56 | 0.01 | 0.11 | 1.16 | 1.18 |
| Butyric acid | 1.33 | 1.76 | 0.01 | 0.01 | 1.38 | 0.96 |
| Cholesterol | 1.15 | 0.62 | 0.03 | 0.54 | −0.65 | −0.19 |
| Docosahexaenoic acid | 1.15 | 0.66 | 0.01 | 0.79 | −1.54 | −0.38 |
| Fructose | 1.34 | 1.53 | 0.02 | 0.08 | −1.34 | −0.72 |
| Fucose | 1.22 | 1.10 | 0.03 | 0.29 | −0.93 | −0.49 |
| Glucaric acid | 1.01 | 0.02 | 0.01 | 0.38 | −1.45 | −0.02 |
| Glutamine | 0.49 | 0.07 | 0.53 | 0.60 | −0.35 | 0.03 |
| Glycerol | 1.4 | 1.74 | 0.01 | 0.05 | −1.44 | −0.86 |
| Glycerol-3-phosphate | 1.14 | 1.10 | 0.00 | 0.09 | −1.05 | −0.49 |
| Hexadecanoic acid | 1.38 | 0.35 | 0.02 | 0.54 | −0.30 | 0.09 |
| Homoserine | 1.42 | 0.61 | 0.02 | 0.43 | −0.94 | −0.24 |
| Isoleucine | 0.71 | 0.33 | 0.04 | 0.06 | −0.34 | −0.11 |
| Isomaltose | 1.29 | 1.64 | 0.00 | 0.02 | −1.66 | −1.07 |
| Isovaleric acid | 1.38 | 0.44 | 0.02 | 0.38 | 1.10 | −0.18 |
| Lactic acid | 0.69 | 1.02 | 0.03 | 0.03 | −2.99 | −2.37 |
| Lactose | 1.15 | 0.85 | 0.03 | 0.48 | −0.86 | −0.32 |
| Leucine | 0.75 | 1.56 | 0.02 | 0.01 | −1.18 | −1.52 |
| Malonic acid | 1.34 | 1.54 | 0.00 | 0.08 | −1.51 | −0.81 |
| Mannose | 1.15 | 1.01 | 0.03 | 0.60 | −1.64 | −0.60 |
| Methionine | 0.58 | 0.71 | 0.40 | 0.38 | −0.27 | −0.19 |
| Nicotinic acid | 1.17 | 0.61 | 0.06 | 0.93 | −0.86 | −0.18 |
| Ornithine | 1.12 | 0.44 | 0.06 | 0.66 | −0.61 | −0.15 |
| Oxalic acid | 1.7 | 3.09 | 0.00 | 0.00 | −1.13 | −1.86 |
| Proline | 0.63 | 0.54 | 0.02 | 0.66 | −0.35 | −0.19 |
| Propionic acid | 1.01 | 1.00 | 0.01 | 0.29 | 1.16 | 0.60 |
| Pyruvic acid | 1.04 | 0.94 | 0.03 | 0.16 | −1.17 | −0.61 |
| Serine | 1.06 | 1.14 | 0.14 | 0.25 | −0.43 | −0.31 |
| Sucrose | 1.39 | 1.04 | 0.01 | 0.16 | −1.05 | −0.41 |
| Threonic acid | 1.39 | 1.57 | 0.01 | 0.08 | −1.69 | −0.96 |
| Threonine | 0.83 | 0.86 | 0.34 | 0.54 | −0.39 | −0.26 |
| Trans-sinapinic acid | 1.16 | 1.09 | 0.00 | 0.05 | −1.35 | −0.70 |
| Valeric acid | 1.25 | 1.17 | 0.00 | 0.08 | 2.02 | 0.85 |
| Valine | 0.65 | 0.33 | 0.04 | 0.03 | −0.27 | 0.22 |
| Xylitol | 1.24 | 1.49 | 0.03 | 0.09 | −1.12 | −0.70 |
| μg/g | NC | POS | MET | p Value |
|---|---|---|---|---|
| Acetic acid | 1505.2 ± 82.92 b | 2208.6 ± 121.56 a | 1718.1 ± 108.93 b | <0.01 |
| Propionic acid | 1244.2 ± 201.89 | 1468.2 ± 291.65 | 1641.5 ± 378.17 | 0.68 |
