Evaluating the Application of Portable Handheld X-ray Fluorescence (XRF) Scanner for Determining Seafood Provenance: A Case Study on Penaeus monodon
Abstract
1. Introduction
2. Experimental Methods
2.1. Materials
2.1.1. Analytical Instruments
2.1.2. Sample Collection and Preparation
2.2. Protocol Methodology
2.2.1. Validation of Methodology
Calibration and Arrangement of the Prawn Elemental Profile
Repeatability
2.2.2. Factors That Influence Measurements
Exposure Time
Sample Moisture
2.3. Preliminary Provenance Modelling
3. Results
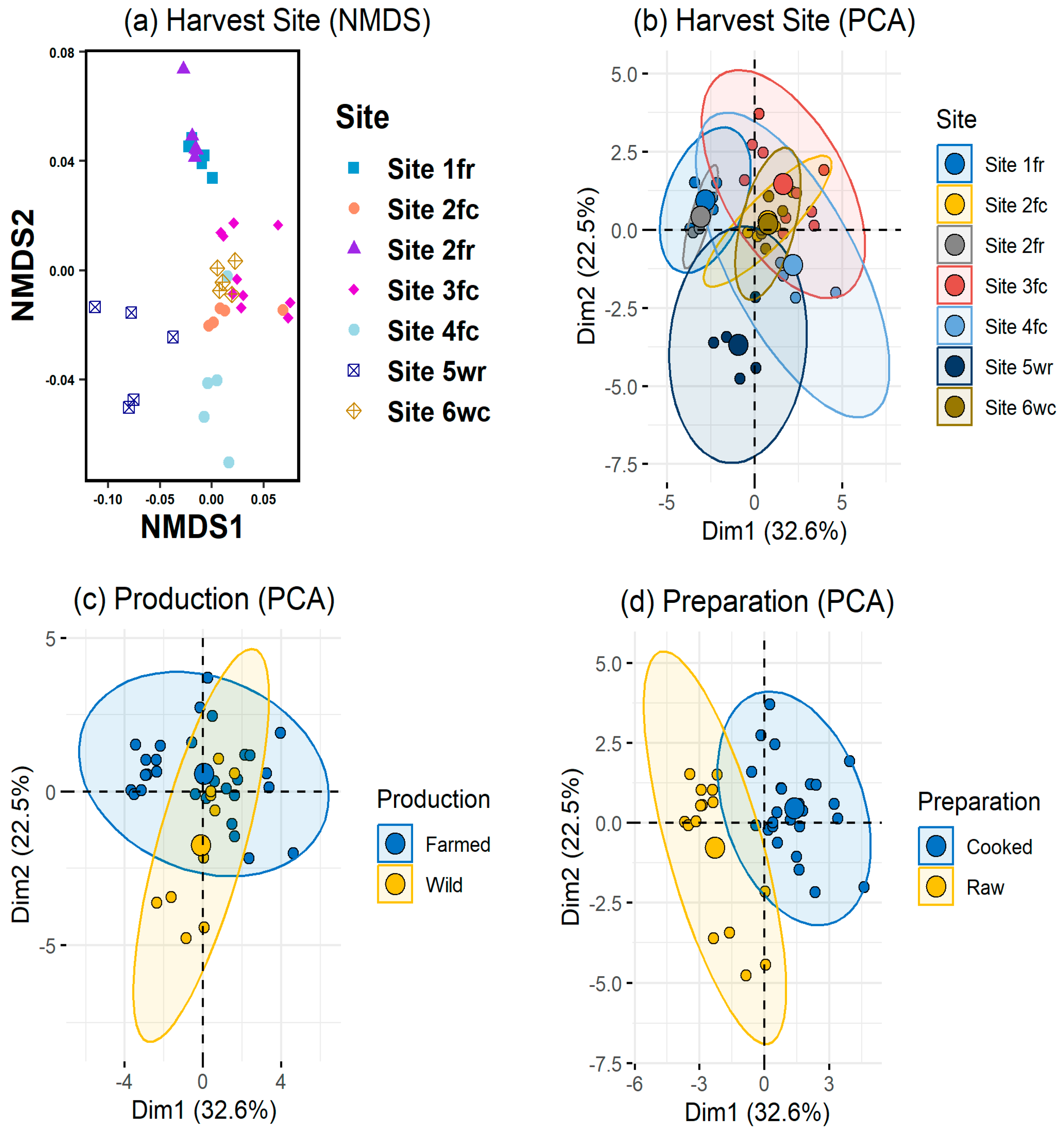
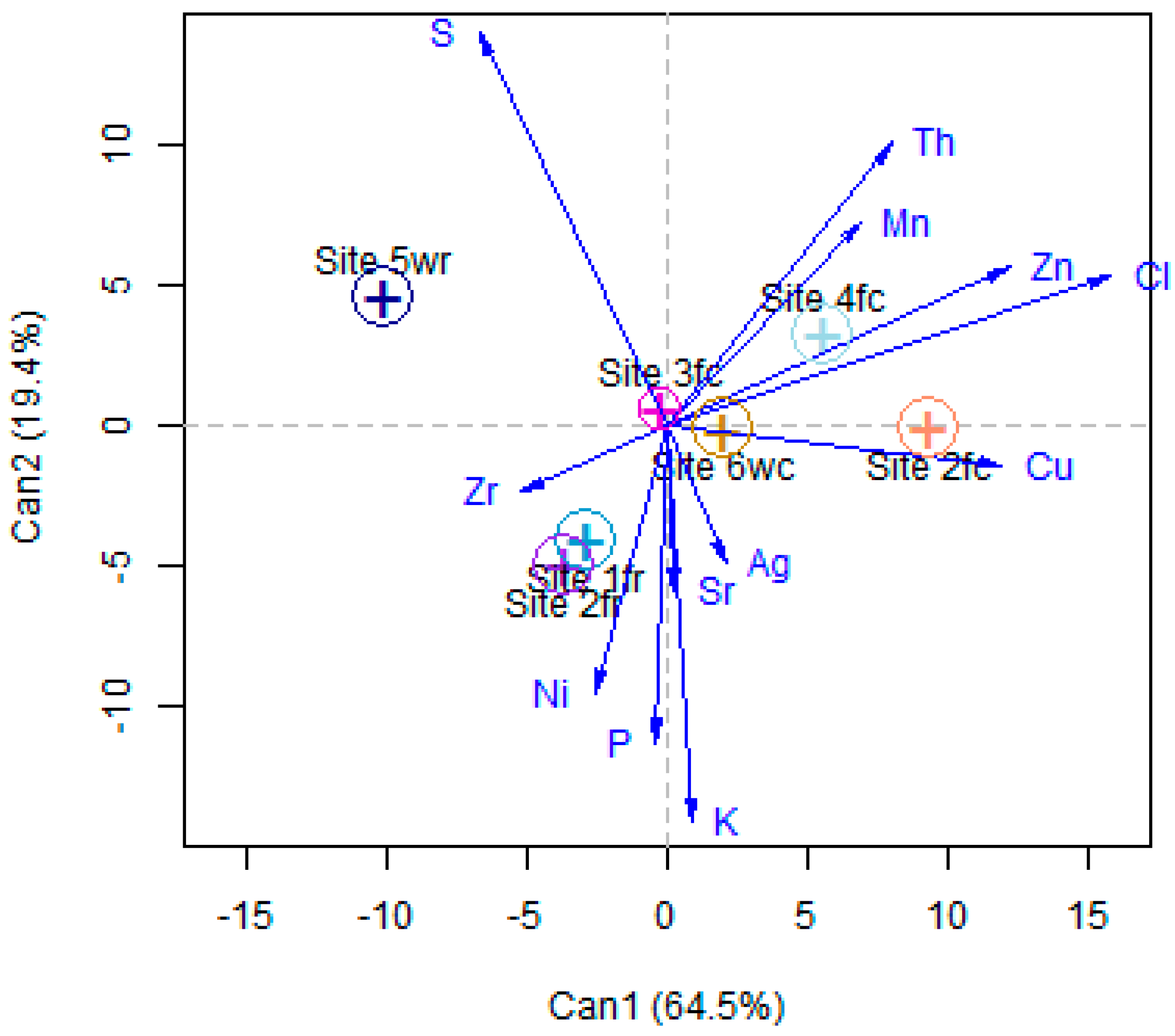
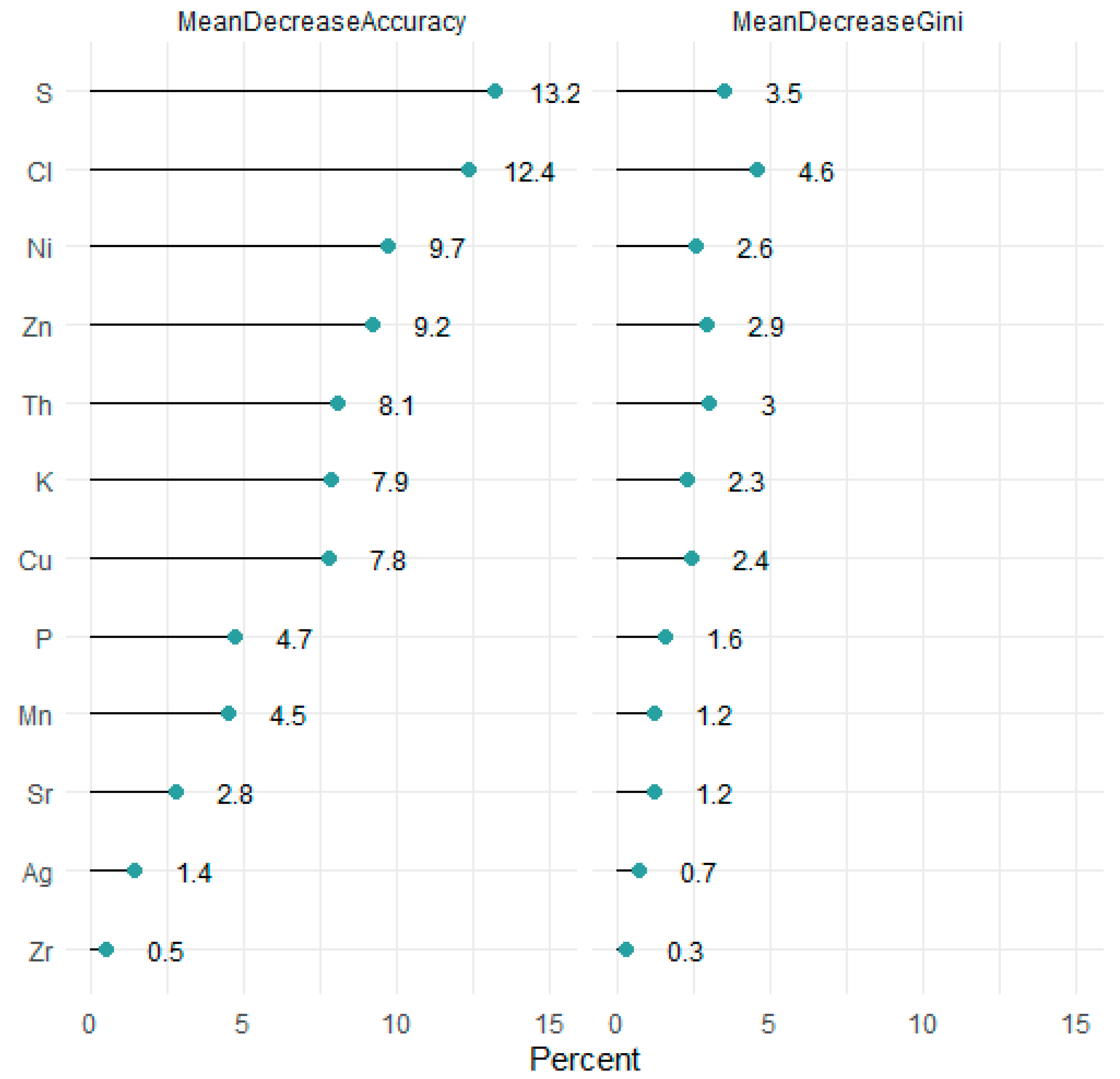
3.1. Protocol Methodology
3.1.1. Calibration and Arrangement of the Prawn Elemental Profile
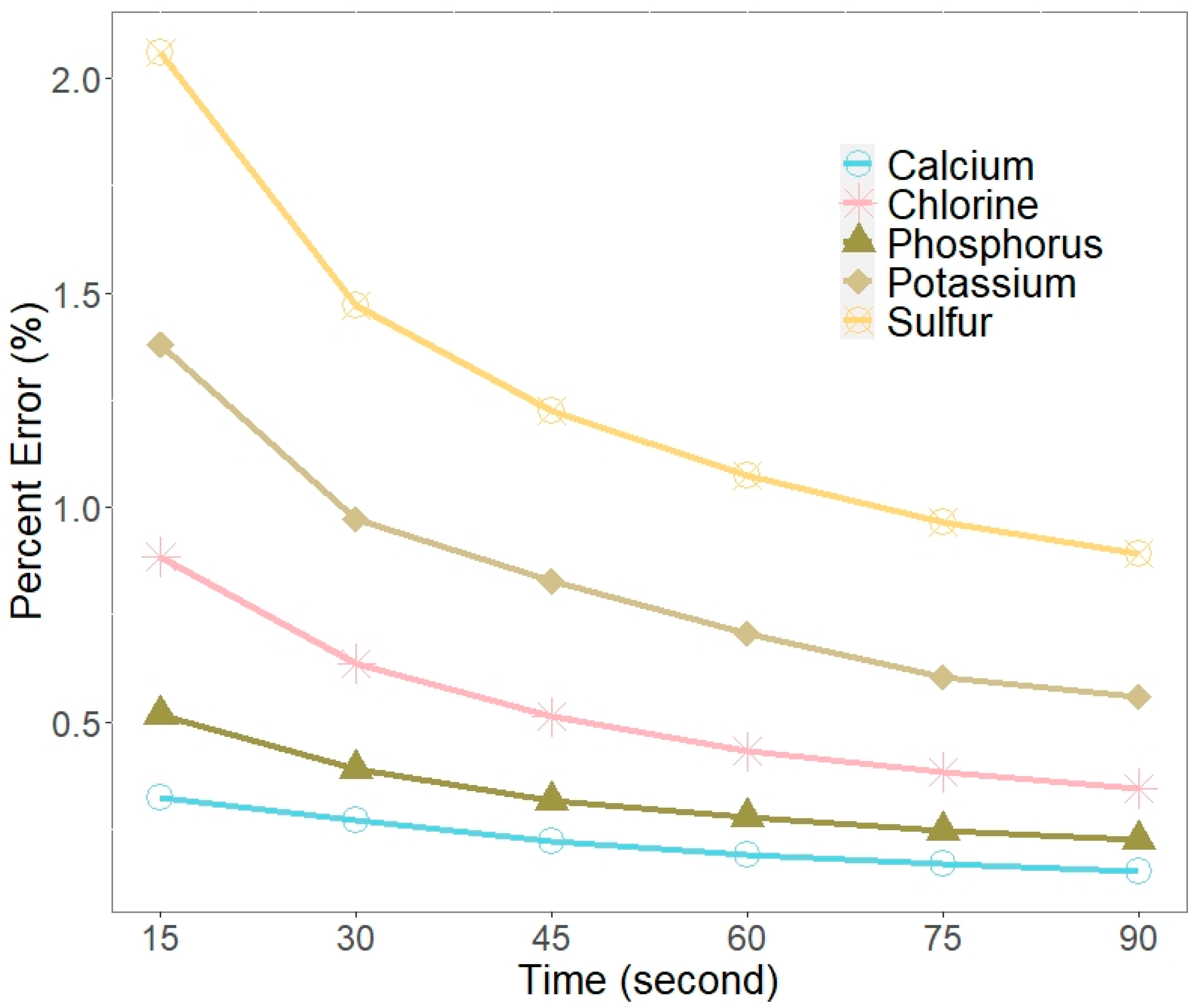
3.1.2. Exposure Time
3.1.3. Sample Moisture
3.1.4. Repeatability
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martino, J.C.; Mazumder, D.; Gadd, P.; Doubleday, Z.A. Tracking the provenance of octopus using isotopic and multi-elemental analysis. Food Chem. 2022, 371, 131133. [Google Scholar] [CrossRef]
- Baki, M.A.; Hossain, M.M.; Akter, J.; Quraishi, S.B.; Haque Shojib, M.F.; Atique Ullah AK, M.; Khan, M.F. Concentration of heavy metals in seafood (fishes, shrimp, lobster and crabs) and human health assessment in Saint Martin Island, Bangladesh. Ecotoxicol. Environ. Saf. 2018, 159, 153–163. [Google Scholar] [CrossRef]
