Quality Assessment and Ripeness Prediction of Table Grapes Using Visible–Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
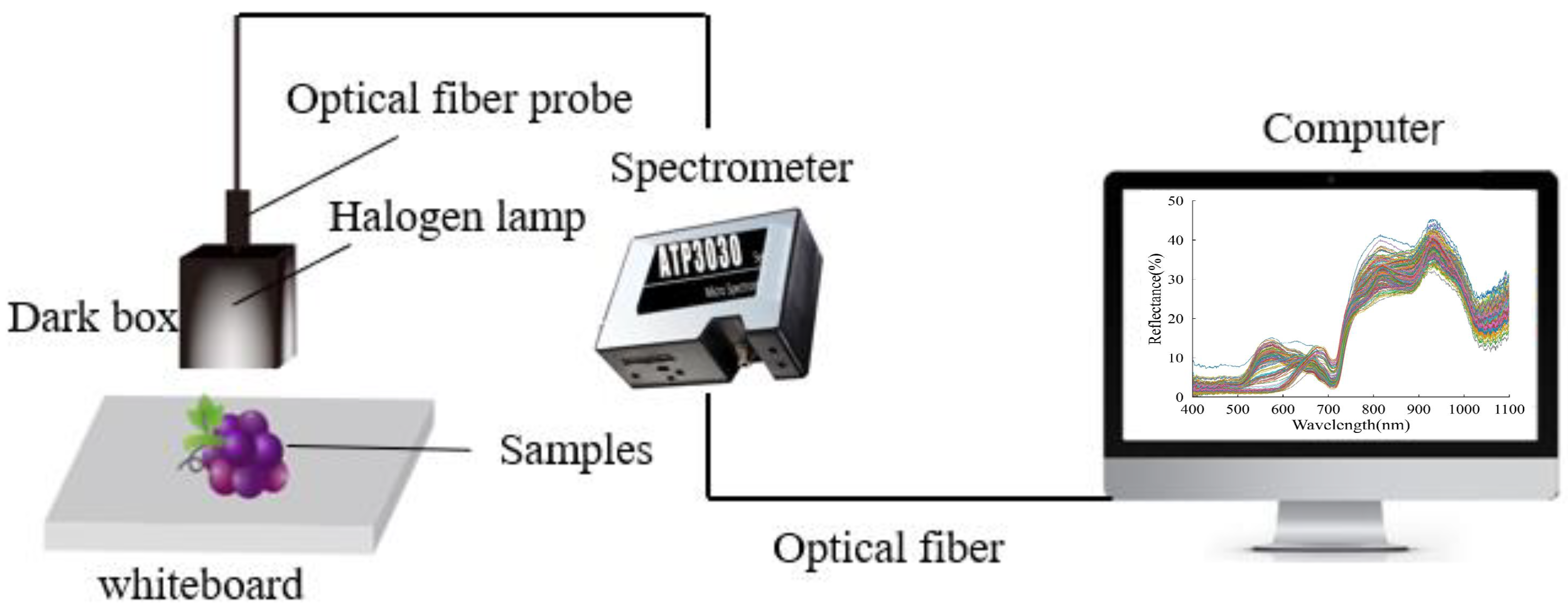
2.2. Vis-NIR Reflectance Spectral Data Acquisition
2.3. Measurement of Physicochemical Parameters
2.3.1. Color Testing
2.3.2. Texture Measurement
2.3.3. SSC and TA Determination
2.4. Data Analysis and Model Establishment
2.4.1. Spectral Data Preprocessing
2.4.2. Effective Wavelength Selection
2.4.3. Model Establishment Method
2.4.4. Model Performance Evaluation
3. Results and Discussion
3.1. Analysis of Physical and Chemical Indicators of Grapes
3.1.1. Color Analysis
3.1.2. Texture Characteristics
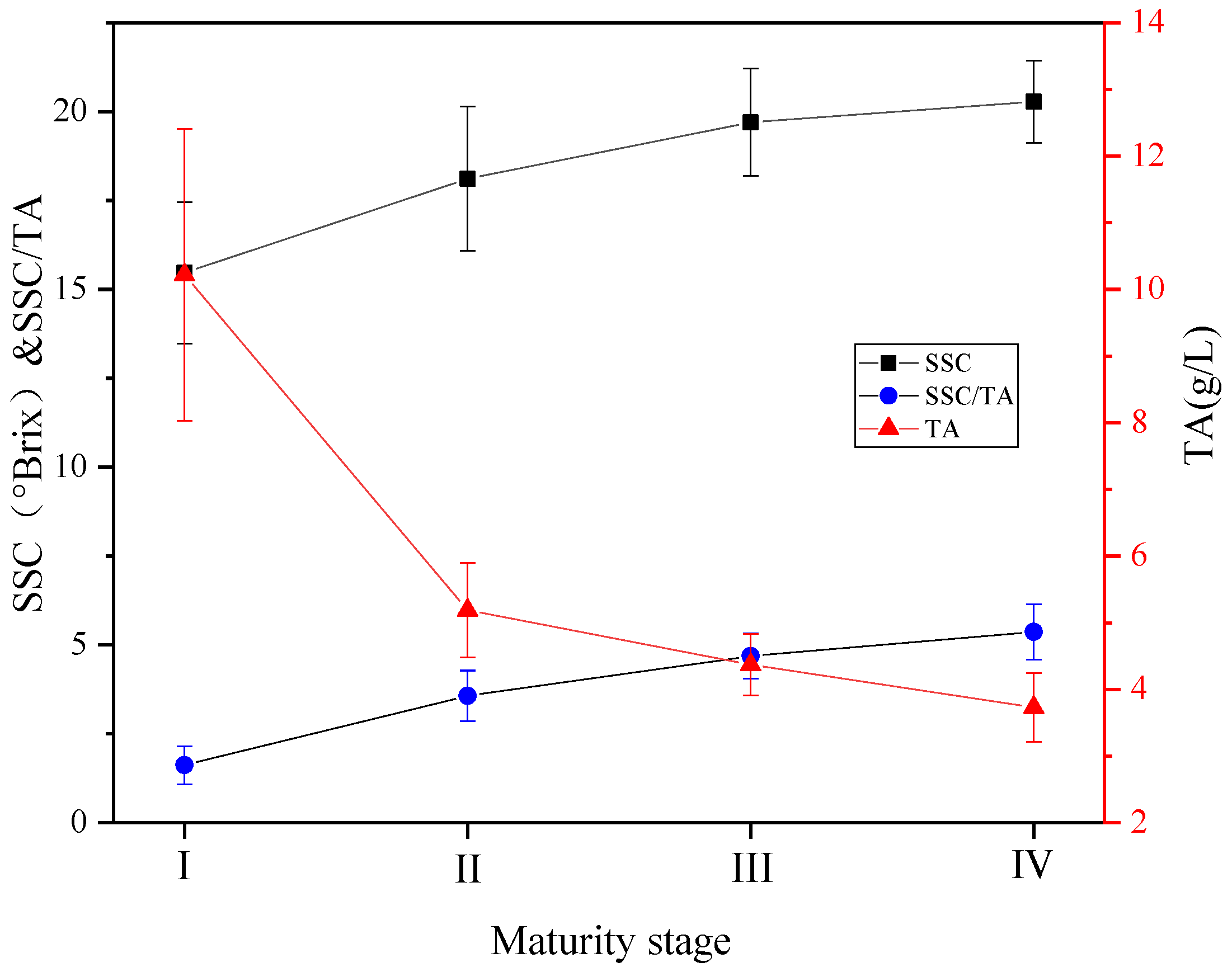
3.1.3. Soluble Solids and Total Acid Analysis
3.2. Spectral Model Establishment
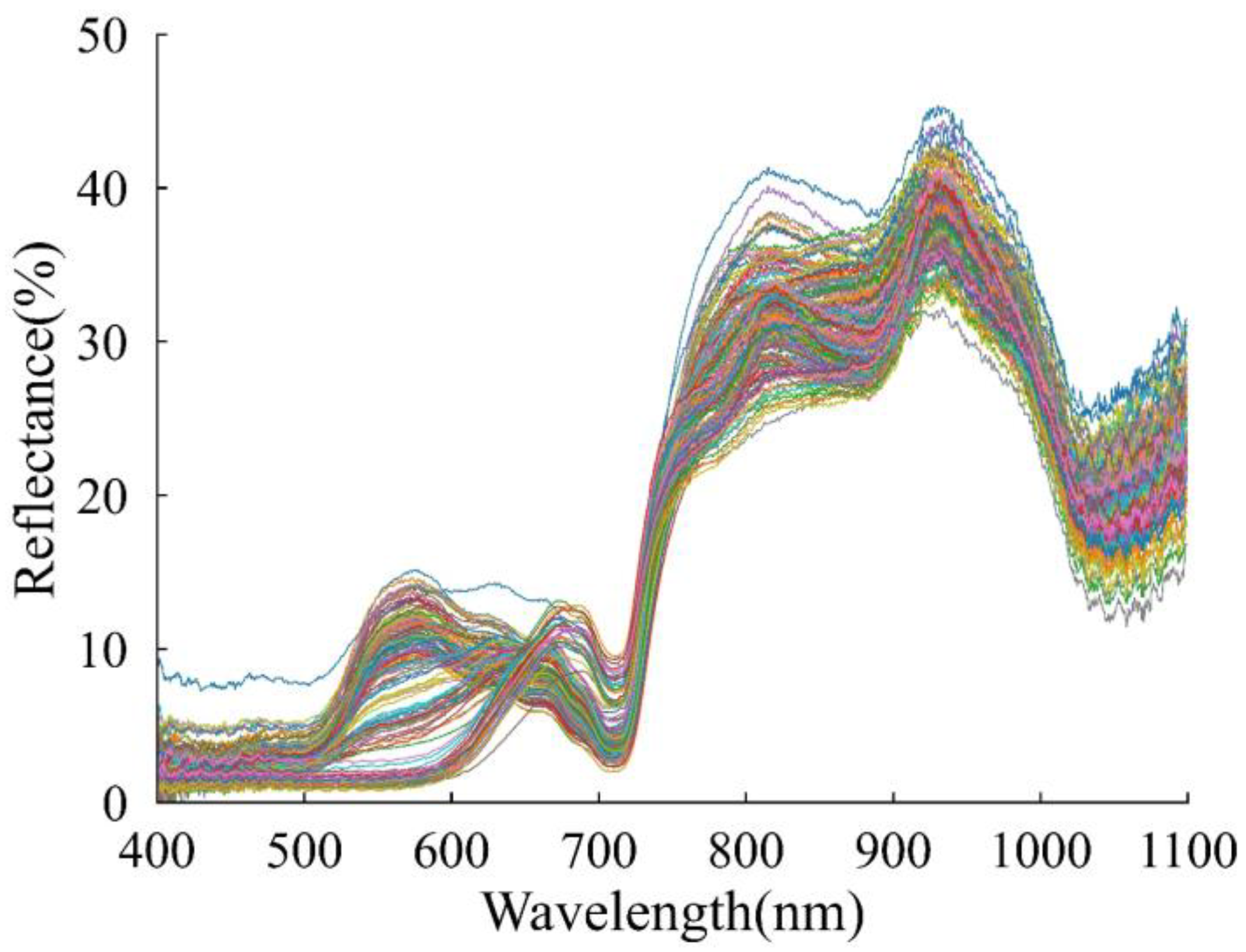
3.2.1. Spectral Profile
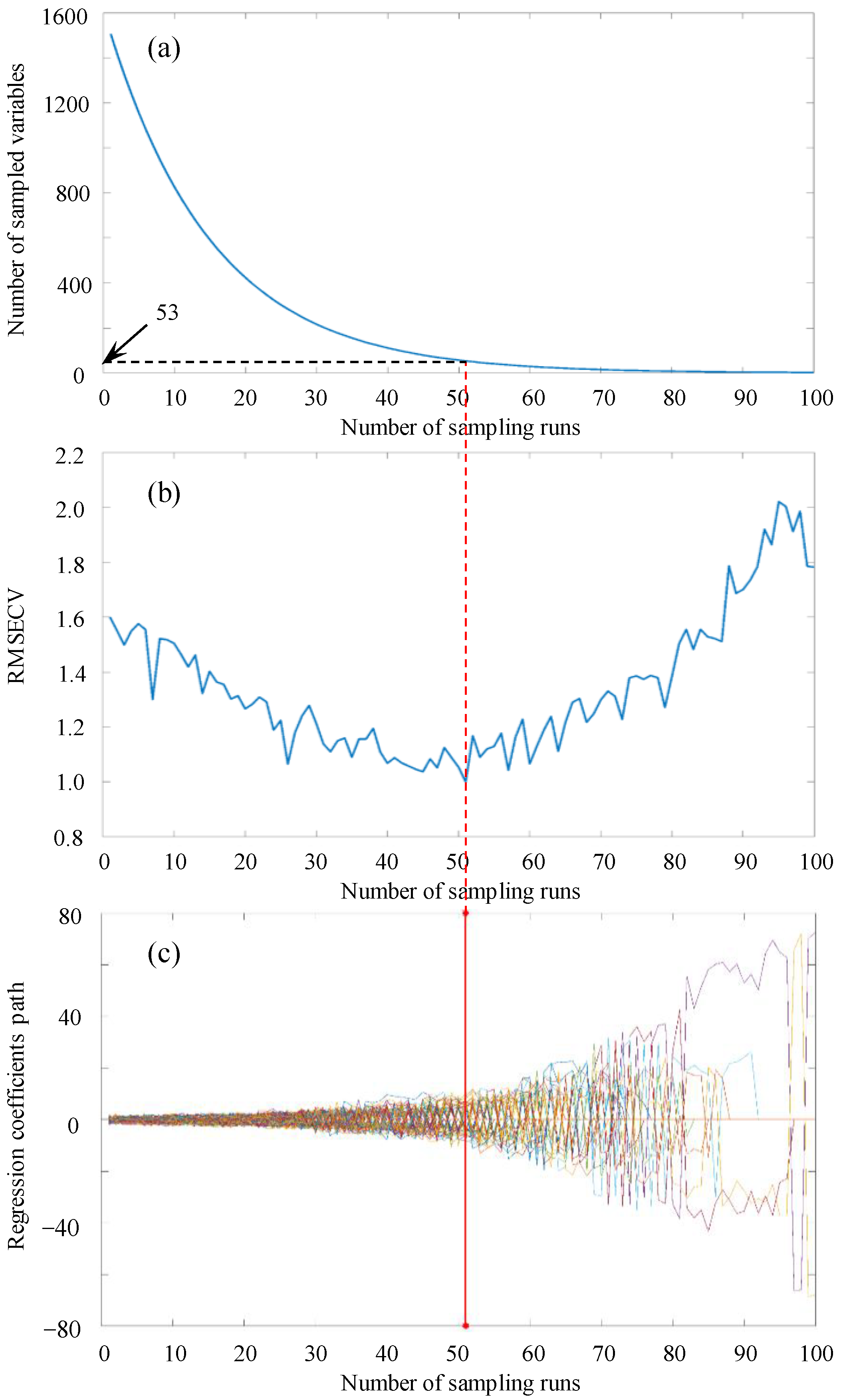
3.2.2. Effective Wavelength Selection
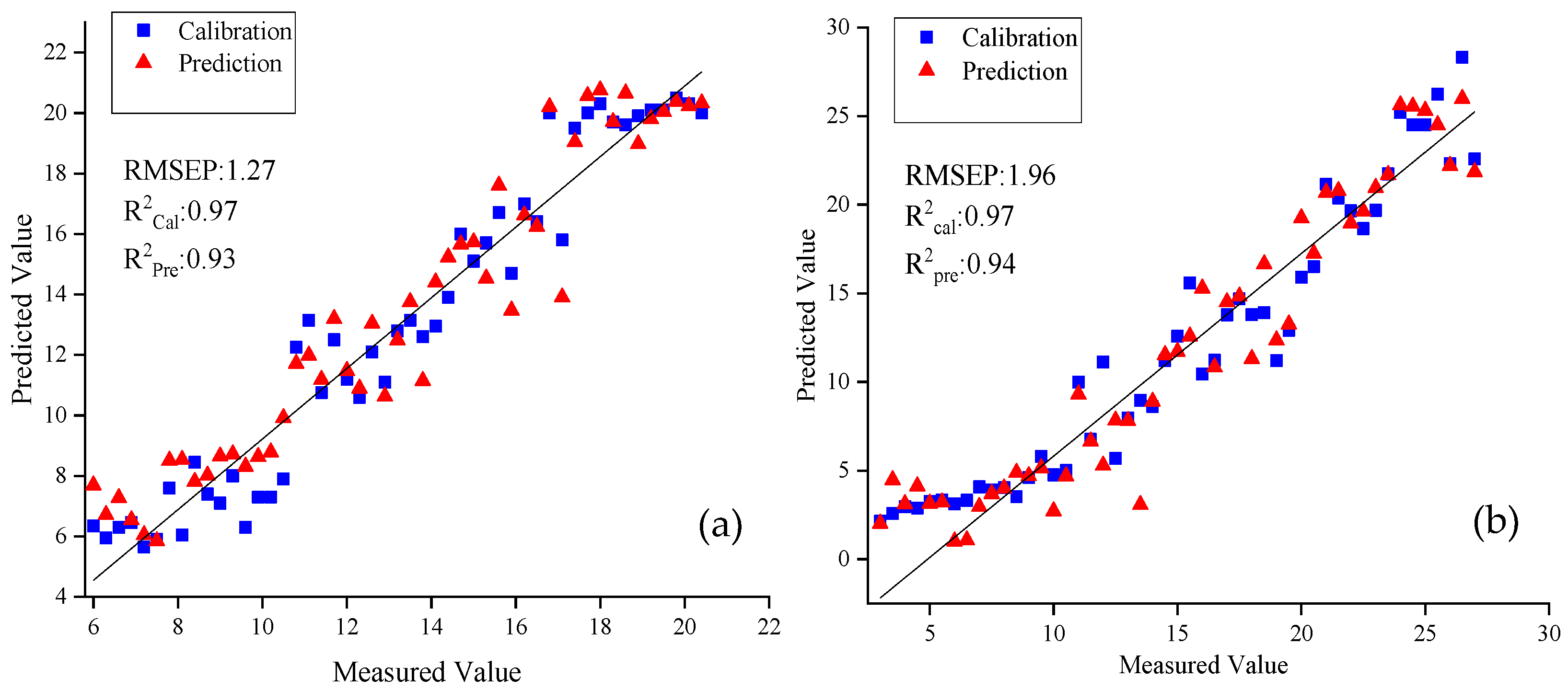
3.2.3. Prediction Model Establishment
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, D.D.; Li, J.H.; Xiong, R.G.; Saimaiti, A.; Huang, S.Y.; Wu, S.X.; Yang, Z.J.; Shang, A.; Zhao, C.N.; Gan, R.Y.; et al. Bioactive Compounds, Health Benefits and Food Applications of Grape. Foods 2022, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Guo, J.; Yin, M.; Liu, Y.; You, Y.; Zhan, J.; Huang, W. Grape Extract Activates Brown Adipose Tissue Through Pathway Involving the Regulation of Gut Microbiota and Bile Acid. Mol. Nutr. Food Res. 2020, 64, 202000149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.J.; Xha, Z.H.; Wu, J. Maturity discrimination of “Red Globe” grape cluster in grapery by improved circle Hough transform. Trans. Chin. Soc. Agric. Eng. 2020, 36, 205–213. [Google Scholar] [CrossRef]
- Yang, S.H.; Zheng, Y.J.; Liu, X.X.; Zhang, T.G.; Zhang, X.S.; Xu, L.M. Cabernet gernischt maturity determination based on near-ground multispectral figures by using UAVs. Spectrosc. Spectr. Anal. 2021, 41, 3220–3226. [Google Scholar]
- Kou, X.H.; Feng, Y.; Yuan, S.; Zhao, X.Y.; Wu, C.; Wang, C.; Xue, Z.H. Different regulatory mechanisms of plant hormones in the ripening of climacteric and non-climacteric fruits: A review. Plant Mol. Biol. 2021, 107, 477–497. [Google Scholar] [CrossRef]
