Metabolomic Differences between Viable but Nonculturable and Recovered Lacticaseibacillus paracasei Zhang
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and MRS Medium Formulation
2.2. Cell Activation and Induction of the VBNC State
2.3. Sorting VBNC L. paracasei Zhang Cells by Flow Cytometry
2.4. Recovery of VBNC Cells
2.5. Assessing the Viability and Activity of Recovered VBNC Cells
2.6. UPLC-Q-TOF-MS/MS Analysis
3. Results and Discussion
3.1. Sorting L. paracasei Zhang VBNC Cells by Flow Cytometry
3.2. Resuscitation of VBNC Cells
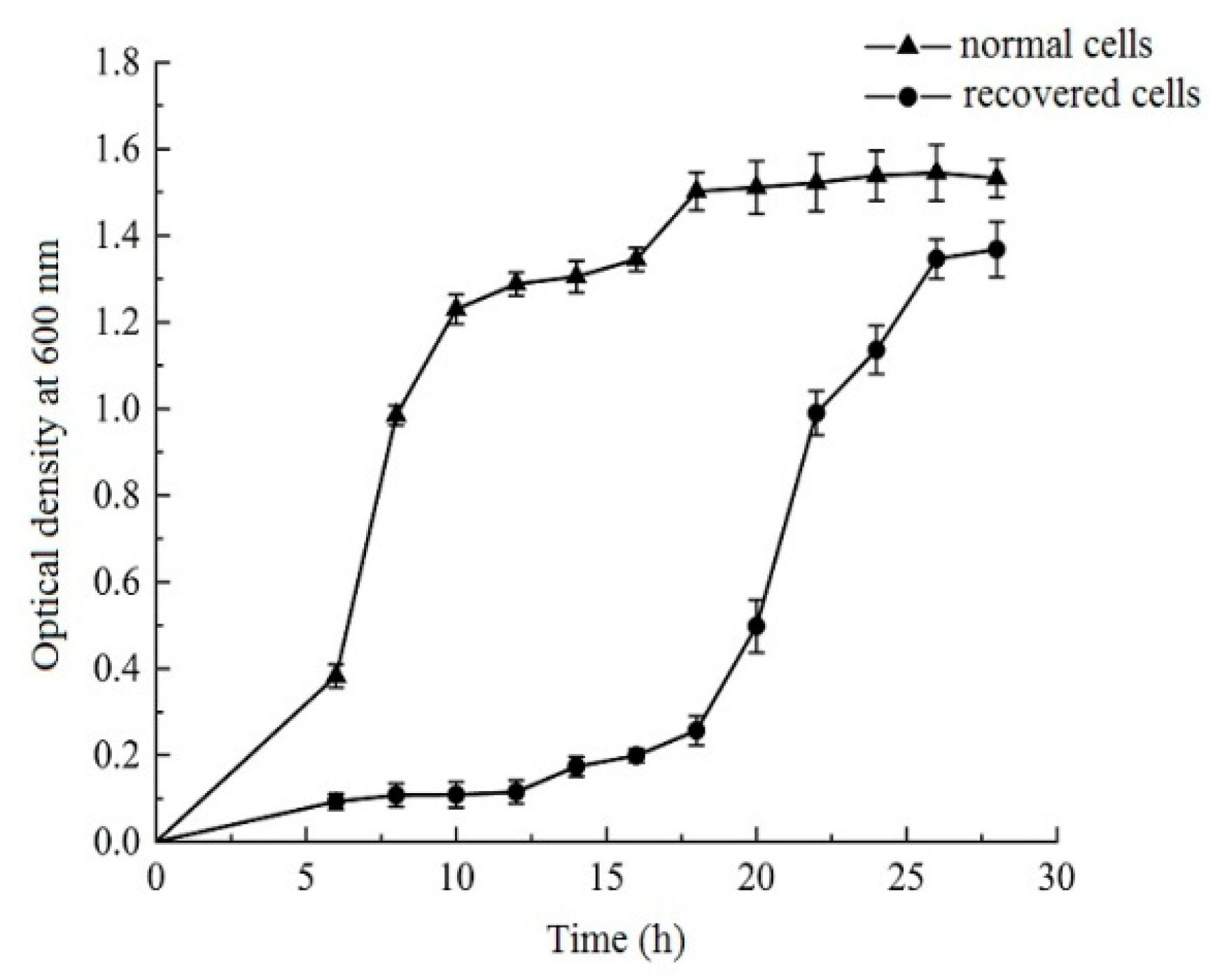
3.3. Growth Curves of Normal and Recovered L. paracasei Zhang
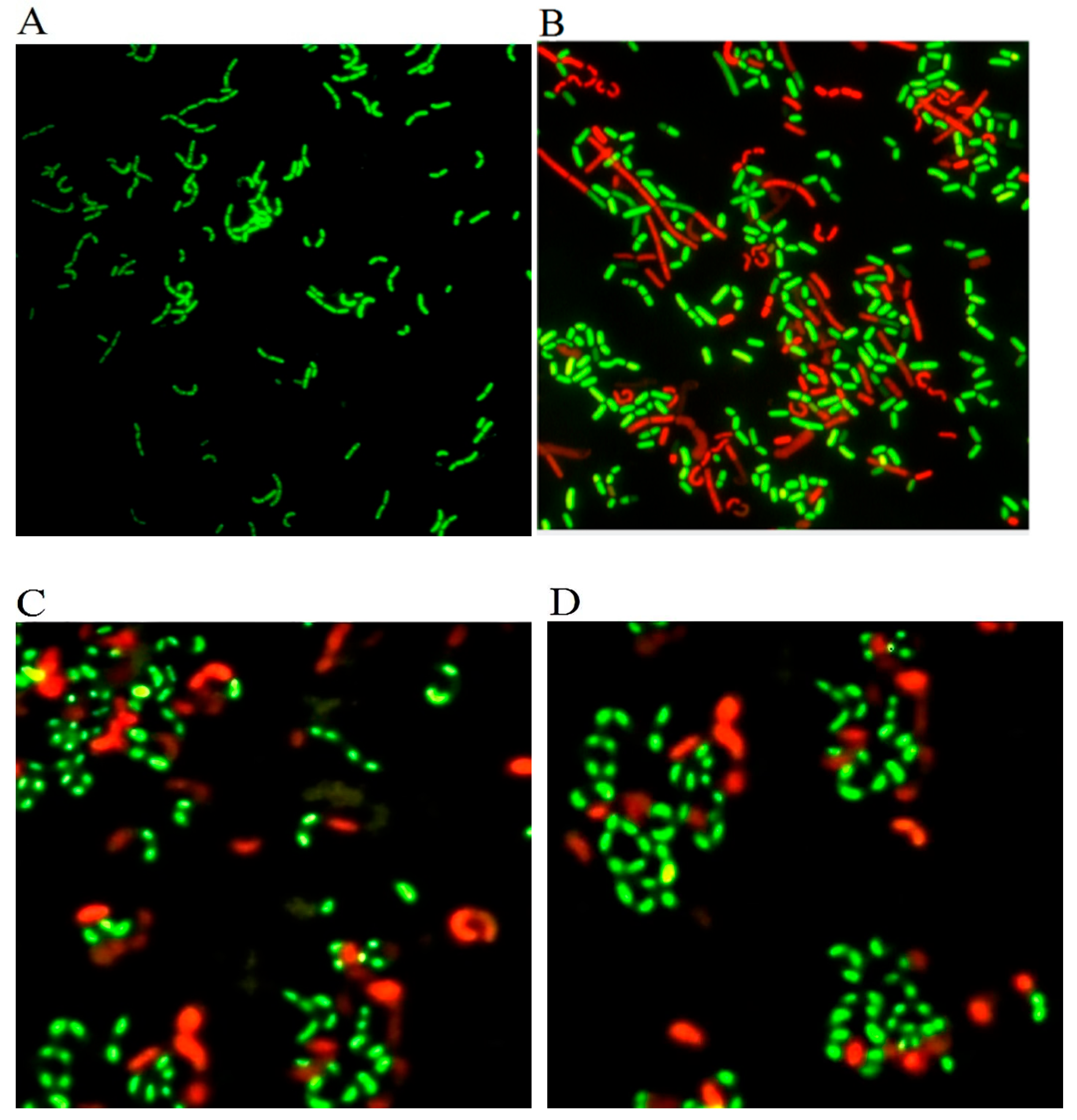
3.4. Evaluation of the Activity of Recovered L. paracasei Zhang by Fluorescent Microscopy
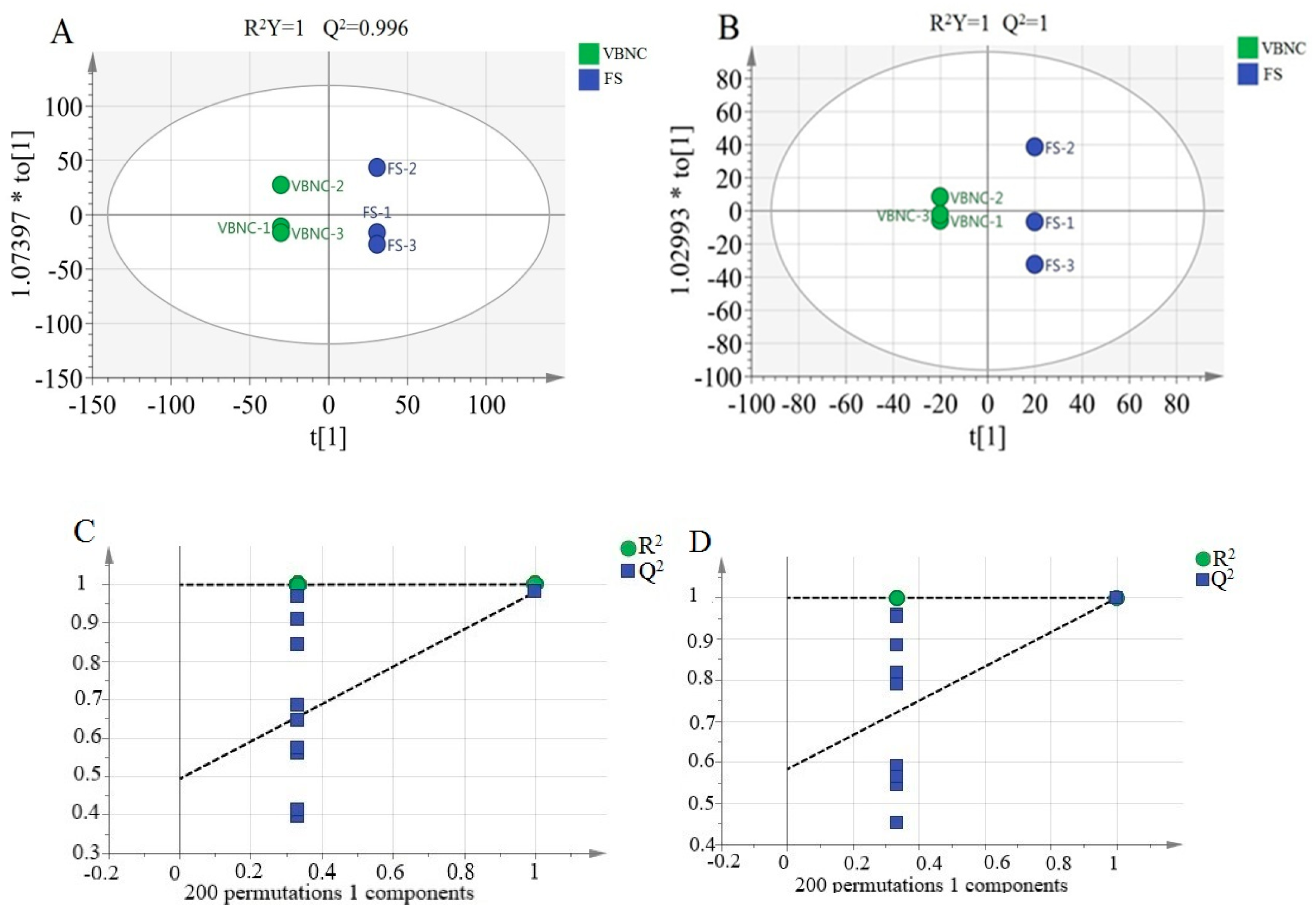
3.5. Metabolomic Differences between VBNC and Resuscitated Cells
3.6. Differential Metabolites between VBNC and Recovered Cells
3.6.1. Amino Acids
3.6.2. Carbohydrates
3.6.3. Lipids
3.6.4. Other Macromolecules
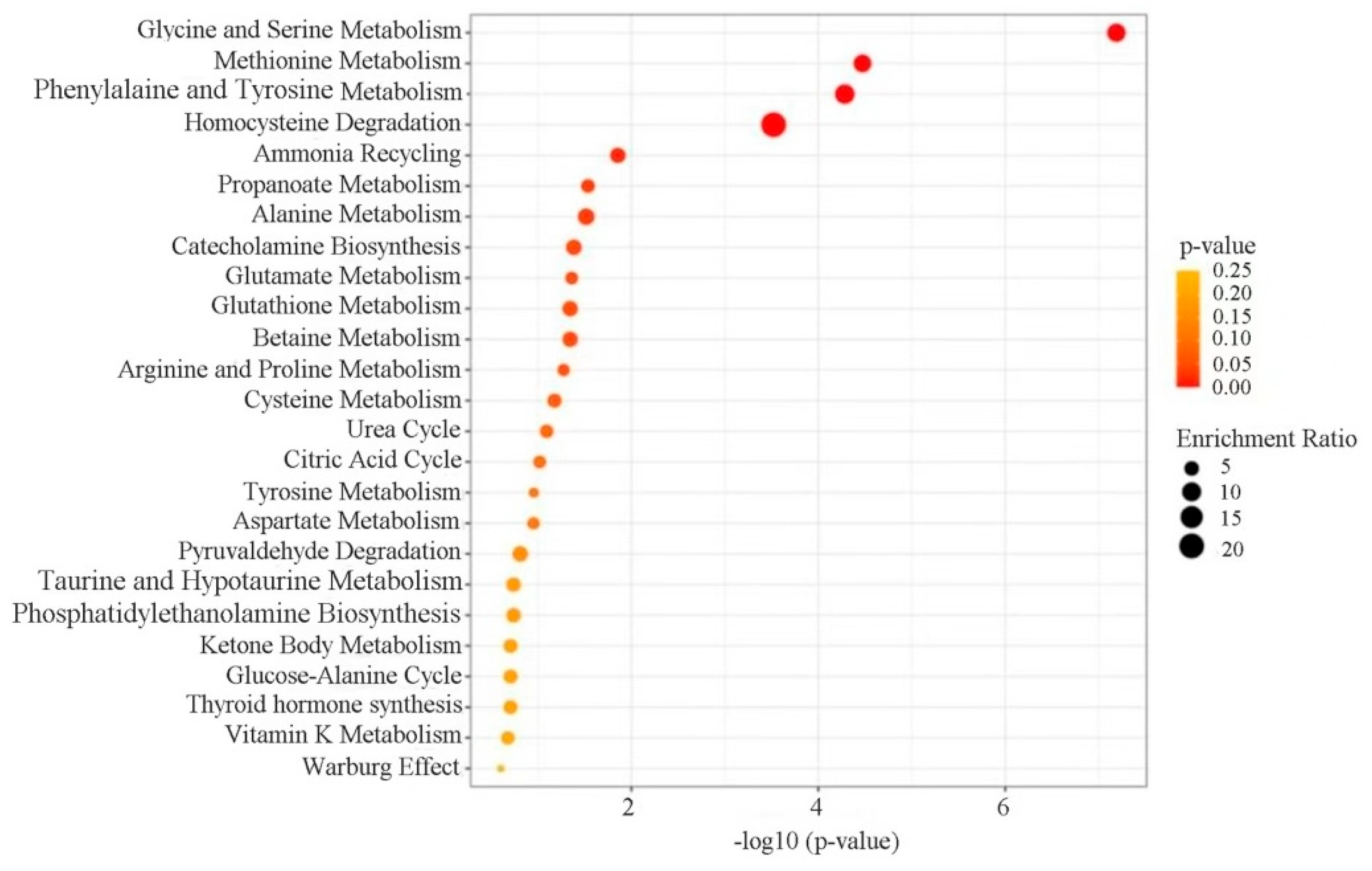
3.7. Enrichment Analysis of Differential Metabolite Pathways
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.Q.; Wu, J.T.; Lv, M.X.; Shao, Z.; Hungwe, M.; Wang, J.J.; Bai, X.J.; Xie, J.L.; Wang, Y.P.; Geng, W.T. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, N.M.; Hussain, M.A.; Britz, M.L. Stress responses in probiotic Lactobacillus casei. Crit. Rev. Food Sci. Nutr. 2015, 55, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, S.; Robben, C.; Alter, T.; Rossmanith, P.; Mester, P. How to evaluate non-growing cells-current strategies for determining antimicrobial resistance of VBNC bacteria. Antibiotics 2021, 10, 115. [Google Scholar] [CrossRef]
- Pan, H.X.; Ren, Q. Wake Up! Resuscitation of Viable but Nonculturable bacteria: Mechanism and potential application. Foods 2022, 12, 82. [Google Scholar] [CrossRef]
- İzgördü, Ö.K.; Darcan, C.; Kariptaş, E. Overview of VBNC, a survival strategy for microorganisms. 3 Biotech. 2022, 12, 307. [Google Scholar] [CrossRef]