- Sammut, J.; Gopi, K.; Saintilan, N.; Mazumder, D. Facing the challenges of food fraud in the global food system. In Food Authentication and Traceability; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Guérin, T.; Chekri, R.; Vastel, C.; Sirot, V.; Volatier, J.L.; Leblanc, J.C.; Noël, L. Determination of 20 trace elements in fish and other seafood from the French market. Food Chem. 2011, 127, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Mouchi, V.; Godbillot, C.; Forest, V.; Ulianov, A.; Lartaud, F.; de Rafélis, M.; Emmanuel, L.; Verrecchia, E.P. Rare earth elements in oyster shells: Provenance discrimination and potential vital effects. Biogeosciences 2020, 17, 2205–2217. [Google Scholar] [CrossRef]
- Salvo, A.; Cicero, N.; Vadalà, R.; Mottese, A.F.; Bua, D.; Mallamace, D.; Giannetto, C.; Dugo, G. Toxic and essential metals determination in commercial seafood: Paracentrotus lividus by ICP-MS. Nat. Prod. Res. 2016, 30, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Bergström, E. Effect of natural and artificial diets on seasonal changes in fatty acid composition and total body lipid content of wild and hatchery-reared Atlantic salmon (Salmo salar L.) parr-smolt. Aquaculture 1989, 82, 205–217. [Google Scholar] [CrossRef]
- Budge, S.M.; Iverson, S.J.; Bowen, W.D.; Ackman, R.G. Among- and within-species variability in fatty acid signatures of marine fish and invertebrates on the Scotian Shelf, Georges Bank, and southern Gulf of St. Lawrence. Can. J. Fish. Aquat. Sci. 2002, 59, 886–898. [Google Scholar] [CrossRef]