- Liu, C.; Yang, S.X.; Deng, L. Determination of internal qualities of Newhall navel oranges based on NIR spectroscopy using machine learning. J. Food Eng. 2015, 161, 16–23. [Google Scholar] [CrossRef]
- Campos, I.; Bataller, R.; Armero, R.; Gandia, J.M.; Soto, J.; Martinez-Manez, R.; Gil-Sanchez, L. Monitoring grape ripeness using a voltammetric electronic tongue. Food Res. Int. 2013, 54, 1369–1375. [Google Scholar] [CrossRef]
- Ma, T.; Xia, Y.; Inagaki, T.; Tsuchikawa, S. Rapid and nondestructive evaluation of soluble solids content (SSC) and firmness in apple using Vis-NIR spatially resolved spectroscopy. Postharvest Biol. Technol. 2021, 173, 12. [Google Scholar] [CrossRef]
- Zhu, M.; Huang, D.; Hu, X.J.; Tong, W.H.; Han, B.L.; Tian, J.P.; Luo, H.B. Application of hyperspectral technology in detection of agricultural products and food: A Review. Food Sci. Nutr. 2020, 8, 5206–5214. [Google Scholar] [CrossRef]
- Zhu, S.S.; Feng, L.; Zhang, C.; Bao, Y.D.; He, Y. Identifying Freshness of Spinach Leaves Stored at Different Temperatures Using Hyperspectral Imaging. Foods 2019, 8, 12. [Google Scholar] [CrossRef]
- Dashti, A.; Mueller-Maatsch, J.; Weesepoel, Y.; Parastar, H.; Kobarfard, F.; Daraei, B.; AliAbadi, M.H.S.; Yazdanpanah, H. The Feasibility of Two Handheld Spectrometers for Meat Speciation Combined with Chemometric Methods and Its Application for Halal Certification. Foods 2022, 11, 17. [Google Scholar] [CrossRef]
- Cirilli, M.; Bellincontro, A.; Urbani, S.; Servili, M.; Esposto, S.; Mencarelli, F.; Muleo, R. On-field monitoring of fruit ripening evolution and quality parameters in olive mutants using a portable NIR-AOTF device. Food Chem. 2016, 199, 96–104. [Google Scholar] [CrossRef]
- Musingarabwi, D.M.; Nieuwoudt, H.H.; Young, P.R.; Eyeghe-Bickong, H.A.; Vivier, M.A. A rapid qualitative and quantitative evaluation of grape berries at various stages of development using Fourier-transform infrared spectroscopy and multivariate data analysis. Food Chem. 2016, 190, 253–262. [Google Scholar] [CrossRef]