- Xu, H.S.; Roberts, N.; Singleton, F.L.; Attwell, R.W.; Grimes, D.J.; Colwell, R.R. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 1982, 8, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Pan, H.X.; Yang, D.; Rao, L.; Zhao, L.; Wang, Y.T.; Liao, X.J. Induction, detection, formation, and resuscitation of viable but non-culturable state microorganisms. Compr. Rev. Food Sci. Food Saf. 2020, 19, 149–183. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Williams, T.; Oliver, J.D. Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol. 2018, 200, e00249-18. [Google Scholar] [CrossRef]
- Oliveira, M.M.; de Almeida, F.A.D.; Baglinière, F.; de Oliveira, L.L.D.; Vanetti, M.C.D. Behavior of Salmonella Enteritidis and Shigella flexneri during induction and recovery of the viable but nonculturable state. FEMS Microbiol. Lett. 2021, 368, fnab087. [Google Scholar] [CrossRef] [PubMed]
- Soto-Beltrá, N.M.; Lee, B.G.; Amézquita-López, B.A.; Quiñones, B. Overview of methodologies for the culturing, recovery and detection of Campylobacter. Int. J. Environ. Health Res. 2023, 33, 307–323. [Google Scholar] [CrossRef]
- Power, A.L.; Barber, D.G.; Groenhof, S.R.M.; Wagley, S.; Liu, P.; Parker, D.A.; Love, J. The application of imaging Flow Cytometry for characterisation and quantification of bacterial phenotypes. Front. Cell Infect. Microbiol. 2021, 11, 716592. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, K.M. Flow Cytometry: An overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, F.; Tenev, T.; Meier, P. Analysis of apoptosis and necroptosis by fluorescence-activated cell sorting. Cold Spring Harb. Protoc. 2016, 2016, pdb-prot087387. [Google Scholar] [CrossRef]
- Bunthof, C.J.; Bloemen, K.; Breeuwer, P.; Rombouts, F.M.; Abee, T. Flow Cytometric assessment of viability of lactic acid bacteria. Appl. Environ. Microbiol. 2001, 67, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, L.L.; Chen, Y.J.; Long, Y. Optimization of staining with SYTO 9/propidium iodide: Interplay, kinetics and impact on Brevibacillus brevis. Biotechniques 2020, 69, 88–98. [Google Scholar] [CrossRef]
- Pozarowski, P.; Darzynkiewicz, Z. Analysis of cell cycle by Flow Cytometry. Methods Mol. Biol. 2004, 281, 301–311. [Google Scholar] [CrossRef]
