Quantitative Detection of Viable but Nonculturable Vibrio parahaemolyticus in Frozen Bivalve Molluscs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Preparation
2.2. Detection and Enumeration of V. parahaemolyticus via Standard Culture Methods
2.3. PMA Assay and Genomic DNA Extraction
2.4. Real-Time qPCR Analysis
2.5. Bacterial Strain and Growth Conditions
2.6. Preparation of Heat-Killed Cell Suspension
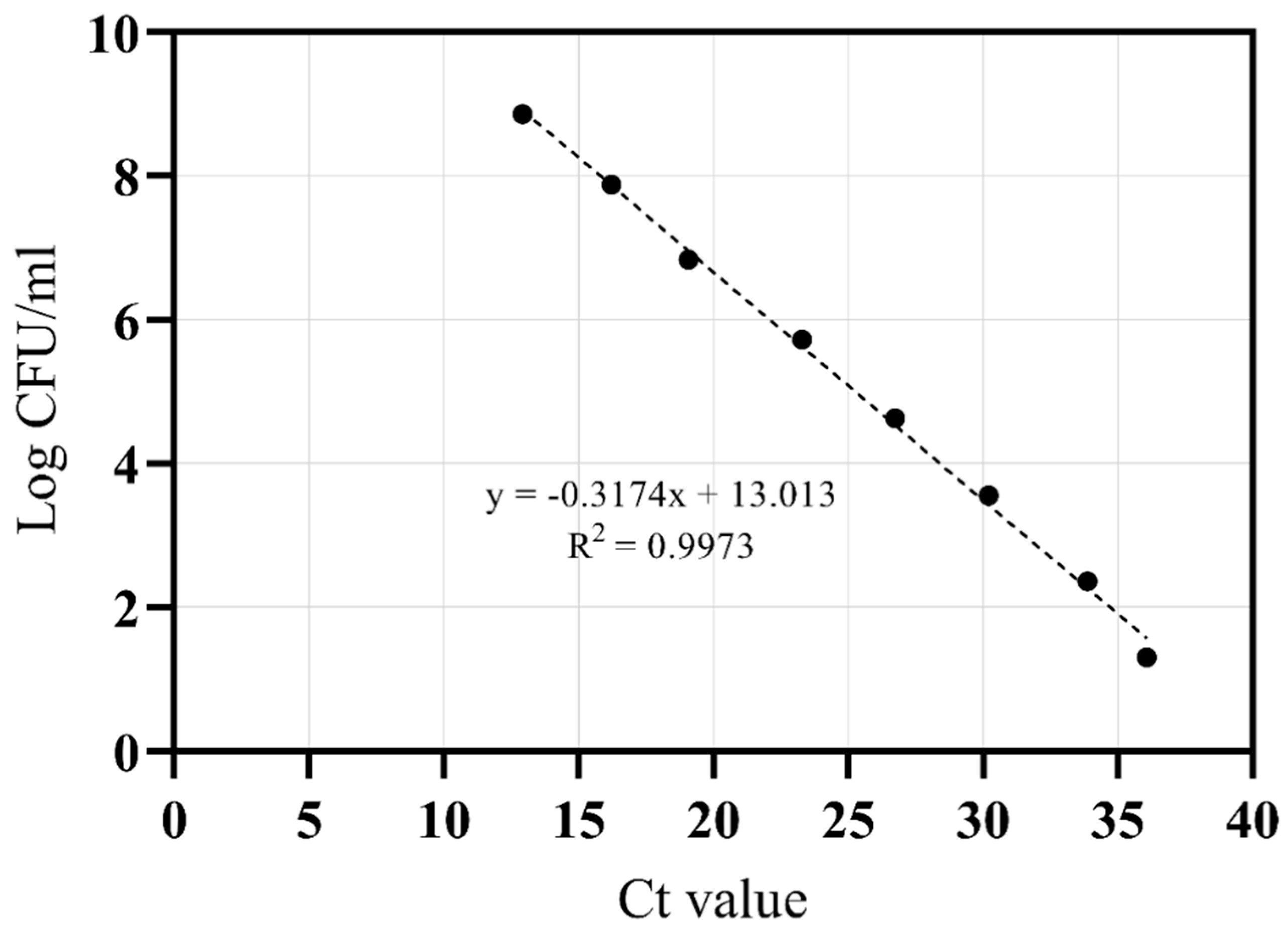
2.7. Standard Curve
2.8. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ansaruzzaman, M.; Lucas, M.; Deen, J.L.; Bhuiyan, N.A.; Wang, X.Y.; Safa, A.; Sultana, M.; Chowdhury, A.; Balakrish Nair, G.; Sack, D.A.; et al. Pandemic Serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus Associated with Diarrhea in Mozambique: Spread of the Pandemic into the African Continent. J. Clin. Microbiol. 2005, 43, 2559–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.C.; Liu, C. Vibrio parahaemolyticus: A Concern of Seafood Safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Balter, S.; Hanson, H.; Kornstein, L.; Lee, L.; Reddy, V.; Sahl, S. Vibrio parahaemolyticus Infections Associated with Consumption of Raw Shellfish—Three States, 2006. Morb. Mortal. Wkly. Rep. 2006, 55, 854–857. [Google Scholar]
- Sferlazzo, G.; Meloni, D.; Lamon, S.; Marceddu, M.; Mureddu, A.; Consolati, S.G.; Pisanu, M.; Virgilio, S. Evaluation of Short Purification Cycles in Naturally Contaminated Mediterranean Mussels (Mytilus galloprovincialis) Harvested in Sardinia (Italy). Food Microbiol. 2018, 74, 86–91. [Google Scholar] [CrossRef]
- Murphree, R.L.; Tamplin, M.L. Uptake and Retention of Vibrio Cholerae O1 in the Eastern Oyster, Crassostrea Virginica. Appl. Environ. Microbiol. 1991, 61, 3656–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.H.; Bae, Y.M.; Lee, S.Y. Effects of Varying Concentrations of Sodium Chloride and Acidic Conditions on the Behavior of Vibrio parahaemolyticus and Vibrio Vulnificus Cold-Starved in Artificial Sea Water Microcosms. Food Sci. Biotechnol. 2017, 26, 829–839. [Google Scholar] [CrossRef]
- Jiang, Y.; Chu, Y.; Xie, G.; Li, F.; Wang, L.; Huang, J.; Zhai, Y.; Yao, L. Antimicrobial Resistance, Virulence and Genetic Relationship of Vibrio parahaemolyticus in Seafood from Coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 2019, 290, 116–124. [Google Scholar] [CrossRef]
- Daniels, N.A.; Mackinnon, L.; Bishop, R.; Altekruse, S.; Ray, B.; Hammond, R.M.; Thompson, S.; Wilson, S.; Bean, N.H.; Griffin, P.M.; et al. Vibrio parahaemolyticus Infections in the United States, 1973–1998. J. Infect. Dis. 2000, 181, 1661–1666. [Google Scholar] [CrossRef] [Green Version]
- Feldhusen, F. The role of seafood in bacterialfoodborne diseases. Microbes Infect. 2000, 2, 1651–1660. [Google Scholar] [CrossRef]
- Lozano-León, A.; Torres, J.; Osorio, C.R.; Martínez-Urtaza, J. Identification of Tdh-Positive Vibrio parahaemolyticus from an Outbreak Associated with Raw Oyster Consumption in Spain. FEMS Microbiol. Lett. 2003, 226, 281–284. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Urtaza, J.; Lozano-Leon, A.; DePaola, A.; Ishibashi, M.; Shimada, K.; Nishibuchi, M.; Liebana, E. Characterization of Pathogenic Vibrio parahaemolyticus Isolates from Clinical Sources in Spain and Comparison with Asian and North American Pandemic Isolates. J. Clin. Microbiol. 2004, 42, 4672–4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Urtaza, J.; Powell, A.; Jansa, J.; Rey, J.L.C.; Montero, O.P.; Campello, M.G.; López, M.J.Z.; Pousa, A.; Valles, M.J.F.; Trinanes, J.; et al. Epidemiological Investigation of a Foodborne Outbreak in Spain Associated with U.S. West Coast Genotypes of Vibrio parahaemolyticus. Springerplus 2016, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Stockley, L.; Rangdale, R.; Martinez-Urtaza, J. Environmental Occurrence and Clinical Impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European Perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- European Commission (EC). Commission Regulation (EC) No. 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs; European Commission (EC): Brussels, Belgium, 2005. [Google Scholar]
- Savichtcheva, O.; Okabe, S. Alternative Indicators of Fecal Pollution: Relations with Pathogens and Conventional Indicators, Current Methodologies for Direct Pathogen Monitoring and Future Application Perspectives. Water Res. 2006, 40, 2463–2476. [Google Scholar] [CrossRef]
- Suffredini, E.; Mioni, R.; Mazzette, R.; Bordin, P.; Serratore, P.; Fois, F.; Piano, A.; Cozzi, L.; Croci, L. Detection and Quantification of Vibrio parahaemolyticus in Shellfish from Italian Production Areas. Int. J. Food. Microbiol. 2014, 184, 14–20. [Google Scholar] [CrossRef]
