Study on the In Silico Screening and Characterization, Inhibition Mechanisms, Zinc-Chelate Activity, and Stability of ACE-Inhibitory Peptides Identified in Naked Oat Bran Albumin Hydrolysates
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of Naked Oat Bran Albumin Hydrolysates (NOBAH)
2.3. Isolation of Peptides with High Inhibition Ability on ACE
2.4. Determination of Zn-Chelating Ability
2.5. Determination of ACE Inhibition Capacity and Inhibition Kinetics
2.6. Identification, In Silico Screening, and Physicochemical Characterization of Peptide Sequences
2.7. Chemical Synthesis
2.8. Toxicity and Allergenicity Evaluation
2.9. Molecular Docking
2.10. Interactions of Zinc Ions with NOBAH ACE-Inhibitory Peptides
2.10.1. Preparation of the Peptide−Zn Complex
2.10.2. Coordination Patterns of NOBAH Peptides with Zinc Ions
2.11. Stabilities of NOBAH Peptides
2.12. Data Analysis
3. Results and Discussion
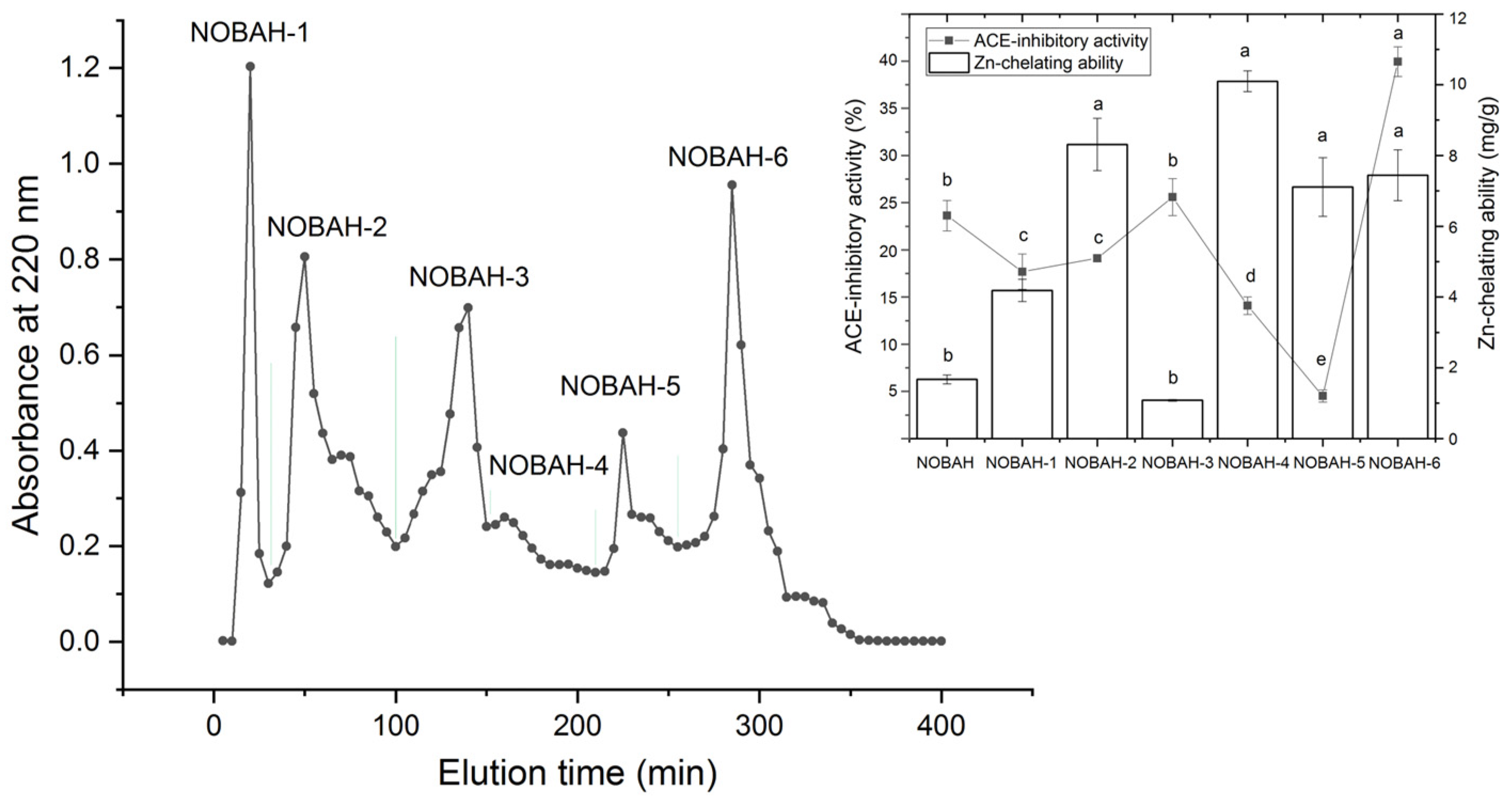
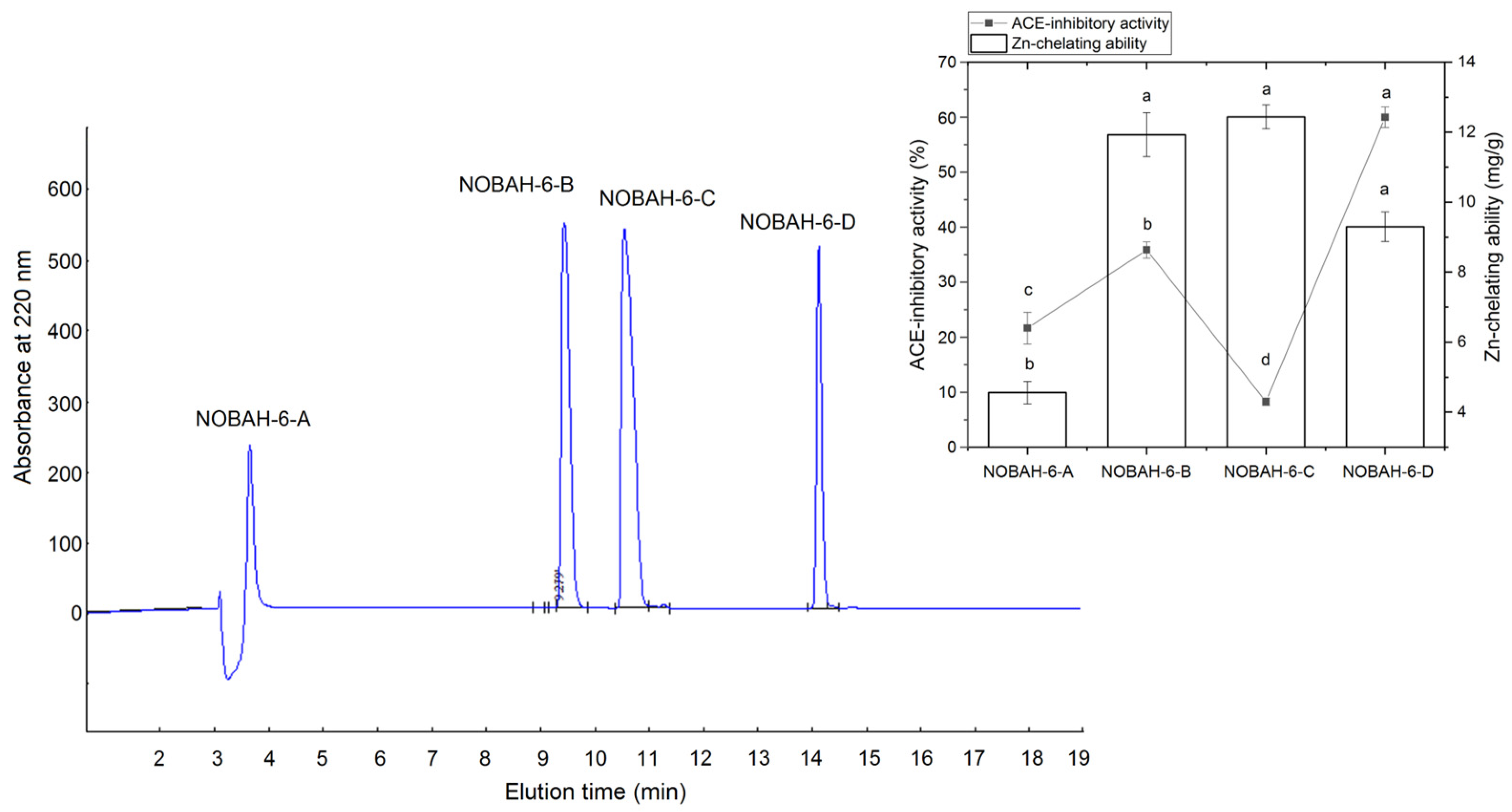
3.1. Selection of Fractions with High ACE-Inhibitory Activity from NOBAH
3.2. Identification and Characterization of Peptides from NOBAH-4-D
3.3. Physicochemical Characterization In Silico
3.4. Security Predictions In Silico
3.5. Inhibition Mechanisms on ACE
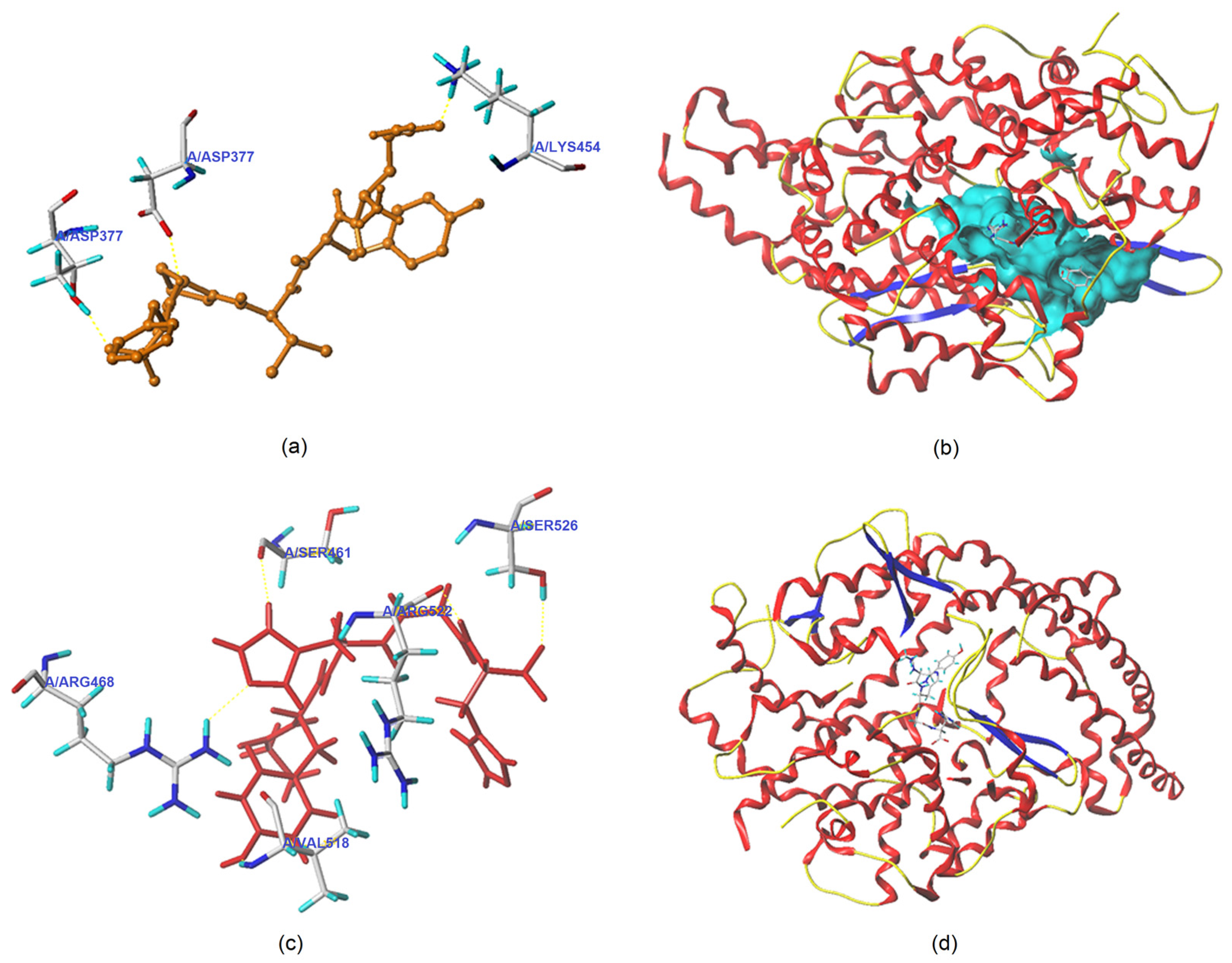
3.5.1. Molecular Docking
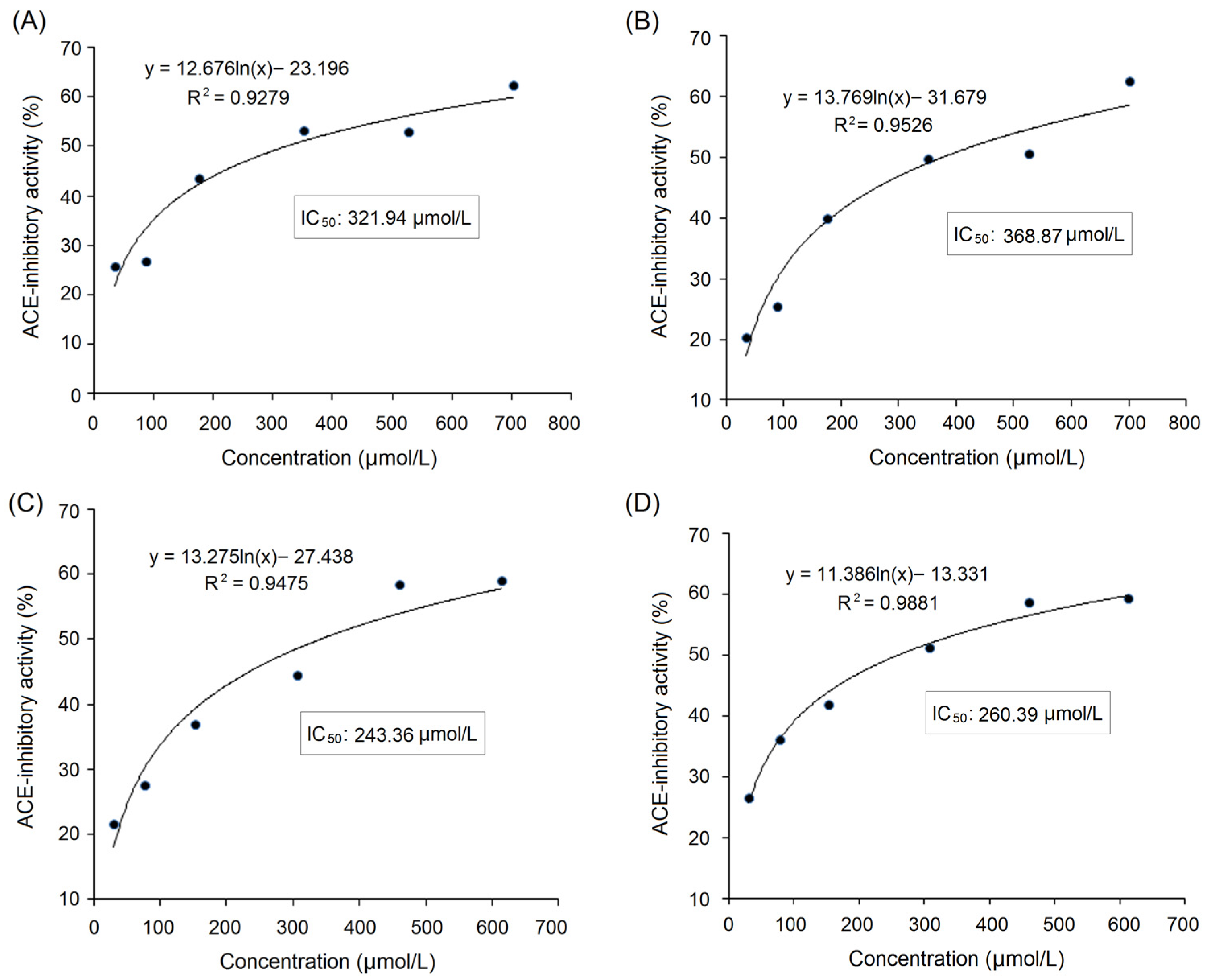
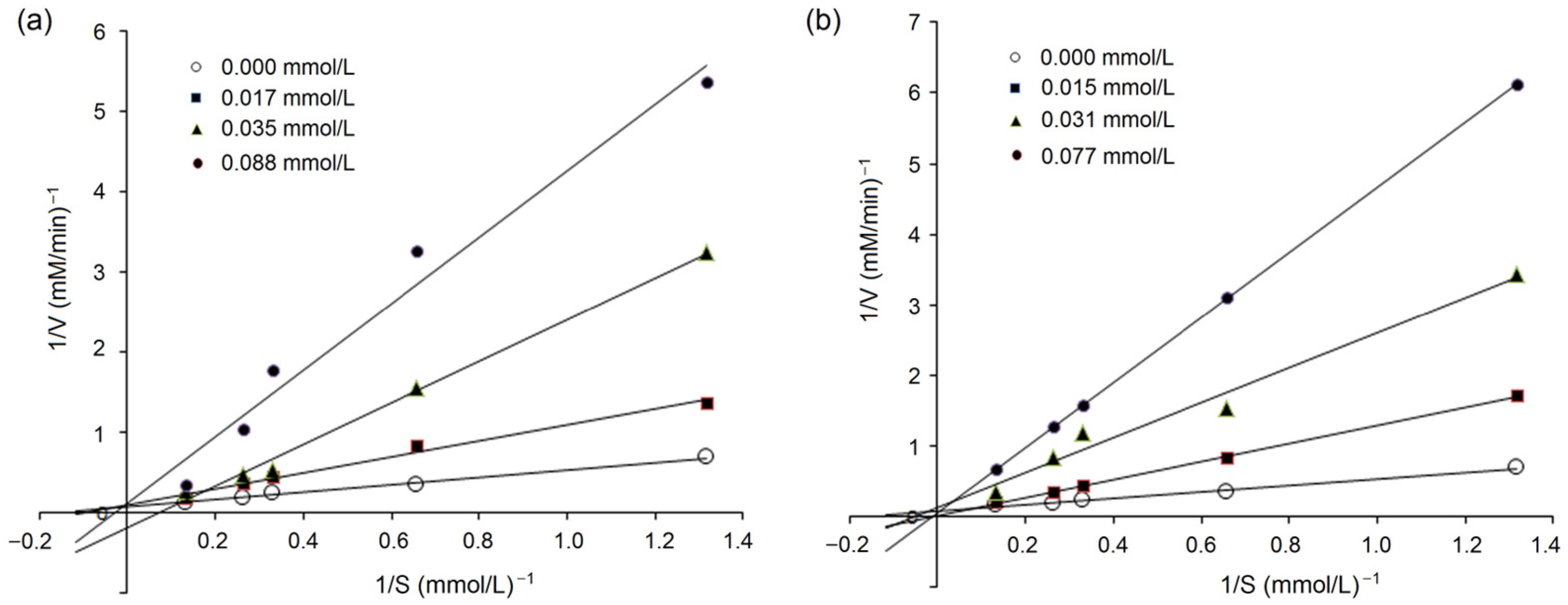
3.5.2. Inhibition Kinetics on ACE
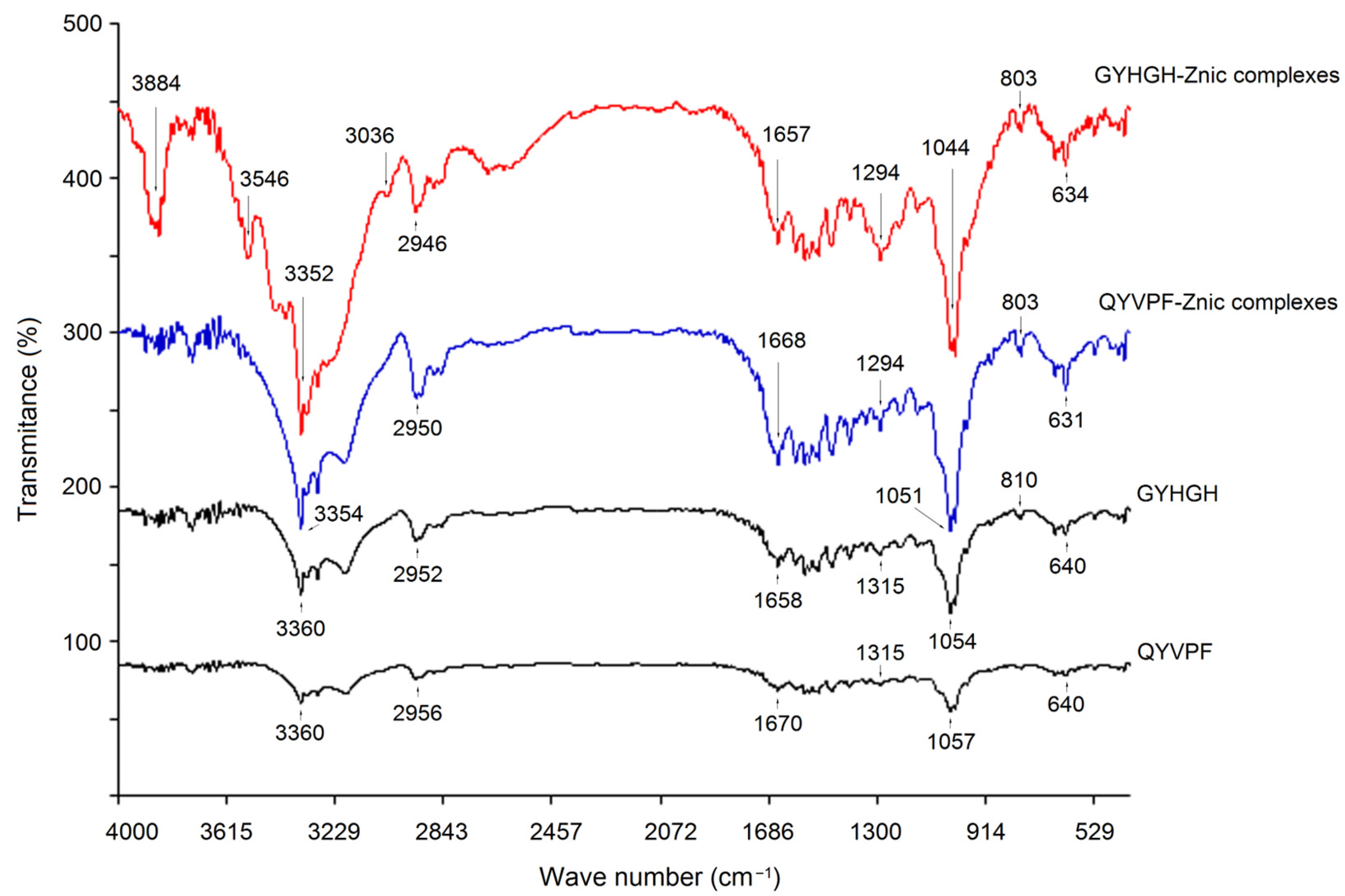