- Burford, M.A.; Preston, N.P.; Minh, T.H.; Hoa TT, T.; Bunn, S.E.; Fry, V.M. Dominant sources of dietary carbon and nitrogen for shrimp reared in extensive rice-shrimp ponds. Aquac. Res. 2004, 35, 194–203. [Google Scholar] [CrossRef][Green Version]
- Busetto, M.L.; Moretti, V.M.; Moreno-Rojas, J.M.; Caprino, F.; Giani, I.; Malandra, R.; Bellagamba, F.; Guillou, C. Authentication of farmed and wild turbot (Psetta maxima) by fatty acid and isotopic analyses combined with chemometrics. J. Agric. Food Chem. 2008, 56, 2742–2750. [Google Scholar] [CrossRef]
- Shiel, A.E.; Weis, D.; Cossa, D.; Orians, K.J. Determining provenance of marine metal pollution in French bivalves using Cd, Zn and Pb isotopes. Geochim. Cosmochim. Acta 2013, 121, 155–167. [Google Scholar] [CrossRef]
- Gopi, K.; Mazumder, D.; Sammut, J.; Saintilan, N.; Crawford, J.; Gadd, P. Isotopic and elemental profiling to trace the geographic origins of farmed and wild-caught Asian seabass (Lates calcarifer). Aquaculture 2019, 502, 56–62. [Google Scholar] [CrossRef]
- Matthies, M.; Giupponi, C.; Ostendorf, B. Environmental decision support systems: Current issues, methods and tools. Environ. Model. Softw. 2007, 22, 123–127. [Google Scholar] [CrossRef]
- Gadd, P.; Gopi, K.; Sammut, J.; Saintilan, N.; Crawford, J.; Mazumder, D. Itrax micro X-ray fluorescence (μXRF) for soft biological tissues. MethodsX 2018, 5, 1267–1271. [Google Scholar] [CrossRef]