- Bonnin-Jusserand, M.; Copin, S.; Bris, C.L.; Brauge, T.; Gay, M.; Brisabois, A.; Grard, T.; Midelet-Bourdin, G. Vibrio Species Involved in Seafood-Borne Outbreaks (Vibrio cholerae, V. parahaemolyticus and V. vulnificus): Review of Microbiological versus Recent Molecular Detection Methods in Seafood Products. Crit. Rev. Food Sci. Nutr. 2019, 59, 597–610. [Google Scholar] [CrossRef]
- Boutin, B.K.; Reyes, A.L.; Peeler, J.T.; Twedt, R.M. Effect of Temperature and Suspending Vehicle on Survival of Vibrio parahaemolyticus and Vibrio vulnificus. J. Food Prot. 1985, 48, 875–878. [Google Scholar] [CrossRef]
- Bryan, P.J.; Steffan, R.J.; Depaola, A.; Foster, J.W.; Bej, A.K. Adaptive Response to Cold Temperatures in Vibrio vulnificus. Curr. Microbiol. 1999, 38, 168–175. [Google Scholar] [CrossRef]
- Howard, C.; Johnson, J.L. Sensitivity of Vibrio parahaemolyticus to Cold in Oysters, Fish Fillets and Crabmeat. J. Food Sci. 1973, 38, 437–441. [Google Scholar] [CrossRef]
- Shen, X.; Cai, Y.; Liu, C.; Liu, W.; Hui, Y.; Su, Y.C. Effect of Temperature on Uptake and Survival of Vibrio parahaemolyticus in Oysters (Crassostrea plicatula). Int. J. Food Microbiol. 2009, 136, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Telli, A.E.; Doğruer, Y. Discrimination of Viable and Dead Vibrio parahaemolyticus Subjected to Low Temperatures Using Propidium Monoazide—Quantitative Loop Mediated Isothermal Amplification (PMA-QLAMP) and PMA-QPCR. Microb. Pathog. 2019, 132, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, P.; Marek, P.; Daigle, S.; Hoagland, T.; Venkitanarayanan’, K.S. Effect of Chilling and Freezing on Survival of Vibrio parahaemolyticus on Fish Fillets. J. Food Saf. 2002, 22, 209–217. [Google Scholar] [CrossRef]
- Wong, H.-C.; Chen, L.-L.; Yu, C.-M. Survival of psychrotrophic Vibrio mimicus, Vibrio fluvialis and Vibrio parahaemolyticus in culture broth at low temperatures. J. Food Prot. 1994, 57, 607–610. [Google Scholar] [CrossRef]
- Caburlotto, G.; Bianchi, F.; Gennari, M.; Ghidini, V.; Socal, G.; Aubry, F.B.; Bastianini, M.; Tafi, M.C.; Lleo, M.M. Integrated Evaluation of Environmental Parameters Influencing Vibrio Occurrence in the Coastal Northern Adriatic Sea (Italy) Facing the Venetian Lagoon. Microb. Ecol. 2012, 63, 20–31. [Google Scholar] [CrossRef]
- Tang, J.Y.H.; Wan-Rosli, W.F.; Abdul-Razak, N.H.; Yeo, C.C.; Bakar, C.A.; Son, R. Incidence and antibiogram of Vibrio parahaemolyticus in processed and frozen bivalve mollusks in Kuala Terengganu, Malaysia. Int. Food Res. J. 2014, 21, 1349. [Google Scholar]
- Panebianco, A.; Ruolo, A.; Giarratana, F.; Ziino, G. Occurrence of halophilic vibrions in frozen bivalve molluscs from retail outlets. LXV Annu. Meet. Ital. Soc. Vet. Sci. 2011, 93, 341–343. [Google Scholar]
- Xu, X.; Wu, Q.; Zhang, J.; Cheng, J.; Zhang, S.; Wu, K. Prevalence, Pathogenicity, and Serotypes of Vibrio parahaemolyticus in Shrimp from Chinese Retail Markets. Food Control 2014, 46, 81–85. [Google Scholar] [CrossRef]
- Yang, Z.Q.; Jiao, X.A.; Zhou, X.H.; Cao, G.X.; Fang, W.M.; Gu, R.X. Isolation and Molecular Characterization of Vibrio parahaemolyticus from Fresh, Low-Temperature Preserved, Dried, and Salted Seafood Products in Two Coastal Areas of Eastern China. Int. J. Food Microbiol. 2008, 125, 279–285. [Google Scholar] [CrossRef]
- Singleton, F.L.; Attwell, R.; Jangi, S.; Colwell, R.R. Effects of Temperature and Salinity on Vibrio Cholerae Growth. Appl. Environ. Microbiol. 1982, 44, 1047–1058. [Google Scholar] [CrossRef] [Green Version]
- Huq, A.; West’, P.A.; Small, E.B.; Imdadul Huq, M.; Colwell’, R.R. Influence of Water Temperature, Salinity, and PH on Survival and Growth of Toxigenic Vibrio Cholerae Serovar 01 Associated with Live Copepods in Laboratory Microcosms. Appl. Environ. Microbiol. 1984, 48, 420–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamplin, M.L.; Colwell, R.R. Effects of Microcosm Salinity and Organic Substrate Concentration on Production of Vibrio cholerae Enterotoxin. Appl. Environ. Microbiol. 1986, 52, 297–301. [Google Scholar] [CrossRef] [Green Version]
- Amako, K.; Shimodori, S.; Imoto, T.; Miake, S.; Umeda’, A. Effects of Chitin and Its Soluble Derivatives on Survival of Vibrio cholerae 01 at Low Temperature. Appl. Environ. Microbiol. 1987, 53, 603–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunasagar, I.; Venugopal, M.N.; Karunasagar, I.; Segar, K. Role of Chitin in the Survival of Vibrio parahaemolyticus at Different Temperatures. Can. J. Microbiol. 1986, 32, 889–891. [Google Scholar] [CrossRef]
- Liu, C.; Lu, J.; Su, Y.-C. Effects of flash freezing, followed by frozen storage, on reducing Vibrio parahaemolyticus in Pacific raw oysters (Crassostrea gigas). J. Food Prot. 2009, 72, 174–177. [Google Scholar] [CrossRef]