- Pu, H.; Liu, D.; Wang, L.; Sun, D.-W. Soluble Solids Content and pH Prediction and Maturity Discrimination of Lychee Fruits Using Visible and Near Infrared Hyperspectral Imaging. Food Anal. Methods 2016, 9, 235–244. [Google Scholar] [CrossRef]
- Escribano, S.; Biasi, W.V.; Lerud, R.; Slaughter, D.C.; Mitcham, E.J. Non-destructive prediction of soluble solids and dry matter content using NIR spectroscopy and its relationship with sensory quality in sweet cherries. Postharvest Biol. Technol. 2017, 128, 112–120. [Google Scholar] [CrossRef]
- Xiao, H.; Li, A.; Li, M.Y.; Sun, Y.; Tu, K.; Wang, S.J.; Pan, L.Q. Quality assessment and discrimination of intact white and red grapes from Vitis vinifera L. at five ripening stages by visible and near-infrared spectroscopy. Sci. Hortic. 2018, 233, 99–107. [Google Scholar] [CrossRef]
- Pourdarbani, R.; Sabzi, S.; Kalantari, D.; Arribas, J.I. Non-destructive visible and short-wave near-infrared spectroscopic data estimation of various physicochemical properties of Fuji apple (Malus pumila) fruits at different maturation stages. Chemom. Intell. Lab. Syst. 2020, 206, 104147. [Google Scholar] [CrossRef]
- Pissard, A.; Marques, E.J.N.; Dardenne, P.; Lateur, M.; Pasquini, C.; Pimentel, M.F.; Pierna, J.A.F.; Baeten, V. Evaluation of a handheld ultra-compact NIR spectrometer for rapid and non-destructive determination of apple fruit quality. Postharvest Biol. Technol. 2021, 172, 10. [Google Scholar] [CrossRef]
- Vega-Castellote, M.; Sanchez, M.T.; Torres, I.; de la Haba, M.J.; Perez-Marin, D. Assessment of watermelon maturity using portable new generation NIR spectrophotometers. Sci. Hortic. 2022, 304, 7. [Google Scholar] [CrossRef]
- Fatchurrahman, D.; Nosrati, M.; Amodio, M.L.; Chaudhry, M.M.A.; de Chiara, M.L.V.; Mastrandrea, L.; Colelli, G. Comparison Performance of Visible-NIR and Near-Infrared Hyperspectral Imaging for Prediction of Nutritional Quality of Goji Berry (Lycium barbarum L.). Foods 2021, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, K.M.; Chen, X.; Hurley, J.E.; Stone, J.S.; Li, M.J. High data rate few-mode transmission over graded-index single-mode fiber using 850 nm single-mode VCSEL. Opt. Express 2019, 27, 21395–21404. [Google Scholar] [CrossRef] [PubMed]