- Shackley, M.S. X-ray Fluorescence Spectrometry (XRF). In The Encyclopedia of Archaeological Sciences; Springer: Cham, Switzerland, 2018; pp. 1–5. [Google Scholar] [CrossRef][Green Version]
- Gopi, K.; Mazumder, D.; Sammut, J.; Saintilan, N. Determining the provenance and authenticity of seafood: A review of current methodologies. Trends Food Sci. Technol. 2019, 91, 294–304. [Google Scholar] [CrossRef]
- Power, A.; Cozzolino, D. How fishy is your fish? Authentication, provenance and traceability in fish and seafood by means of vibrational spectroscopy. Appl. Sci. 2020, 10, 4150. [Google Scholar] [CrossRef]
- Watson, R.A.; Green, B.S.; Tracey, S.R.; Farmery, A.; Pitcher, T.J. Provenance of global seafood. Fish Fish. 2016, 17, 585–595. [Google Scholar] [CrossRef]
- Han, C.; Li, L.; Zhang, G.; Dong, S.; Tian, X. Assessment of elemental profiling combined with chemometrics for authenticating the geographical origins of Pacific white shrimp (Litopenaeus vannamei). Food Control 2021, 130, 108231. [Google Scholar] [CrossRef]
- Li, L.; Boyd, C.E.; Racine, P.; McNevin, A.A.; Somridhivej, B.; Minh, H.N.; Tinh, H.Q.; Godumala, R. Assessment of elemental profiling for distinguishing geographic origin of aquacultured shrimp from India, Thailand and Vietnam. Food Control 2017, 80, 162–169. [Google Scholar] [CrossRef]
- Nguyen-Quang, T.; Bui-Quang, M.; Truong-Ngoc, M. Rapid Identification of Geographical Origin of Commercial Soybean Marketed in Vietnam by ICP-MS. J. Anal. Methods Chem. 2021, 2021, 5583860. [Google Scholar] [CrossRef]