- Yoon, J.H.; Moon, S.K.; Choi, C.; Ryu, B.Y.; Lee, S.Y. Detection of Viable but Nonculturable Vibrio parahaemolyticus Induced by Prolonged Cold-Starvation Using Propidium Monoazide Real-Time Polymerase Chain Reaction. Lett. Appl. Microbiol. 2019, 68, 537–545. [Google Scholar] [CrossRef]
- Wagley, S.; Morcrette, H.; Kovacs-Simon, A.; Yang, Z.R.; Power, A.; Tennant, R.K.; Love, J.; Murray, N.; Titball, R.W.; Butler, C.S. Bacterial Dormancy: A Subpopulation of Viable but Non-Culturable Cells Demonstrates Better Fitness for Revival. PLoS Pathog. 2021, 17, e1009194. [Google Scholar] [CrossRef]
- Oliver, J.D. The Viable but Nonculturable State in Bacteria. J. Microbiol. 2005, 43, 93–100. [Google Scholar] [PubMed]
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.W.; Ding, T. Current Perspectives on Viable but Non-Culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef] [Green Version]
- Dong, K.; Pan, H.; Yang, D.; Rao, L.; Zhao, L.; Wang, Y.; Liao, X. Induction, Detection, Formation, and Resuscitation of Viable but Non-Culturable State Microorganisms. Compr. Rev. Food Sci. Food Saf. 2020, 19, 149–183. [Google Scholar] [CrossRef] [Green Version]
- Cole, K.M.; Supan, J.; Ramirez, A.; Johnson, C.N. Suspension of Oysters Reduces the Populations of Vibrio parahaemolyticus and Vibrio vulnificus. Lett. Appl. Microbiol. 2015, 61, 209–213. [Google Scholar] [CrossRef] [Green Version]
- Ferro, S.; Amorico, T.; Deo, P. Role of Food Sanitising Treatments in Inducing the ‘Viable but Nonculturable’ State of Microorganisms. Food Control 2018, 91, 321–329. [Google Scholar] [CrossRef]
- Schottroff, F.; Fröhling, A.; Zunabovic-Pichler, M.; Krottenthaler, A.; Schlüter, O.; Jäger, H. Sublethal Injury and Viable but Non-Culturable (VBNC) State in Microorganisms during Preservation of Food and Biological Materials by Non-Thermal Processes. Front. Microbiol. 2018, 9, 2773. [Google Scholar] [CrossRef] [PubMed]
- Shamloei, S.; Nabavi-Rad, A.; Nazem, H.; Yadegar, A. Current Perspectives on Viable but Non-Culturable Bacteria in Food Safety and Public Health. Avicenna J. Clin. Microbiol. Infect. 2022, 9, 31–40. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Oliver, J.D. The Viable but Non-Culturable State and Its Relevance in Food Safety. Curr. Opin. Food Sci. 2016, 8, 127–133. [Google Scholar] [CrossRef]
- Gao, R.; Liao, X.; Zhao, X.; Liu, D.; Ding, T. The Diagnostic Tools for Viable but Nonculturable Pathogens in the Food Industry: Current Status and Future Prospects. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2146–2175. [Google Scholar] [CrossRef] [PubMed]
- İzgördü, Ö.K.; Darcan, C.; Kariptaş, E. Overview of VBNC, a Survival Strategy for Microorganisms. 3 Biotech 2022, 12, 307. [Google Scholar] [CrossRef] [PubMed]
- Vattakaven, T.; Bond, P.; Bradley, G.; Munn, C.B. Differential Effects of Temperature and Starvation on Induction of the Viable-but-Nonculturable State in the Coral Pathogens Vibrio shiloi and Vibrio tasmaniensis. Appl. Environ. Microbiol. 2006, 72, 6508–6513. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J.D. Recent Findings on the Viable but Nonculturable State in Pathogenic Bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The Importance of the Viable but Non-Culturable State in Human Bacterial Pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Bae, Y.M.; Jo, S.; Moon, S.K.; Oh, S.W.; Lee, S.Y. Optimization of Resuscitation-Promoting Broths for the Revival of Vibrio parahaemolyticus from a Viable but Nonculturable State. Food Sci. Biotechnol. 2021, 30, 159–169. [Google Scholar] [CrossRef]
- Baffone, W.; Citterio, B.; Vittoria, E.; Casaroli, A.; Campana, R.; Falzano, L.; Donelli, G. Retention of Virulence in Viable but Non-Culturable Halophilic Vibrio Spp. Int. J. Food Microbiol. 2003, 89, 31–39. [Google Scholar] [CrossRef]
- Zhang, X.H.; Ahmad, W.; Zhu, X.Y.; Chen, J.; Austin, B. Viable but Nonculturable Bacteria and Their Resuscitation: Implications for Cultivating Uncultured Marine Microorganisms. Mar. Life Sci. Technol. 2021, 3, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Ren, Q. Wake Up! Resuscitation of Viable but Nonculturable Bacteria: Mechanism and Potential Application. Foods 2023, 12, 82. [Google Scholar] [CrossRef]
- Wu, B.; Liang, W.; Kan, B. Enumeration of Viable Non-Culturable Vibrio cholerae Using Propidium Monoazide Combined with Quantitative PCR. J. Microbiol. Methods 2015, 115, 147–152. [Google Scholar] [CrossRef]
- Wong, H.C.; Wang, P. Induction of Viable but Nonculturable State in Vibrio parahaemolyticus and Its Susceptibility to Environmental Stresses. J. Appl. Microbiol. 2004, 96, 359–366. [Google Scholar] [CrossRef] [Green Version]