- Tugnolo, A.; Giovenzana, V.; Beghi, R.; Grassi, S.; Alamprese, C.; Casson, A.; Casiraghi, E.; Guidetti, R. A diagnostic visible/near infrared tool for a fully automated olive ripeness evaluation in a view of a simplified optical system. Comput. Electron. Agric. 2021, 180, 7. [Google Scholar] [CrossRef]
- Deng, L.Z.; Pan, Z.L.; Zhang, Q.; Liu, Z.L.; Zhang, Y.; Meng, J.S.; Gao, Z.J.; Xiao, H.W. Effects of ripening stage on physicochemical properties, drying kinetics, pectin polysaccharides contents and nanostructure of apricots. Carbohydr. Polym. 2019, 222, 8. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wu, Q.; Ng, L.Y.; Wang, S. Effects of Vacuum Impregnation with Calcium Lactate and Pectin Methylesterase on Quality Attributes and Chelate-Soluble Pectin Morphology of Fresh-Cut Papayas. Food Bioprocess Technol. 2017, 10, 901–913. [Google Scholar] [CrossRef]
- Wang, J.; Tian, T.; Wang, H.; Cui, J.; Zhu, Y.; Zhang, W.; Tong, X.; Zhou, T.; Yang, Z.; Sun, J. Estimating cotton leaf nitrogen by combining the bands sensitive to nitrogen concentration and oxidase activities using hyperspectral imaging. Comput. Electron. Agric. 2021, 189, 106390. [Google Scholar] [CrossRef]
- Su, W.H.; Bakalis, S.; Sun, D.W. Chemometrics in tandem with near infrared (NIR) hyperspectral imaging and Fourier transform mid infrared (FT-MIR) microspectroscopy for variety identification and cooking loss determination of sweet potato. Biosyst. Eng. 2019, 180, 70–86. [Google Scholar] [CrossRef]
- Zhao, N.F.; Xu, Q.S.; Tang, M.L.; Wang, H. Variable Screening for Near Infrared (NIR) Spectroscopy Data Based on Ridge Partial Least Squares Regression. Comb. Chem. High Throughput Screen 2020, 23, 740–756. [Google Scholar] [CrossRef]
- Mishra, P.; Nikzad-Langerodi, R. Partial least square regression versus domain invariant partial least square regression with application to near-infrared spectroscopy of fresh fruit. Infrared Phys. Technol. 2020, 111, 7. [Google Scholar] [CrossRef]