- Nguyen-Quang, T.; Do-Hoang, G.; Truong-Ngoc, M. Multielement Analysis of Pakchoi (Brassica rapa L. ssp. chinensis) by ICP-MS and Their Classification according to Different Small Geographical Origins. J. Anal. Methods Chem. 2021, 2021, 8860852. [Google Scholar] [CrossRef]
- Gopi, K.; Mazumder, D.; Sammut, J.; Saintilan, N.; Crawford, J.; Gadd, P. Combined use of stable isotope analysis and elemental profiling to determine provenance of black tiger prawns (Penaeus monodon). Food Control 2019, 95, 242–248. [Google Scholar] [CrossRef]
- Pohar, M.; Blas, M.; Turk, S. Comparison of logistic regression and linear discriminant analysis. Adv. Methodol. Stat. 2004, 1, 143. [Google Scholar] [CrossRef]
- Biau, G.; Scornet, E. A random forest guided tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Weindorf, D.C.; Bakr, N.; Zhu, Y. Advances in portable X-ray fluorescence (PXRF) for environmental, pedological, and agronomic applications. Adv. Agron. 2014, 128, 1–45. [Google Scholar] [CrossRef]
- Kalcsits, L.A. Non-destructive measurement of calcium and potassium in apple and pear using handheld X-ray fluorescence. Front. Plant Sci. 2016, 7, 442. [Google Scholar] [CrossRef]
- McLaren, T.I.; Guppy, C.N.; Tighe, M.K. A Rapid and Nondestructive Plant Nutrient Analysis using Portable X-Ray Fluorescence. Soil Sci. Soc. Am. J. 2012, 76, 1446–1453. [Google Scholar] [CrossRef]