- Van Dijk, M.A.; Gregori, G.; Hoogveld, H.L.; Rijkeboer, M.; Denis, M.; Malkassian, A.; Gons, H.J. Optimizing the setup of a Flow Cytometric cell sorter for efficient quantitative sorting of long filamentous cyanobacteria. Cytometry A 2010, 77, 911–924. [Google Scholar] [CrossRef]
- Baffone, W.; Casaroli, A.; Citterio, B.; Pierfelici, L.; Campana, R.; Vittoria, E.; Guaglianone, E.; Donelli, G. Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int. J. Food Microbiol. 2006, 107, 83–91. [Google Scholar] [CrossRef]
- Tang, J. Microbial metabolomics. Curr. Genom. 2011, 12, 391–403. [Google Scholar] [CrossRef]
- Chumachenko, M.S.; Waseem, T.V.; Fedorovich, S.V. Metabolomics and metabolites in ischemic stroke. Rev. Neurosci. 2021, 33, 181–205. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, F.; Li, P.; He, C.W.; Wang, R.B.; Su, H.X.; Wan, J.B. An improved pseudotargeted metabolomics approach using multiple ion monitoring with time-staggered ion lists based on ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Anal. Chim. Acta 2016, 927, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yan, F.F.; Lu, Q.; Tang, C.; Wang, X.H.; Liu, R. UPLC-Q-TOF-MS and NMR identification of structurally different A-type procyanidins from peanut skin and their inhibitory effect on acrylamide. J. Sci. Food Agric. 2022, 102, 7062–7071. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, M.; Wang, Y.; Cao, Y.W.; Sun, Z.L.; Chen, M.C.; Tian, X.T.; Wan, J.B.; Huang, C.G. Metabonomics study on the hepatoprotective effect of Panax notoginseng leaf saponins using UPLC/Q-TOF-MS analysis. Am. J. Chin. Med. 2019, 47, 1–17. [Google Scholar] [CrossRef]
- Ma, X.B.; Wang, L.N.; Dai, L.X.; Kwok, L.Y.; Bao, Q.H. Rapid detection of the activity of Lacticaseibacillus casei Zhang by Flow Cytometry. Foods 2023, 12, 1208. [Google Scholar] [CrossRef]
- Bai, M.; Huang, T.; Guo, S.; Wang, Y.; Wang, J.C.; Kwok, L.Y.; Dan, T.; Zhang, H.P.; Bilige, M. Probiotic Lactobacillus casei Zhang improved the properties of stirred yogurt. Food Biosci. 2020, 37, 100718. [Google Scholar] [CrossRef]