3.5.3. Coordination Patterns of QYVPF and GYHGH with Zinc Ions
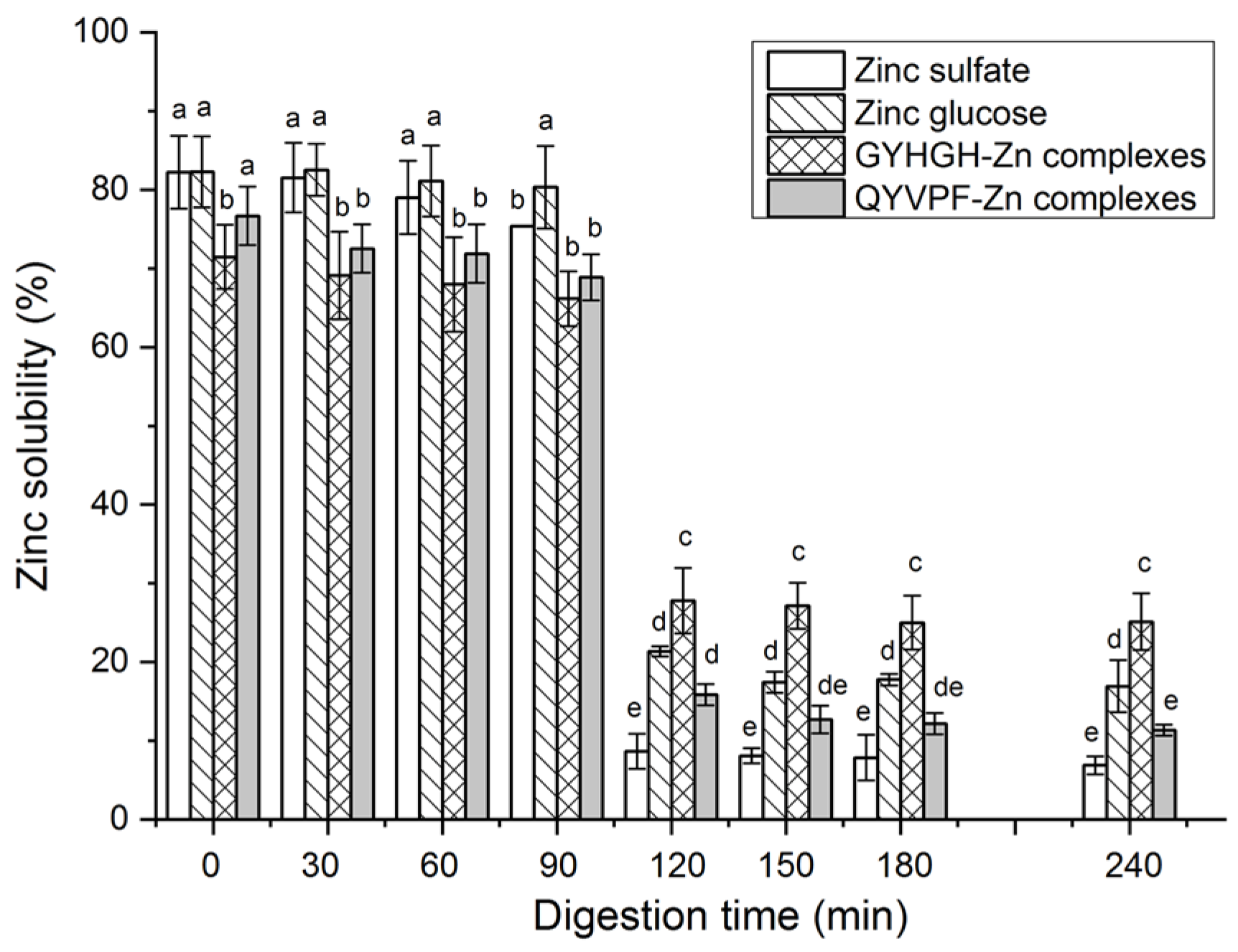
3.6. Stability against Gastrointestinal Digestion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: A pooled analysis of 1201 population-representative studies with 104 million participants. Lancet 2021, 398, 957–980. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wu, J. Purification and identification of novel ACE inhibitory and ACE2 upregulating peptides from spent hen muscle proteins. Food Chem. 2021, 345, 128867. [Google Scholar] [CrossRef]
- Wu, J.; Liao, W.; Udenigwe, C.C. Revisiting the mechanisms of ACE inhibitory peptides from food proteins. Trends Food Sci. Technol. 2017, 69, 214–219. [Google Scholar] [CrossRef]