- Towett, E.K.; Shepherd, K.D.; Lee Drake, B. Plant elemental composition and portable X-ray fluorescence (pXRF) spectroscopy: Quantification under different analytical parameters. X-Ray Spectrom. 2015, 45, 117–124. [Google Scholar] [CrossRef]
- Turner, A.; Poon, H.; Taylor, A.; Brown, M.T. In situ determination of trace elements in Fucus spp. by field-portable-XRF. Sci. Total Environ. 2017, 593–594, 227–235. [Google Scholar] [CrossRef]
- Zhou, S.; Cheng, Q.; Weindorf, D.C.; Yuan, Z.; Yang, B.; Sun, Q.; Zhang, Z.; Yang, J.; Zhao, M. Elemental assessment of dried and ground samples of leeches: Via portable X-ray fluorescence. J. Anal. At. Spectrom. 2020, 35, 2573–2581. [Google Scholar] [CrossRef]
- Tjallingii, R.; Röhl, U.; Kölling, M.; Bickert, T. Influence of the water content on X-ray fluorescence corescanning measurements in soft marine sediments. Geochem. Geophys. Geosyst. 2007, 8. [Google Scholar] [CrossRef]
- FAO. Fishery and aquaculture statistics 2018. In Fisheries and Aquaculture Department; FAO yearbook; FAO: Rome, Italy, 2020; pp. 26–28. [Google Scholar]
- Sitko, R. Quantitative X-ray fluorescence analysis of samples of less than “infinite thickness”: Difficulties and possibilities. Spectrochim. Acta—Part B At. Spectrosc. 2009, 64, 1161–1172. [Google Scholar] [CrossRef]
- Grave, P.; Robinson, E.; Barbetti, M.; Yu, Z.; Bailey, G.; Bird, R. Analysis of South Italian Pottery by PIXE-PIGME. Mediterr. Archaeol. 1996, 9/10, 113–125. Available online: http://www.jstor.org/stable/24667886 (accessed on 4 August 2022).