- Lu, B.; Liu, N.H.; Li, H.L.; Yang, K.F.; Hu, C.; Wang, X.F.; Li, Z.X.; Shen, Z.X.; Tang, X.Y. Quantitative determination and characteristic wavelength selection of available nitrogen in coco-peat by NIR spectroscopy. Soil Tillage Res. 2019, 191, 266–274. [Google Scholar] [CrossRef]
- Kaur, S.; Randhawa, S.; Malhi, A. An efficient ANFIS based pre-harvest ripeness estimation technique for fruits. Multimed. Tools Appl. 2021, 80, 19459–19489. [Google Scholar] [CrossRef]
- Wang, A.Q.; Chen, D.Y.; Ma, Q.Y.; Rose, J.K.C.; Fei, Z.J.; Liu, Y.S.; Giovannoni, J.J. The tomato HIGH PIGMENT1/DAMAGED DNA BINDING PROTEIN 1 gene contributes to regulation of fruit ripening. Hortic. Res. 2019, 6, 10. [Google Scholar] [CrossRef]
- Hu, B.; Lai, B.; Wang, D.; Li, J.Q.; Chen, L.H.; Qin, Y.Q.; Wang, H.C.; Qin, Y.H.; Hu, G.B.; Zhao, J.T. Three LcABFs are Involved in the Regulation of Chlorophyll Degradation and Anthocyanin Biosynthesis During Fruit Ripening in Litchi chinensis. Plant Cell Physiol. 2019, 60, 448–461. [Google Scholar] [CrossRef]
- Temnani, A.; Berrios, P.; Conesa, M.R.; Perez-Pastor, A. Modelling the Impact of Water Stress during Post-Veraison on Berry Quality of Table Grapes. Agronomy 2022, 12, 15. [Google Scholar] [CrossRef]
- Padda, M.S.; do Amarante, C.V.T.; Garcia, R.M.; Slaughter, D.C.; Mitcham, E.J. Methods to analyze physico-chemical changes during mango ripening: A multivariate approach. Postharvest Biol. Technol. 2011, 62, 267–274. [Google Scholar] [CrossRef]
- Pathare, P.B.; Opara, U.L.; Al-Said, F.A. Colour Measurement and Analysis in Fresh and Processed Foods: A Review. Food Bioprocess Technol. 2013, 6, 36–60. [Google Scholar] [CrossRef]
- Nambi, V.E.; Thangavel, K.; Rajeswari, K.A.; Manickavasagan, A.; Geetha, V. Texture and rheological changes of Indian mango cultivars during ripening. Postharvest Biol. Technol. 2016, 117, 152–160. [Google Scholar] [CrossRef]
- Liu, S.; Huang, H.; Huber, D.J.; Pan, Y.; Shi, X.; Zhang, Z. Delay of ripening and softening in ‘Guifei’ mango fruit by postharvest application of melatonin. Postharvest Biol. Technol. 2020, 163, 111136. [Google Scholar] [CrossRef]
- Pan, H.X.; Wang, L.M.; Wang, R.; Xie, F.; Cao, J.K. Modifications of cell wall pectin in chilling-injured ‘Friar’ plum fruit subjected to intermediate storage temperatures. Food Chem. 2018, 242, 538–547. [Google Scholar] [CrossRef]