- Zhu, H.; Cao, C.J.; Wu, Z.C.; Zhang, H.P.; Sun, Z.H.; Wang, M.; Xu, H.Z.; Zhao, Z.; Wang, Y.X.; Pei, G.C.; et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab. 2021, 33, 1926–1942.e8. [Google Scholar] [CrossRef] [PubMed]
- He, Q.W.; Hou, Q.; Wang, Y.; Shen, L.; Sun, Z.H.; Zhang, H.P.; Liong, M.T.; Kwok, L.Y. Long-term administration of Lactobacillus casei Zhang stabilized gut microbiota of adults and reduced gut microbiota age index of older adults. J. Funct. Foods 2020, 64, 103682. [Google Scholar] [CrossRef]
- Wang, J.C.; Bai, X.Y.; Peng, C.T.; Yu, Z.J.; Li, B.H.; Zhang, W.Y.; Sun, Z.H.; Zhang, H.P. Fermented milk containing Lactobacillus casei Zhang and Bifidobacterium animalis ssp. lactis V9 alleviated constipation symptoms through regulation of intestinal microbiota, inflammation, and metabolic pathways. J. Dairy Sci. 2020, 103, 11025–11038. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Yan, X.; Zhang, W.W.; Liu, Y.Y.; Han, D.P.; Teng, K.D.; Ma, Y.F. Lactobacillus casei Zhang prevents jejunal epithelial damage to early-weaned piglets induced by Escherichia coli K88 via regulation of intestinal mucosal integrity, tight junction proteins and immune factor expression. J. Microbiol. Biotechnol. 2019, 29, 863–876. [Google Scholar] [CrossRef]
- Bao, Q.H.; Bo, X.Y.; Chen, L.; Ren, Y.; Wang, H.Y.; Kwok, L.Y.; Liu, W.J. Comparative analysis using raman spectroscopy of the cellular constituents of Lacticaseibacillus paracasei Zhang in a normal and viable but nonculturable state. Microorganisms 2023, 11, 1266. [Google Scholar] [CrossRef]
- Hayek, S.A.; Gyawali, R.; Aljaloud, S.O.; Krastanov, A.; Ibrahim, S.A. Cultivation media for lactic acid bacteria used in dairy products. J. Dairy Res. 2019, 86, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Shailaja, A.; Bruce, T.F.; Gerard, P.; Powell, R.R.; Pettigrew, C.A.; Kerrigan, J.L. Comparison of cell viability assessment and visualization of Aspergillus niger biofilm with two fluorescent probe staining methods. Biofilm 2022, 4, 100090. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.R.; Peng, C.T.; Wang, J.C.; Sun, H.T.; Guo, S.; Zhang, H.P. Metabolic footprint analysis of volatile metabolites to discriminate between different key time points in the fermentation and storage of starter cultures and probiotic Lactobacillus casei Zhang milk. J. Dairy Sci. 2021, 104, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.Q.; Guo, Q.X.; Zhang, J.; Huang, J.; Xu, W.; Gong, L.; Su, H.; Luo, Y.G.; Li, J.H.; Qiu, X.H.; et al. Metabolic profile of dendrobine in rats determined by Ultra-high-performance Liquid Chromatography/Quadrupole Time-of-flight Mass Spectrometry. Comb. Chem. High. Throughput Screen. 2021, 24, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Ciosek, A.; Fulara, K.; Hrabia, O.; Satora, P.; Poreda, A. Chemical composition of sour beer resulting from supplementation the fermentation medium with magnesium and zinc Ions. Biomolecules 2020, 10, 1599. [Google Scholar] [CrossRef]
- Fera, M.T.; Maugeri, T.L.; La Camera, E.; Lentini, V.; Favaloro, A.; Bonanno, D.; Carbone, M. Induction and resuscitation of viable nonculturable Arcobacter butzleri cells. Appl. Environ. Microbiol. 2008, 74, 3266–3268. [Google Scholar] [CrossRef]
- Wei, C.J.; Zhao, X.H. Induction of viable but nonculturable Escherichia coli O157:H7 by low temperature and its resuscitation. Front. Microbiol. 2018, 9, 2728. [Google Scholar] [CrossRef]
- Adebo, O.A.; Kayitesi, E.; Tugizimana, F.; Njobeh, P.B. Differential metabolic signatures in naturally and lactic acid bacteria (LAB) fermented ting (a Southern African food) with different tannin content, as revealed by gas chromatography mass spectrometry (GC-MS)-based metabolomics. Food Res. Int. 2019, 121, 326–335. [Google Scholar] [CrossRef]
- Miyajima, M. Amino acids: Key sources for immunometabolites and immunotransmitters. Int. Immunol. 2020, 32, 435–446. [Google Scholar] [CrossRef]
- Yin, J.; Ren, W.K.; Yang, G.; Duan, J.L.; Huang, X.G.; Fang, R.J.; Li, C.Y.; Li, T.J.; Yin, Y.L.; Hou, Y.Q.; et al. L-Cysteine metabolism and its nutritional implications. Mol. Nutr. Food Res. 2016, 60, 134–146. [Google Scholar] [CrossRef]