- Grahl-Nielsen, O.; Jacobsen, A.; Christophersen, G.; Magnesen, T. Fatty acid composition in adductor muscle of juvenile scallops (Pecten maximus) from five Norwegian populations reared in the same environment. Biochem. Syst. Ecol. 2010, 38, 478–488. [Google Scholar] [CrossRef]
- Anderson, K.A.; Hobbie, K.A.; Smith, B.W. Chemical profiling with modeling differentiates wild and farm-raised salmon. J. Agric. Food Chem. 2010, 58, 11768–11774. [Google Scholar] [CrossRef] [PubMed]
- Kruskal, J.B. Multidimensional scaling by optimizing goodness-of-fit to a nonmetric hypothesis. Psychometrika 1964, 29, 1–28. [Google Scholar] [CrossRef]
- Gabriel, K.R. The biplot graphical display of matrices with applications to principal component analysis. Biometrika 1971, 58, 453–467. [Google Scholar] [CrossRef]
- Gittins, R. Canonical Analysis: A Review with Applications in Ecology; Springer: Berlin/Heidelberg, Germany, 1985. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 2 June 2021).
- Conrey, R.M.; Goodman-Elgar, M.; Bettencourt, N.; Seyfarth, A.; Van Hoose, A.; Wolff, J.A. Calibration of a portable X-ray fluorescence spectrometer in the analysis of archaeological samples using influence coefficients. Geochem. Explor. Environ. Anal. 2014, 14, 291. [Google Scholar] [CrossRef]
- Liu, H.-C.; You, C.-F.; Chen, C.-Y.; Liu, Y.-C.; Chung, M.-T. Geographic determination of coffee beans using multi-element analysis and isotope ratios of boron and strontium. Food Chem. 2014, 142, 439–445. [Google Scholar] [CrossRef]
- Kashparov, V.; Colle, C.; Levchuk, S.; Yoschenko, V.; Svydynuk, N. Transfer of chlorine from the environment to agricultural foodstuffs. J. Environ. Radioact. 2007, 94, 1–15. [Google Scholar] [CrossRef]
- Hosseini, H.; Mahmoudzadeh, M.; Rezaei, M.; Mahmoudzadeh, L.; Khaksar, R.; Khosroshahi, N.K.; Babakhani, A. Effect of different cooking methods on minerals, vitamins and nutritional quality indices of kutum roach (Rutilus frisii kutum). Food Chem. 2014, 148, 86–91. [Google Scholar] [CrossRef]
- Batista, R.; Japur, C.; Prestes, I.; Silva, J.; Cavanha, M.; Pena, G. Potassium reduction in food by preparation technique for the dietetic management of patients with chronic kidney disease: A review. J. Hum. Nutr. Diet. 2021, 34, 736–746. [Google Scholar] [CrossRef]
- Abd-Elghany, S.M.; Zaher, H.A.; Elgazzar, M.M.; Sallam, K.I. Effect of boiling and grilling on some heavy metal residues in crabs and shrimps from the Mediterranean Coast at Damietta region with their probabilistic health risk assessment. J. Food Compos. Anal. 2020, 93, 103606. [Google Scholar] [CrossRef]
- Gopi, K.; Mazumder, D.; Crawford, J.; Gadd, P.; Tadros, C.V.; Atanacio, A.; Saintilan, N.; Sammut, J. Developing a My SQL Database for the Provenance of Black Tiger Prawns (Penaeus Monodon). Foods 2023, 12, 2677. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Margui Grabulosa, E.; Pili, E.; Floor, G.H.; Roman-Ross, G.; Charlet, L. Quantification of trace arsenic in soils by field-portable X-ray fluorescence spectrometry: Considerations for sample preparation and measurement conditions. J. Hazard. Mater. 2013, 262, 1213–1222. [Google Scholar] [CrossRef]