- Lohani, S.; Trivedi, P.K.; Nath, P. Changes in activities of cell wall hydrolases during ethylene-induced ripening in banana: Effect of 1-MCP, ABA and IAA. Postharvest Biol. Technol. 2004, 31, 119–126. [Google Scholar] [CrossRef]
- Kiumarsi, M.; Shahbazi, M.; Yeganehzad, S.; Majchrzak, D.; Lieleg, O.; Winkeljann, B. Relation between structural, mechanical and sensory properties of gluten-free bread as affected by modified dietary fibers. Food Chem. 2019, 277, 664–673. [Google Scholar] [CrossRef]
- Amoriello, T.; Ciccoritti, R.; Paliotta, M.; Carbone, K. Classification and prediction of early-to-late ripening apricot quality using spectroscopic techniques combined with chemometric tools. Sci. Hortic. 2018, 240, 310–317. [Google Scholar] [CrossRef]
- Lima, F.V.; de Moura, E.A.; Mendonca, V.; de Oliveira, L.M.; Melo, M.F.; Celedonio, W.F.; Mendonca, L.F.D.; Alves, A.A.; Irineu, T.H.S.; Figueiredo, F.R.A.; et al. Nitrogen and organic fertilizer on the postharvest quality of Isabel Precoce grapes. J. Plant Nutr. 2021, 44, 1693–1704. [Google Scholar] [CrossRef]
- Li, J.B.; Zhang, H.L.; Zhan, B.S.; Wang, Z.L.; Jiang, Y.L. Determination of SSC in pears by establishing the multi-cultivar models based on visible-NIR spectroscopy. Infrared Phys. Technol. 2019, 102, 10. [Google Scholar] [CrossRef]
- Shutova, V.V.; Tyutyaev, E.V.; Churin, A.A.; Ponomarev, V.Y.; Belyakova, G.A.; Maksimov, G.V. IR and Raman spectroscopy in the study of carotenoids of Cladophora rivularis algae. Biophysics 2016, 61, 601–605. [Google Scholar] [CrossRef]
- Flynn, K.C.; Frazier, A.E.; Admas, S. Performance of chlorophyll prediction indices forEragrostis tefat Sentinel-2 MSI and Landsat-8 OLI spectral resolutions. Precis. Agric. 2020, 21, 1057–1071. [Google Scholar] [CrossRef]
- Hernandez-Sanchez, N.; Gomez-del-Campo, M. From NIR spectra to singular wavelengths for the estimation of the oil and water contents in olive fruits. Grasas Aceites 2018, 69, 13. [Google Scholar] [CrossRef]