- Nocker, A.; Cheung, C.Y.; Camper, A.K. Comparison of Propidium Monoazide with Ethidium Monoazide for Differentiation of Live vs. Dead Bacteria by Selective Removal of DNA from Dead Cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Sheet, O.H.; Grabowski, N.T.; Klein, G.; Abdulmawjood, A. Development and Validation of a Loop Mediated Isothermal Amplification (LAMP) Assay for the Detection of Staphylococcus aureus in Bovine Mastitis Milk Samples. Mol. Cell Probes 2016, 30, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhang, Z.; Sun, X.; Pan, Y.; Zhao, Y. Development of a Quantitative Real-Time PCR Assay for Viable salmonella Spp. without Enrichment. Food Control 2015, 57, 185–189. [Google Scholar] [CrossRef]
- Yoon, J.H.; Wei, S.; Oh, D.H. A Highly Selective Enrichment Broth Combined with Real-Time PCR for Detection of Staphylococcus aureus in Food Samples. LWT 2018, 94, 103–110. [Google Scholar] [CrossRef]
- Cao, X.; Zhao, L.; Zhang, J.; Chen, X.; Shi, L.; Fang, X.; Xie, H.; Chang, Y.; Wang, L. Detection of Viable but Nonculturable Vibrio parahaemolyticus in Shrimp Samples Using Improved Real-Time PCR and Real-Time LAMP Methods. Food Control 2019, 103, 145–152. [Google Scholar] [CrossRef]
- Ling, N.; Shen, J.; Guo, J.; Zeng, D.; Ren, J.; Sun, L.; Jiang, Y.; Xue, F.; Dai, J.; Li, B. Rapid and Accurate Detection of Viable Vibrio parahaemolyticus by Sodium Deoxycholate-Propidium Monoazide-QPCR in Shrimp. Food Control 2020, 109, 106883. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, Q.; Wang, J.; Lei, S. Enumeration of Vibrio parahaemolyticus in VBNC State by PMA-Combined Real-Time Quantitative PCR Coupled with Confirmation of Respiratory Activity. Food Control 2018, 91, 85–91. [Google Scholar] [CrossRef]
- Zhong, Q.; Tian, J.; Wang, B.; Wang, L. PMA Based Real-Time Fluorescent LAMP for Detection of Vibrio parahaemolyticus in Viable but Nonculturable State. Food Control 2016, 63, 230–238. [Google Scholar] [CrossRef]
- Zhu, R.G.; Li, T.P.; Jia, Y.F.; Song, L.F. Quantitative Study of Viable Vibrio parahaemolyticus Cells in Raw Seafood Using Propidium Monoazide in Combination with Quantitative PCR. J. Microbiol. Methods 2012, 90, 262–266. [Google Scholar] [CrossRef]
- Zhao, L.; Lv, X.; Cao, X.; Zhang, J.; Gu, X.; Zeng, H.; Wang, L. Improved Quantitative Detection of VBNC Vibrio parahaemolyticus Using Immunomagnetic Separation and PMAxx-QPCR. Food Control 2020, 110, 106962. [Google Scholar] [CrossRef]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in Understanding Preferential Detection of Live Cells Using Viability Dyes in Combination with DNA Amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef]
- Kobayashi, H.; Oethinger, M.; Tuohy, M.J.; Hall, G.S.; Bauer, T.W. Improving Clinical Significance of PCR: Use of Propidium Monoazide to Distinguish Viable from Dead Staphylococcus aureus and Staphylococcus epidermidis. J. Orthop. Res. 2009, 27, 1243–1247. [Google Scholar] [CrossRef]
- Yang, X.; Badoni, M.; Gill, C.O. Use of Propidium Monoazide and Quantitative PCR for Differentiation of Viable Escherichia Coli from E. Coli Killed by Mild or Pasteurizing Heat Treatments. Food Microbiol. 2011, 28, 1478–1482. [Google Scholar] [CrossRef]
- Lee, J.L.; Levin, R.E. A Comparative Study of the Ability of EMA and PMA to Distinguish Viable from Heat Killed Mixed Bacterial Flora from Fish Fillets. J. Microbiol. Methods 2009, 76, 93–96. [Google Scholar] [CrossRef]
- Dinu, L.D.; Bach, S. Detection of Viable but Non-Culturable Escherichia coli O157: H7 from Vegetable Samples Using Quantitative PCR with Propidium Monoazide and Immunological Assays. Food Control 2013, 31, 268–273. [Google Scholar] [CrossRef]
- Xiao, X.; Tian, C.; Yu, Y.; Wu, H. Detection of Viable but Nonculturable Escherichia coli O157:H7 Using Propidium Monoazide Treatments and QPCR. Can. J. Microbiol. 2013, 59, 157–163. [Google Scholar] [CrossRef]
- Lv, X.; Wang, L.; Zhang, J.; Zeng, H.; Chen, X.; Shi, L.; Cui, H.; He, X.; Zhao, L. Rapid and Sensitive Detection of VBNC Escherichia coli O157: H7 in Beef by PMAxx and Real-Time LAMP. Food Control 2020, 115, 107292. [Google Scholar] [CrossRef]
- Zhang, H.-N.; Hou, P.-B.; Chen, Y.-Z.; Ma, Y.; Li, X.-P.; Lv, H.; Wang, M.; Tan, H.-L.; Bi, Z.-W. Prevalence of Foodborne Pathogens in Cooked Meat and Seafood from 2010 to 2013 in Shandong Province, China. Iran. J. Public Health 2016, 45, 1577. [Google Scholar]
- El-Aziz, N.K.A.; Tartor, Y.H.; El-Aziz Gharib, A.A.; Ammar, A.M. Propidium Monoazide Quantitative Real-Time Polymerase Chain Reaction for Enumeration of Some Viable but Nonculturable Foodborne Bacteria in Meat and Meat Products. Foodborne Pathog. Dis. 2018, 15, 226–234. [Google Scholar] [CrossRef]
- Rowan, N.J. Viable but Non-Culturable Forms of Food and Waterborne Bacteria: Quo Vadis? Trends Food Sci. Technol. 2004, 15, 462–467. [Google Scholar] [CrossRef]