- van der Sman, R.G.M. Moisture transport during cooking of meat: An analysis based on Flory-Rehner theory. Meat Sci. 2007, 76, 730–738. [Google Scholar] [CrossRef]
- Devesa, V.; Macho, M.L.; Jalón, M.; Urieta, I.; Muñoz, O.; Súñer, M.A.; López, F.; Vélez, D.; Montoro, R. Arsenic in cooked seafood products: Study on the effect of cooking on total and inorganic arsenic contents. J. Agric. Food Chem. 2001, 49, 4132–4140. [Google Scholar] [CrossRef]
- Boonsumrej, S.; Chaiwanichsiri, S.; Tantratian, S.; Suzuki, T.; Takai, R. Effects of freezing and thawing on the quality changes of tiger shrimp (Penaeus monodon) frozen by air-blast and cryogenic freezing. J. Food Eng. 2007, 80, 292–299. [Google Scholar] [CrossRef]
| Site | State | Production | Cooking | Collection Date |
|---|---|---|---|---|
| 1fr | Queensland | Farmed | Uncooked | 21 February |
| 2fr | Queensland | Farmed | Uncooked | 21 February |
| 2fc | Queensland | Cooked | 21 February | |
| 3fc | Queensland | Farmed | Cooked | 21 January |
| 3fc | Queensland | Cooked | 21 February | |
| 4fc | Queensland | Farmed | Cooked | 21 January |
| 5wr | New South Wales | Wild-caught | Uncooked | 21 January |
| 6wc | New South Wales | Wild-caught | Cooked | 21 January |
| Element | Correction Factor |
|---|---|
| P | 0.7196 |
| S | 0.3138 |
| Cl | 0.6894 |
| K | 0.7977 |
| Ca | 1.1036 |
| Mn | 2.2247 |
| Fe | 0.4202 |
| Ni | −1.625 |
| Cu | 0.4114 |
| Zn | 0.1765 |
| As | −21.486 |
| Sr | 1.3978 |
| Zr | N/A |
| Th | N/A |
| Element | p-Value | |
|---|---|---|
| Raw | Cooked | |
| P | 0.06 | 0.92 |
| S | 0.31 | 0.85 |
| Cl | 0.06 | 0.77 |
| K | 0.31 | 0.49 |
| Ca | 0.06 | 1.00 |
| Mn | 0.20 | 0.10 |
| Fe | 0.19 | 0.17 |
| Ni | 1.00 | 0.18 |
| Cu | 0.77 | 0.63 |
| Zn | 0.09 | 0.15 |
| As | 0.35 | * |
| Sr | 0.77 | 0.92 |
| Zr | 0.35 | 0.77 |
| Th | 1.00 | 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malo, N.; Mazumder, D.; Crawford, J.; Gadd, P.; Gopi, K.; Saintilan, N.; Sammut, J. Evaluating the Application of Portable Handheld X-ray Fluorescence (XRF) Scanner for Determining Seafood Provenance: A Case Study on Penaeus monodon. Foods 2023, 12, 2874. https://doi.org/10.3390/foods12152874
Malo N, Mazumder D, Crawford J, Gadd P, Gopi K, Saintilan N, Sammut J. Evaluating the Application of Portable Handheld X-ray Fluorescence (XRF) Scanner for Determining Seafood Provenance: A Case Study on Penaeus monodon. Foods. 2023; 12(15):2874. https://doi.org/10.3390/foods12152874
Chicago/Turabian StyleMalo, Nondita, Debashish Mazumder, Jagoda Crawford, Patricia Gadd, Karthik Gopi, Neil Saintilan, and Jesmond Sammut. 2023. "Evaluating the Application of Portable Handheld X-ray Fluorescence (XRF) Scanner for Determining Seafood Provenance: A Case Study on Penaeus monodon" Foods 12, no. 15: 2874. https://doi.org/10.3390/foods12152874
APA StyleMalo, N., Mazumder, D., Crawford, J., Gadd, P., Gopi, K., Saintilan, N., & Sammut, J. (2023). Evaluating the Application of Portable Handheld X-ray Fluorescence (XRF) Scanner for Determining Seafood Provenance: A Case Study on Penaeus monodon. Foods, 12(15), 2874. https://doi.org/10.3390/foods12152874