- Qiao, Y.L.; Liu, G.F.; Leng, C.; Zhang, Y.J.; Lv, X.P.; Chen, H.Y.; Sun, J.H.; Feng, Z. Metabolic profiles of cysteine, methionine, glutamate, glutamine, arginine, aspartate, asparagine, alanine and glutathione in Streptococcus thermophilus during pH-controlled batch fermentations. Sci. Rep. 2018, 8, 12441. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, M.L.; Luo, S.; Zhang, R.M.; Han, P.; Hu, W. Metabolic responses to ethanol in Saccharomyces cerevisiae using a gas chromatography tandem mass spectrometry-based metabolomics approach. Int. J. Biochem. Cell Biol. 2012, 44, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Almeida, V.; Almeida Santos, M.; Chambel, L. Resuscitation of Escherichia coli VBNC cells depends on a variety of environmental or chemical stimuli. J. Appl. Microbiol. 2011, 110, 1601–1611. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.K.; Yue, Z.H.; Liu, E.; Li, X.F.; Li, C.W. Assessment of the bifidogenic and antibacterial activities of xylooligosaccharide. Front. Nutr. 2022, 9, 858949. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.P.; Summanen, P.H.; Komoriya, T.; Finegold, S.M. In vitro study of the prebiotic xylooligosaccharide (XOS) on the growth of Bifidobacterium spp. and Lactobacillus spp. Int. J. Food Sci. Nutr. 2015, 66, 919–922. [Google Scholar] [CrossRef]
- Zhao, X.; Song, X.X.; Li, Y.P.; Yu, C.X.; Zhao, Y.; Gong, M.; Shen, X.X.; Chen, M.J. Gene expression related to trehalose metabolism and its effect on Volvariella volvacea under low temperature stress. Sci. Rep. 2018, 8, 11011. [Google Scholar] [CrossRef]
- Soper, A.K.; Ricci, M.A.; Bruni, F.; Rhys, N.H.; McLain, S.E. Trehalose in water revisited. J. Phys. Chem. B 2018, 122, 7365–7374. [Google Scholar] [CrossRef]
- Wu, P.Y.; An, J.; Chen, L.; Zhu, Q.Y.; Li, Y.J.; Mei, Y.X.; Chen, Z.M.; Liang, Y.X. Differential analysis of stress tolerance and transcriptome of probiotic Lacticaseibacillus casei Zhang produced from Solid-State (SSF-SW) and Liquid-State (LSF-MRS) fermentations. Microorganisms 2020, 8, 1656. [Google Scholar] [CrossRef]
- Wu, C.D.; Zhang, J.; Wang, M.; Du, G.C.; Chen, J. Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J. Ind. Microbiol. Biotechnol. 2012, 39, 1031–1039. [Google Scholar] [CrossRef]
- Marquet, A.; Bui, B.T.; Florentin, D. Biosynthesis of biotin and lipoic acid. Vitam. Horm. 2001, 61, 51–101. [Google Scholar] [CrossRef]