- Fakruddin, M.; Mannan, K.S.B.; Andrews, S. Viable but Nonculturable Bacteria: Food Safety and Public Health Perspective. ISRN Microbiol. 2013, 2013, 703813. [Google Scholar] [CrossRef] [PubMed]
- EN ISO 21872-1: 2017; Microbiology of the Food Chian-Horizontal Method for the Determination of Vibrio spp. Part 1: Detection of Potentially Enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. International Organization for Standardization: Geneva, Switzerland, 2017.
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.C.L.; Jones, D.D.; Kaysner, C.A. Detection of Total and Hemoly-sin-Producing Vibrio parahaemolyticus in Shellfish Using Multiplex PCR Amplification of Tl, Tdh and Trh. J. Microbiol. Methods 1999, 36, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR; Springer: Berlin/Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar]
- Caraguel, C.G.B.; Stryhn, H.; Gagné, N.; Dohoo, I.R.; Hammell, K.L. Selection of a Cutoff Value for Real-Time Polymerase Chain Reaction Results to Fit a Diagnostic Purpose: Analytical and Epidemiologic Approaches. J. Vet. Diagn. Investig. 2011, 23, 2–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.B.; Okuda, J.U.N.; Matsumoto, C.; Takahashi, N.; Hashimoto, S.; Nishibuchi, M. Identification of Vibrio parahaemolyticus Strains at the Species Level by PCR Targeted to the ToxR Gene. J. Clin. Microbiol. 1999, 37, 1173–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tada, J.; Ohashi, T.; Nishimura, N.; Shirasaki, Y.; Ozaki, H.; Fukushima, S.; Takano, J.; Nishibuchi, M.; Takeda, Y. Detection of the Thermostable Direct Hemolysin Gene (Tdh) and the Thermostable Direct Hemolysin-Related Hemolysin Gene (Trh) of Vibrio parahaemolyticus by Polymerase Chain Reaction. Mol. Cell Probes 1992, 6, 477–487. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Pan, S.-F.; Chen, C.-H. Sequence of a Cloned PR72H Fragment and Its Use for Detection of Vibrio parahaemolyticus in Shellfish with the PCR. Appl. Environ. Microbiol 1995, 61, 1311–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkateswaran, K.; Dohmoto, N.; Harayama, S. Cloning and Nucleotide Sequence of the GyrB Gene of Vibrio parahaemolyticus and Its Application in Detection of This Pathogen in Shrimp. Appl. Environ. Microbiol. 1998, 64, 681–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, K.; Oshima, K.; Kurokawa, K.; Yokoyama, K.; Uda, T.; Tagomori, K.; Iijima, Y.; Najima, M.; Nakano, M.; Yamashita, A.; et al. Genome Sequence of Vibrio parahaemolyticus: A Pathogenic Mechanism Distinct from That of V Cholerae. Lancet 2003, 361, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.L.; Lüdeke, C.H.M.; Bowers, J.C.; Garrett, N.; Fischer, M.; Parsons, M.B.; Bopp, C.A.; DePaola, A. Biochemical, Serological, and Virulence Characterization of Clinical and Oyster Vibrio parahaemolyticus Isolates. J. Clin. Microbiol. 2012, 50, 2343–2352. [Google Scholar] [CrossRef] [Green Version]
- Nocker, A.; Camper, A.K. Novel Approaches toward Preferential Detection of Viable Cells Using Nucleic Acid Amplification Techniques. FEMS Microbiol. Lett. 2009, 291, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Lv, R.; Wang, K.; Feng, J.; Heeney, D.D.; Liu, D.; Lu, X. Detection and Quantification of Viable but Non-Culturable Campylobacter Jejuni. Front. Microbiol. 2020, 10, 2920. [Google Scholar] [CrossRef]
- Wang, R.; Zhong, Y.; Gu, X.; Yuan, J.; Saeed, A.F.; Wang, S. The Pathogenesis, Detection, and Prevention of Vibrio parahaemolyticus. Front. Microbiol. 2015, 6, 144. [Google Scholar] [CrossRef]
- Chahorm, K.; Prakitchaiwattana, C. Application of Reverse Transcriptase-PCR-DGGE as a Rapid Method for Routine Determination of Vibrio Spp. in Foods. Int. J. Food Microbiol. 2018, 264, 46–52. [Google Scholar] [CrossRef]
- Shimodori, S.; Moriya, T.; Kohashi, O.; Faming, D.; Amako, K. Extraction from Prawn Shells of Substances Cryoprotective for Vibrio Cholerae. Appl. Environ. Microbiol. 1989, 55, 2726–2728. [Google Scholar] [CrossRef] [Green Version]
- Bang, W.; Drake, M.A. Resistance of cold-and starvation-stressed Vibrio vulnificus to heat and freeze-thaw exposure. J. Food Prot. 2002, 65, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Johnston, M.D. An Investigation into the Changed Physiological State of Vibrio Bacteria as a Survival Mechanism in Response to Cold Temperatures and Studies on Their Sensitivity to Heating and Freezing. J. Appl. Microbiol. 2002, 92, 1066–1077. [Google Scholar] [CrossRef]