- Badaro, A.T.; Morimitsu, F.L.; Ferreira, A.R.; Clerici, M.; Barbin, D.F. Identification of fiber added to semolina by near infrared (NIR) spectral techniques. Food Chem. 2019, 289, 195–203. [Google Scholar] [CrossRef]
- Li, H.D.; Liang, Y.Z.; Xu, Q.S.; Cao, D.S. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef]
- Puertas, G.; Vazquez, M. Fraud detection in hen housing system declared on the eggs’ label: An accuracy method based on UV-VIS-NIR spectroscopy and chemometrics. Food Chem. 2019, 288, 8–14. [Google Scholar] [CrossRef]

| Ripening Stage | L* | a* | b* | C* | h* |
|---|---|---|---|---|---|
| I | 35.80 ± 3.87 a | 0.77 ± 2.08 c | 7.31 ± 1.51 a | 5.77 ± 1.59 c | 83.24 ± 14.36 a |
| II | 32.31 ± 2.85 b | 1.90 ± 2.55 c | 4.80 ± 1.66 b | 6.26 ± 0.98 c | 66.30 ± 25.84 b |
| III | 27.69 ± 2.88 c | 5.82 ± 2.54 b | 3.26 ± 1.68 c | 7.24 ± 1.32 b | 32.84 ± 23.34 c |
| IV | 25.15 ± 1.46 d | 8.02 ± 1.36 a | 2.07 ± 0.81 d | 8.32 ± 1.46 a | 14.24 ± 4.14 d |
| Ripening Stage | Hardness | Chewiness | Flexibility |
|---|---|---|---|
| I | 1435.87 ± 285.19 a | 238.64 ± 100.00 a | 0.60 ± 0.05 b |
| II | 1075.31 ± 110.70 b | 196.79 ± 38.98 b | 0.62 ± 0.03 a |
| III | 982.84 ± 134.65 bc | 192.09 ± 55.96 b | 0.60 ± 0.06 ab |
| IV | 967.49 ± 99.82 c | 165.17 ± 22.56 b | 0.57 ± 0.02 c |
| Method | Effective Wavelengths (nm) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CARS | 38 | 61 | 62 | 73 | 306 | 307 | 403 | 404 | 405 |
| 425 | 426 | 427 | 702 | 755 | 756 | 757 | 758 | 759 | |
| 775 | 787 | 788 | 834 | 835 | 836 | 908 | 909 | 910 | |
| 958 | 1087 | 1099 | 1100 | 1108 | 1123 | 1127 | 1131 | 1150 | |
| 1189 | 1262 | 1275 | 1297 | 1298 | 1312 | 1326 | 1344 | 1358 | |
| 1390 | 1391 | 1403 | 1417 | 1431 | 1453 | 1468 | 1499 | ||
| Parameter | Pre-Processing Methods | Calibration | Prediction | RPD | ||
|---|---|---|---|---|---|---|
| RMSEC | RMSEP | |||||
| SSC | SNV | 0.91 | 1.13 | 0.96 | 0.88 | 5.44 |
| MSC | 0.91 | 1.13 | 0.96 | 0.88 | 5.33 | |
| 1st derivative | 0.97 | 0.62 | 0.93 | 1.27 | 4.09 | |
| 2nd derivative | 0.94 | 0.80 | 0.82 | 1.94 | 2.70 | |
| S-G Smoothing | 0.89 | 1.31 | 0.94 | 1.06 | 4.43 | |
| S-G Smoothing + 1st derivative | 0.95 | 0.78 | 0.92 | 1.33 | 3.95 | |