- Renjuan, L.; Xiuli, Z.; Liping, S.; Yongliang, Z. Identification, in silico screening, and molecular docking of novel ACE inhibitory peptides isolated from the edible symbiot Boletus griseus-Hypomyces chrysospermus. LWT-Food Sci. Technol. 2022, 169, 114008. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef]
- Urbizo-Reyes, U.; Liceaga, A.M.; Reddivari, L.; Kim, K.-H.; Anderson, J.M. Enzyme kinetics, molecular docking, and in silico characterization of canary seed (Phalaris canariensis L.) peptides with ACE and pancreatic lipase inhibitory activity. J. Funct. Foods 2022, 88, 104892. [Google Scholar] [CrossRef]
- Sun, R.; Liu, X.; Yu, Y.; Miao, J.; Leng, K.; Gao, H. Preparation process optimization, structural characterization and in vitro digestion stability analysis of Antarctic krill (Euphausia superba) peptides-zinc chelate. Food Chem. 2021, 340, 128056. [Google Scholar] [CrossRef]
- Khan, S.T.; Malik, A.; Alwarthan, A.; Shaik, M.R. The enormity of the zinc deficiency problem and available solutions; an overview. Arab. J. Chem. 2022, 15, 103668. [Google Scholar] [CrossRef]
- Ke, X.; Hu, X.; Li, L.; Yang, X.; Chen, S.; Wu, Y.; Xue, C. A novel zinc-binding peptide identified from tilapia (Oreochromis niloticus) skin collagen and transport pathway across Caco-2 monolayers. Food Biosci. 2022, 42, 101127. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, F.; Liu, X.; Zhao, M. Particulate nanocomposite from oyster (Crassostrea rivularis) hydrolysates