- Yu, H.; Fang, J.; Ma, B.; Li, J.; Zhang, M. One Real-Time Fluorescent Loop-Mediated Isothermal Amplification Combined with Propidium Monoazide for Detection of Viable Vibrio parahaemolyticus in Seafood. Am. J. Biochem. Biotechnol. 2019, 15, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Niu, B.; Hong, B.; Zhang, Z.; Mu, L.; Malakar, P.K.; Liu, H.; Pan, Y.; Zhao, Y. A Novel QPCR Method for Simultaneous Detection and Quantification of Viable Pathogenic and Non-Pathogenic Vibrio parahaemolyticus (Tlh+, Tdh+, and UreR+). Front. Microbiol. 2018, 9, 1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randazzo, W.; Khezri, M.; Ollivier, J.; Le Guyader, F.S.; Rodríguez-Díaz, J.; Aznar, R.; Sánchez, G. Optimization of PMAxx Pretreatment to Distinguish between Human Norovirus with Intact and Altered Capsids in Shellfish and Sewage Samples. Int. J. Food Microbiol. 2018, 266, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouju-Albert, A.; Saltaji, S.; Dousset, X.; Prévost, H.; Jaffrès, E. Quantification of Viable Brochothrix Thermosphacta in Cold-Smoked Salmon Using PMA/PMAxx-QPCR. Front. Microbiol. 2021, 12, 654178. [Google Scholar] [CrossRef]
- Lamon, S.; Bastardo, A.; Meloni, D.; Consolati, S.G.; Fois, F.; Porcheddu, G.; Agus, V.; Pes, M.; Cambula, M.G.; Mureddu, A.; et al. Clonal Relationship among Vibrio parahaemolyticus Isolated from Mediterranean Mussels (Mytilus galloprovincialis) and Grooved Carpet Shells (Ruditapes decussatus) Harvested in Sardinia (Italy). Food Microbiol. 2019, 84, 103258. [Google Scholar] [CrossRef]
- Bacian, C.; Verdugo, C.; García, K.; Perez-Larruscain, J.; de Blas, I.; Cachicas, V.; Lopez-Joven, C. Longitudinal Study of Total and Pathogenic Vibrio parahaemolyticus (Tdh+ and/or Trh+) in Two Natural Extraction Areas of Mytilus Chilensis in Southern Chile. Front. Microbiol. 2021, 12, 621737. [Google Scholar] [CrossRef]
- Lamon, S.; Consolati, S.G.; Fois, F.; Cambula, M.G.; Pes, M.; Porcheddu, G.; Agus, V.; Esposito, G.; Mureddu, A.; Meloni, D. Occurrence, Seasonal Distribution, and Molecular Characterization of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in Shellfish (Mytilus galloprovincialis and Ruditapes decussatus) Collected in Sardinia (Italy). J. Food Prot. 2019, 82, 1851–1856. [Google Scholar] [CrossRef]
- Lopatek, M.; Wieczorek, K.; Osek, J. Prevalence and Antimicrobial Resistance of Vibrio parahaemolyticus Isolated from Raw Shellfish in Poland. J. Food Prot. 2015, 78, 1029–1033. [Google Scholar] [CrossRef]
- Roque, A.; Lopez-Joven, C.; Lacuesta, B.; Elandaloussi, L.; Wagley, S.; Furones, M.D.; Ruiz-Zarzuela, I.; De Blas, I.; Rangdale, R.; Gomez-Gil, B. Detection and Identification of Tdh- And Trh-Positive Vibrio parahaemolyticus Strains from Four Species of Cultured Bivalve Molluscs on the Spanish Mediterranean Coast. Appl. Environ. Microbiol. 2009, 75, 7574–7577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.N.; Bowers, J.C.; Griffitt, K.J.; Molina, V.; Clostio, R.W.; Pei, S.; Laws, E.; Paranjpye, R.N.; Strom, M.S.; Chen, A.; et al. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the Coastal and Estuarine Waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microbiol. 2012, 78, 7249–7257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Froelich, B.; Oliver, J. Increases in the Amounts of Vibrio Spp. in Oysters upon Addition of Exogenous Bacteria. Appl. Environ. Microbiol. 2013, 79, 5208–5213. [Google Scholar] [CrossRef] [Green Version]
- Randa, M.A.; Polz, M.F.; Lim, E. Effects of Temperature and Salinity on Vibrio vulnificus Population Dynamics as Assessed by Quantitative PCR. Appl. Environ. Microbiol. 2004, 70, 5469–5476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motes, M.L.; Depaola, A.; Cook, D.W.; Veazey, J.E.; Hunsucker, J.C.; Garthright, W.E.; Blodgett, R.J.; Chirtel, S.J. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 1998, 64, 1459–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, P.W.; Oliver, J.D. Temperature Effects on the Viable but Non-culturable State of Vibrio vulnificus. FEMS Microbiol. Lett. 1992, 101, 33–39. [Google Scholar] [CrossRef]
- Makino, S.-I.; Kii, T.; Asakura, H.; Shirahata, T.; Ikeda, T.; Takeshi, K.; Itoh, K. Does Enterohemorrhagic Escherichia coli O157:H7 Enter the Viable but Nonculturable State in Salted Salmon Roe? Appl. Environ. Microbiol. 2000, 66, 5536–5539. [Google Scholar] [CrossRef] [Green Version]
- Asakura, H.; Panutdaporn, N.; Kawamoto, K.; Igimi, S.; Yamamoto, S.; Makino, S.I. Proteomic characterization of enter-ohemorrhagic Escherichia coli O157: H7 in the oxidation-induced viable but non-culturable state. Microbiol. Immunol. 2007, 51, 875–881. [Google Scholar] [CrossRef]
- Aurass, P.; Prager, R.; Flieger, A. EHEC/EAEC O104:H4 Strain Linked with the 2011 German Outbreak of Haemolytic Uremic Syndrome Enters into the Viable but Non-Culturable State in Response to Various Stresses and Resuscitates upon Stress Relief. Environ. Microbiol. 2011, 13, 3139–3148. [Google Scholar] [CrossRef]