- Tang, Z.W.; Ye, W.R.; Chen, H.T.; Kuang, X.W.; Guo, J.; Xiang, M.M.; Peng, C.; Chen, X.; Liu, H. Role of purines in regulation of metabolic reprogramming. Purinergic Signal. 2019, 15, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Maan, K.; Baghel, R.; Dhariwal, S.; Sharma, A.; Bakhshi, R.; Rana, P. Metabolomics and transcriptomics based multi-omics integration reveals radiation-induced altered pathway networking and underlying mechanism. NPJ Syst. Biol. Appl. 2023, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Cicero, A.F.G.; Fogacci, F.; Di Micoli, V.; Angeloni, C.; Giovannini, M.; Borghi, C. Purine metabolism dysfunctions: Experimental methods of detection and diagnostic potential. Int. J. Mol. Sci. 2023, 24, 7027. [Google Scholar] [CrossRef] [PubMed]
- Razak, M.A.; Begum, P.S.; Viswanath, B.; Rajagopal, S. Multifarious beneficial effect of nonessential amino acid, glycine: A review. Oxid. Med. Cell Longev. 2017, 2017, 1716701. [Google Scholar] [CrossRef]

| Class | Metabolite Identity | Molecular Formula | Retention Time (min) | m/z Ratio | VIP Value | Log2(FC) | p-Value |
|---|---|---|---|---|---|---|---|
| Amino acid | L-Cysteine | C3H7NO2S | 0.80 | 267.07 | 2.27 | 3.53 | 1.12 × 10−5 |
| L-Alanine | C3H7NO2 | 0.83 | 134.05 | 2.23 | 2.63 | 3.72 × 10−6 | |
| L-Glutamic acid | C5H9NO4 | 3.65 | 118.05 | 2.19 | 3.03 | 1.98 × 10−7 | |
| L-Lysine | C6H14N2O2 | 15.75 | 109.77 | 2.16 | 6.68 | 2.70 × 10−5 | |
| L-3-Cyanophenylalanine | C10H10N2O2 | 1.09 | 137.03 | 2.27 | 2.21 | 2.20 × 10−5 | |
| L-Glutamine | C5H10N2O3 | 3.64 | 144.07 | 2.26 | 5.84 | 4.52 × 10−5 | |
| L-Arginine | C6H14N4O2 | 14.56 | 139.10 | 2.40 | 5.62 | 8.97 × 10−5 | |
| L-Asparagine | C9H16N2O5 | 3.64 | 159.08 | 2.02 | 3.21 | 7.26 × 10−5 | |
| L-tyrosine | C9H11NO3 | 3.24 | 180.07 | 2.31 | −3.14 | 1.44 × 10−6 | |
| Carbohydrate | Cellulose | (C6H10O5)n | 0.93 | 295.03 | 2.10 | −2.79 | 1.02 × 10−5 |
| N-Acetyl-D-glucosamine1-phosphate | C8H16NO9P | 0.77 | 304.06 | 2.36 | −2.58 | 3.90 × 10−5 | |
| Xylooligosaccharide | C5H12O5 | 1.12 | 152.06 | 2.19 | 3.12 | 7.70 × 10−5 | |