| TA | SNV | 0.95 | 1.25 | 0.96 | 1.57 | 5.43 |
| MSC | 0.95 | 1.44 | 0.96 | 1.45 | 5.22 | |
| 1st derivative | 0.97 | 0.88 | 0.94 | 1.96 | 4.55 | |
| 2nd derivative | 0.95 | 1.16 | 0.93 | 2.18 | 4.10 | |
| S-G Smoothing | 0.92 | 1.80 | 0.96 | 1.24 | 5.43 | |
| S-G Smoothing + 1st derivative | 0.95 | 1.25 | 0.96 | 1.56 | 5.43 | |
| Parameter | Pre-Processing Methods | Calibration | Prediction | RPD | ||
|---|---|---|---|---|---|---|
| RMSEC | RMSEP | |||||
| SSC | SNV | 0.93 | 1.18 | 0.88 | 1.26 | 4.13 |
| MSC | 0.92 | 0.84 | 0.90 | 1.10 | 5.66 | |
| 1st derivative | 0.88 | 1.26 | 0.89 | 1.45 | 3.52 | |
| 2nd derivative | 0.84 | 1.40 | 0.79 | 2.12 | 2.45 | |
| S-G Smoothing | 0.87 | 1.43 | 0.91 | 1.28 | 3.68 | |
| S-G Smoothing + 1st derivative | 0.95 | 0.81 | 0.92 | 1.01 | 5.47 | |
| TA | SNV | 0.91 | 1.93 | 0.88 | 2.32 | 3.11 |
| MSC | 0.94 | 1.59 | 0.94 | 1.78 | 4.16 | |
| 1st derivative | 0.81 | 2.46 | 0.78 | 3.40 | 2.38 | |
| 2nd derivative | 0.88 | 2.15 | 0.84 | 2.92 | 2.86 | |
| S-G Smoothing | 0.90 | 1.79 | 0.92 | 2.18 | 3.15 | |
| S-G Smoothing + 1st derivative | 0.93 | 1.81 | 0.89 | 2.12 | 4.70 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ping, F.; Yang, J.; Zhou, X.; Su, Y.; Ju, Y.; Fang, Y.; Bai, X.; Liu, W. Quality Assessment and Ripeness Prediction of Table Grapes Using Visible–Near-Infrared Spectroscopy. Foods 2023, 12, 2364. https://doi.org/10.3390/foods12122364
Ping F, Yang J, Zhou X, Su Y, Ju Y, Fang Y, Bai X, Liu W. Quality Assessment and Ripeness Prediction of Table Grapes Using Visible–Near-Infrared Spectroscopy. Foods. 2023; 12(12):2364. https://doi.org/10.3390/foods12122364
Chicago/Turabian StylePing, Fengjiao, Jihong Yang, Xuejian Zhou, Yuan Su, Yanlun Ju, Yulin Fang, Xuebing Bai, and Wenzheng Liu. 2023. "Quality Assessment and Ripeness Prediction of Table Grapes Using Visible–Near-Infrared Spectroscopy" Foods 12, no. 12: 2364. https://doi.org/10.3390/foods12122364
APA StylePing, F., Yang, J., Zhou, X., Su, Y., Ju, Y., Fang, Y., Bai, X., & Liu, W. (2023). Quality Assessment and Ripeness Prediction of Table Grapes Using Visible–Near-Infrared Spectroscopy. Foods, 12(12), 2364. https://doi.org/10.3390/foods12122364