- Nicolò, M.S.; Guglielmino, S.P.P. Viable but nonculturable bacteria in food. In Public Health–Methodology, Environmental and Systems Issues; Maddock, J., Ed.; InTech: Rjeka, Croatia, 2012; pp. 189–216. [Google Scholar] [CrossRef]
- Panebianco, F.; Nava, V.; Giarratana, F.; Gervasi, T.; Cicero, N. Assessment of Heavy- and Semi-Metals Contamination in Edible Seaweed and Dried Fish Sold in Ethnic Food Stores on the Italian Market. J. Food Compos. Anal. 2021, 104, 104150. [Google Scholar] [CrossRef]
- Panebianco, F.; Giusti, A.; Giarratana, F.; Armani, A. Ethnic Seafood Products Sold on the Italian Market: Labelling Assessment and Biological, Chemical and Physical Risk Characterization. Food Control 2019, 105, 198–208. [Google Scholar] [CrossRef]
| Sample ID | Species | Type | FAO Fishing Area | Sampling Date |
|---|---|---|---|---|
| 1 | Paphia undulata | shelled clams | 71 | 10 February 2020 |
| 2 | Mytilus chilensis | shelled mussels | 87 | 10 February 2020 |
| 3 | Paphia undulata | shelled clams | 71 | 10 February 2020 |
| 4 | Mytilus chilensis | half shell mussels | 87 | 10 February 2020 |
| 5 | Mytilus chilensis | shelled mussels | 87 | 10 February 2020 |
| 6 | Chamelea gallina | shelled clams | 71 | 10 February 2020 |
| 7 | Mytilus chilensis | shelled mussels | 87 | 10 February 2020 |
| 8 | Paphia undulata | shelled clams | 71 | 10 February 2020 |
| 9 | Mytilus chilensis | shelled mussels | 87 | 16 March 2020 |
| 10 | Mytilus chilensis | shelled mussels | 87 | 16 March 2020 |
| 11 | Paphia undulata | shelled clams | 71 | 16 March 2020 |
| 12 | Mytilus chilensis | shelled mussels | 87 | 16 March 2020 |
| 13 | Paphia undulata | whole shell clams | 71 | 16 March 2020 |
| 14 | Chamelea gallina | shelled clams | 37 | 16 March 2020 |
| 15 | Mytilus chilensis | shelled mussels | 87 | 16 March 2020 |
| 16 | Chamelea gallina | shelled clams | 71 | 16 March 2020 |
| 17 | Chamelea gallina | shelled clams | 71 | 16 March 2020 |
| 18 | Mytilus chilensis | shelled mussels | 87 | 20 April 2020 |
| 19 | Chamelea gallina | shelled clams | 71 | 20 April 2020 |
| 20 | Paphia textile | shelled clams | 61 | 20 April 2020 |
| 21 | Chamelea gallina | shelled clams | 71 | 20 April 2020 |
| 22 | Paphia textile | shelled clams | 61 | 20 April 2020 |
| 23 | Chamelea gallina | shelled clams | 71 | 20 April 2020 |
| 24 | Mytilus chilensis | shelled mussels | 87 | 20 April 2020 |
| 25 | Chamelea gallina | shelled clams | 71 | 20 April 2020 |
| 26 | Mytilus chilensis | shelled mussels | 87 | 20 April 2020 |
| 27 | Mytilus chilensis | shelled mussels | 87 | 22 June 2020 |
| 28 | Mytilus chilensis | shelled mussels | 87 | 22 June 2020 |
| 29 | Meretix lyrata | whole shell clams | 61 | 22 June 2020 |
| 30 | Paphia undulata | shelled clams | 71 | 22 June 2020 |
| 31 | Mytilus chilensis | whole shell mussels | 87 | 22 June 2020 |
| 32 | Mytilus chilensis | shelled mussels | 87 | 22 June 2020 |
| 33 | Paphia undulata | shelled clams | 71 | 22 June 2020 |
| 34 | Mytilus chilensis | shelled mussels | 87 | 22 June 2020 |
| 35 | Mytilus chilensis | shelled mussels | 87 | 27 July 2020 |
| 36 | Paphia undulata | shelled clams | 71 | 27 July 2020 |
| 37 | Paphia textile | whole shell clams | 61 | 27 July 2020 |
| 38 | Mytilus chilensis | shelled mussels | 87 | 27 July 2020 |
| 39 | Paphia undulata | shelled clams | 71 | 27 July 2020 |
| 40 | Mytilus chilensis | shelled mussels | 87 | 27 July 2020 |
| 41 | Paphia textile | shelled clams | 37 | 27 July 2020 |
| 42 | Paphia textile | shelled clams | 61 | 27 July 2020 |
| 43 | Paphia textile | shelled clams | 61 | 27 July 2020 |
| 44 | Mytilus chilensis | shelled mussels | 87 | 7 September 2020 |
| 45 | Paphia undulata | shelled clams | 71 | 7 September 2020 |
| 46 | Mytilus chilensis | shelled mussels | 87 | 7 September 2020 |
| 47 | Chamelea gallina | shelled clams | 71 | 7 September 2020 |
| 48 | Mytilus chilensis | shelled mussels | 87 | 7 September 2020 |
| 49 | Mytilus chilensis | shelled mussels | 87 | 7 September 2020 |
| 50 | Meretrix meretrix | whole shell clams | 61 | 7 September 2020 |
| 51 | Paphia undulata | shelled clams | 71 | 7 September 2020 |
| 52 | Paphia undulata | shelled clams | 71 | 13 October 2020 |
| 53 | Mytilus chilensis | whole shell mussels | 87 | 13 October 2020 |
| 54 | Meretrix lyrata | whole shell clams | 61 | 13 October 2020 |
| 55 | Paphia undulata | shelled clams | 71 | 13 October 2020 |
| 56 | Mytilus chilensis | shelled mussels | 87 | 13 October 2020 |
| 57 | Paphia textile | shelled clams | 61 | 13 October 2020 |
| 58 | Mytilus chilensis | shelled mussels | 87 | 13 October 2020 |
| 59 | Meretrix lyrata | shelled clams | 61 | 13 October 2020 |
| 60 | Mytilus chilensis | shelled mussels | 87 | 13 October 2020 |
| 61 | Mytilus chilensis | shelled mussels | 87 | 14 December 2020 |