| D-Trehalose anhydrous | C12H22O11 | 2.32 | 253.64 | 2.14 | −2.87 | 1.05 × 10−8 | |
| Lipid | Palmitoleic acid | C16H32O2 | 5.97 | 271.23 | 2.30 | −2.23 | 6.36 × 10−5 |
| N-Anthranilate | C12H16NO9P | 0.77 | 304.06 | 2.36 | −2.58 | 7.75 × 10−5 | |
| Methyl-2-methylvalerate | C7H12O4 | 0.77 | 207.04 | 2.26 | −2.86 | 5.67 × 10−5 | |
| Vitamin | Desthiobiotin | C10H18N2O3 | 1.11 | 214.13 | 2.29 | 3.12 | 6.81 × 10−5 |
| Folinic acid | C20H23N7O7 | 5.18 | 229.09 | 2.39 | −7.60 | 5.80 × 10−5 | |
| Nicotinamide | C6H6N2O | 4.52 | 167.04 | 2.28 | 2.31 | 2.48 × 10−5 | |
| Purine and pyrimidine | Thymidine | C10H13N5O5 | 3.41 | 243.08 | 2.18 | 2.93 | 1.05 × 10−8 |
| Guanine | C5H5N5O | 4.67 | 144.07 | 2.35 | −2.53 | 1.54 × 10−5 | |
| 2-Methylpyrimidine | C5H6N2 | 14.56 | 139.10 | 2.40 | −5.62 | 7.43 × 10−6 | |
| Others | 3-Oxohexanoyl-CoA | C27H44N7O18P3S | 15.75 | 938.13 | 2.34 | 5.33 | 3.44 × 10−5 |
| Uric acid | C5H4N4O3 | 0.8 | 176.05 | 2.27 | 3.56 | 8.95 × 10−6 | |
| Hydroxy methylglutaryl coenzyme A | C27H44N7O20P3S | 8.75 | 950.12 | 2.03 | −5.59 | 9.77 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zhang, Y.; Dai, L.; Bo, X.; Liu, X.; Zhao, X.; Yu, J.; Kwok, L.-Y.; Bao, Q. Metabolomic Differences between Viable but Nonculturable and Recovered Lacticaseibacillus paracasei Zhang. Foods 2023, 12, 3472. https://doi.org/10.3390/foods12183472
Wang H, Zhang Y, Dai L, Bo X, Liu X, Zhao X, Yu J, Kwok L-Y, Bao Q. Metabolomic Differences between Viable but Nonculturable and Recovered Lacticaseibacillus paracasei Zhang. Foods. 2023; 12(18):3472. https://doi.org/10.3390/foods12183472
Chicago/Turabian StyleWang, Huiying, Yuhong Zhang, Lixia Dai, Xiaoyu Bo, Xiangyun Liu, Xin Zhao, Jie Yu, Lai-Yu Kwok, and Qiuhua Bao. 2023. "Metabolomic Differences between Viable but Nonculturable and Recovered Lacticaseibacillus paracasei Zhang" Foods 12, no. 18: 3472. https://doi.org/10.3390/foods12183472
APA StyleWang, H., Zhang, Y., Dai, L., Bo, X., Liu, X., Zhao, X., Yu, J., Kwok, L.-Y., & Bao, Q. (2023). Metabolomic Differences between Viable but Nonculturable and Recovered Lacticaseibacillus paracasei Zhang. Foods, 12(18), 3472. https://doi.org/10.3390/foods12183472