| Isobutyric acid | 60.25 ± 8.24 | 90.68 ± 15.58 | 82.38 ± 16.68 | 0.34 |
| Butyric acid | 352.86 ± 34.96 c | 899.42 ± 61.75 a | 589.92 ± 89.32 b | <0.01 |
| Isovaleric acid | 46.88 ± 4.88 b | 97.52 ± 12.60 a | 82.11 ± 6.11 a | 0.06 |
| Valeric acid | 90.86 ± 20.00 | 143.72 ± 18.63 | 125.36 ± 11.96 | 0.13 |
| Caproic acid | 32.85 ± 0.18 | 33.38 ± 0.51 | 32.78 ± 0.27 | 0.50 |
| Total SCFAs | 3333.1 ± 312.19 b | 4941.5 ± 320.58 a | 4272.2 ± 507.34 ab | 0.03 |
| NC | POS | MET | p Value | |
|---|---|---|---|---|
| FFA2 | 1.00 ± 0.00 c | 4.97 ± 0.23 a | 2.73 ± 0.18 b | <0.01 |
| IL-6 | 1.00 ± 0.00 a | 0.57 ± 0.02 b | 0.17 ± 0.0.03 c | <0.01 |
| TNF-α | 1.00 ± 0.00 a | 0.61 ± 0.03 b | 0.21 ± 0.03 c | <0.01 |
| PDK1 | 1.00 ± 0.00 | 1.13 ± 0.06 | 1.09 ± 0.04 | 0.32 |
| PDX1 | 1.00 ± 0.00 | 1.18 ± 0.05 | 1.21 ± 0.04 | 0.58 |
| PI3K | 1.00 ± 0.00 b | 2.41 ± 0.12 a | 1.97 ± 0.16 a | <0.01 |
| Akt | 1.00 ± 0.00 b | 2.62 ± 0.15 a | 2.51 ± 0.17 a | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Yu, B.; Li, J.; Zhang, Z.; Tao, H.; Zhao, H.; Lu, Y.; Yuan, C.; Li, Q.; Cui, B. Short-Chain Fatty-Acid-Producing Micro-Organisms Regulate the Pancreatic FFA2-Akt/PI3K Signaling Pathway in a Diabetic Rat Model Affected by Pumpkin Oligosaccharides. Foods 2023, 12, 3559. https://doi.org/10.3390/foods12193559
Liu G, Yu B, Li J, Zhang Z, Tao H, Zhao H, Lu Y, Yuan C, Li Q, Cui B. Short-Chain Fatty-Acid-Producing Micro-Organisms Regulate the Pancreatic FFA2-Akt/PI3K Signaling Pathway in a Diabetic Rat Model Affected by Pumpkin Oligosaccharides. Foods. 2023; 12(19):3559. https://doi.org/10.3390/foods12193559
Chicago/Turabian StyleLiu, Guimei, Bin Yu, Jianpeng Li, Zheng Zhang, Haiteng Tao, Haibo Zhao, Yanmin Lu, Chao Yuan, Quanhong Li, and Bo Cui. 2023. "Short-Chain Fatty-Acid-Producing Micro-Organisms Regulate the Pancreatic FFA2-Akt/PI3K Signaling Pathway in a Diabetic Rat Model Affected by Pumpkin Oligosaccharides" Foods 12, no. 19: 3559. https://doi.org/10.3390/foods12193559
APA StyleLiu, G., Yu, B., Li, J., Zhang, Z., Tao, H., Zhao, H., Lu, Y., Yuan, C., Li, Q., & Cui, B. (2023). Short-Chain Fatty-Acid-Producing Micro-Organisms Regulate the Pancreatic FFA2-Akt/PI3K Signaling Pathway in a Diabetic Rat Model Affected by Pumpkin Oligosaccharides. Foods, 12(19), 3559. https://doi.org/10.3390/foods12193559