| 62 | Chamelea gallina | shelled clams | 71 | 14 December 2020 |
| 63 | Chamelea gallina | shelled clams | 71 | 14 December 2020 |
| 64 | Mytilus chilensis | shelled mussels | 87 | 14 December 2020 |
| 65 | Mytilus chilensis | shelled mussels | 87 | 14 December 2020 |
| 66 | Chamelea gallina | shelled clams | 71 | 14 December 2020 |
| 67 | Chamelea gallina | shelled clams | 71 | 14 December 2020 |
| 68 | Paphia textile | shelled clams | 61 | 14 December 2020 |
| 69 | Meretrix lyrata | whole shell clams | 61 | 11 January 2021 |
| 70 | Mytilus chilensis | shelled mussels | 87 | 11 January 2021 |
| 71 | Mytilus chilensis | shelled mussels | 87 | 11 January 2021 |
| 72 | Mytilus chilensis | shelled mussels | 87 | 11 January 2021 |
| 73 | Paphia textile | shelled clams | 61 | 11 January 2021 |
| 74 | Mytilus chilensis | shelled mussels | 87 | 11 January 2021 |
| 75 | Mytilus chilensis | shelled mussels | 87 | 11 January 2021 |
| 76 | Paphia textile | shelled clams | 61 | 11 January 2021 |
| 77 | Meretrix meretrix | whole shell clams | 61 | 11 January 2021 |
| No. | Species | Strain | Source | qPCR Result |
|---|---|---|---|---|
| 1 | V. parahaemolyticus | ATCC 17802 | Shirasu food poisoning, Japan | + |
| 2 | V. parahaemolyticus | ATCC 33847 | human clinical isolate | + |
| 3 | V. parahaemolyticus | CCUG 43363 | unknown | + |
| 4 | V. parahaemolyticus | MELAB 772 | mussels | + |
| 5 | V. parahaemolyticus | MELAB 777 | mussels | + |
| 6 | V. parahaemolyticus | MELAB 778 | mussels | + |
| 7 | V. parahaemolyticus | MELAB 547 | fish | + |
| 8 | V. alginolyticus | ATCC 17749 | fish | − |
| 9 | V. vulnificus | ATCC 27562 | blood | − |
| 10 | V. cholerae | CCUG 37531 | unknown | − |
| 11 | V. mimicus | CCUG 13624 | human clinical isolate | − |
| 12 | A. hydrophila | ATCC 7966T | tin of milk with fishy odor | − |
| 13 | A. molluscorum | CECT 5864 | wedge shells | − |
| 14 | A. sobria | CECT 4245T | fish | − |
| 15 | E. coli | ATCC 8739 | feces | − |
| 16 | L. monocytogenes | ATCC 13932 | human clinical isolate | − |
| 17 | P. aeruginosa | ATCC 15442 | water | − |
| Sample ID | Type | Species | FAO Fishing Area | Sampling Period | Plate Count | CT Values | Log CFU/g Predicted | ||
|---|---|---|---|---|---|---|---|---|---|
| qPCR (Dead + VBNC) | PMA-qPCR (VBNC) | qPRC (Dead + VBNC) | PMA-qPCR (VBNC) | ||||||
| 6 | shelled clams | Chamelea gallina | 71 | February 2020 | UD | 29.05 a | UD | 3.79 a | UD |
| 8 | shelled clams | Paphia undulata | 71 | March 2020 | UD | 31.18 b | 34.54 a | 3.12 b | 2.05 a |
| 11 | shelled clams | Paphia undulata | 71 | March 2020 | UD | 29.26 c | 35.1 b | 3.73 c | 1.87 b |
| 16 | shelled clams | Chamelea gallina | 71 | March 2020 | UD | 32.08 d | 35.73 c | 2.83 d | 1.67 c |
| 19 | shelled clams | Chamelea gallina | 71 | April 2020 | UD | 33.37 e | 34.74 d | 2.42 e | 1.99 d |
| 20 | shelled clams | Paphia textile | 61 | April 2020 | UD | 30.21 f | 33.78 e | 3.42 f | 2.29 e |
| 23 | shelled clams | Chamelea gallina | 71 | April 2020 | UD | 35.21 g | UD | 1.84 g | UD |
| 33 | shelled clams | Paphia undulata | 71 | June 2020 | UD | 29.74 h | 35.58 f | 3.57 h | 1.72 f |
| 36 | shelled clams | Paphia undulata | 71 | July 2020 | UD | 32.86 i | UD | 2.58 i | UD |
| 45 | shelled clams | Paphia undulata | 71 | September 2020 | UD | 27.32 l | 34.62 g | 4.34 l | 2.03 a |
| 51 | shelled clams | Paphia undulata | 71 | September 2020 | UD | 35.67 m | UD | 1.69 m | UD |
| 62 | shelled clams | Chamelea gallina | 71 | December 2020 | UD | 35.59 n | UD | 1.72 n | UD |
| 63 | shelled clams | Chamelea gallina | 71 | December 2020 | UD | 31.19 b | 35.51 h | 3.11 b | 1.74 f |
| 66 | shelled clams | Chamelea gallina | 71 | December 2020 | UD | 33.65 o | 35.48 i | 2.33 o | 1.75 fg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Salvo, E.; Panebianco, F.; Panebianco, A.; Ziino, G. Quantitative Detection of Viable but Nonculturable Vibrio parahaemolyticus in Frozen Bivalve Molluscs. Foods 2023, 12, 2373. https://doi.org/10.3390/foods12122373
Di Salvo E, Panebianco F, Panebianco A, Ziino G. Quantitative Detection of Viable but Nonculturable Vibrio parahaemolyticus in Frozen Bivalve Molluscs. Foods. 2023; 12(12):2373. https://doi.org/10.3390/foods12122373
Chicago/Turabian StyleDi Salvo, Eleonora, Felice Panebianco, Antonio Panebianco, and Graziella Ziino. 2023. "Quantitative Detection of Viable but Nonculturable Vibrio parahaemolyticus in Frozen Bivalve Molluscs" Foods 12, no. 12: 2373. https://doi.org/10.3390/foods12122373
APA StyleDi Salvo, E., Panebianco, F., Panebianco, A., & Ziino, G. (2023). Quantitative Detection of Viable but Nonculturable Vibrio parahaemolyticus in Frozen Bivalve Molluscs. Foods, 12(12), 2373. https://doi.org/10.3390/foods12122373