via zinc chelation improves zinc solubility and peptide activity. Food Chem. 2018, 258, 269–277. [Google Scholar] [CrossRef]
- Udechukwu, M.C.; Downey, B.; Udenigwe, C.C. Influence of structural and surface properties of whey-derived peptides on zinc-chelating capacity, and in vitro gastric stability and bioaccessibility of the zinc-peptide complexes. Food Chem. 2018, 240, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.P.; Vij, S. In vitro stability of bioactive peptides derived from fermented soy milk against heat treatment, pH and gastrointestinal enzymes. LWT-Food Sci. Technol. 2018, 91, 303–307. [Google Scholar] [CrossRef]
- Wong, F.-C.; Xiao, J.; Ong, M.G.-L.; Pang, M.-J.; Wong, S.-J.; Teh, L.K.; Chai, T.-T. Identification and characterization of antioxidant peptides from hydrolysate of blue-spotted stingray and their stability against thermal, pH and simulated gastrointestinal digestion treatments. Food Chem. 2019, 271, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Luo, Q.; Hong, H.; Liu, H.; Luo, Y. Novel antioxidant and ACE inhibitory peptide identified from Arthrospira platensis protein and stability against thermal/pH treatments and simulated gastrointestinal digestion. Food Res. Int. 2021, 139, 109908. [Google Scholar] [CrossRef] [PubMed]
- Sahni, P.; Sharma, S.; Surasani, V.K.R. Influence of processing and pH on amino acid profile, morphology, electrophoretic pattern, bioactive potential and functional characteristics of alfalfa protein isolates. Food Chem. 2020, 333, 127503. [Google Scholar] [CrossRef] [PubMed]
- Sterna, V.; Zute, S.; Brunava, L. Oat Grain Composition and its Nutrition Benefice. Agric. Agric. Sci. Procedia 2016, 8, 252–256. [Google Scholar] [CrossRef]
- Hu, X.-Z.; Zheng, J.-M.; Li, X.-P.; Xu, C.; Zhao, Q. Chemical composition and sensory characteristics of oat flakes: A comparative study of naked oat flakes from China and hulled oat flakes from western countries. J. Cereal Sci. 2014, 60, 297–301. [Google Scholar] [CrossRef]
- Walters, M.E.; Udenigwe, C.C.; Tsopmo, A. Structural Characterization and Functional Properties of Proteins from Oat Milling Fractions. J. Am. Oil Chem. Soc. 2018, 95, 991–1000. [Google Scholar] [CrossRef]
- Kumar, L.; Sehrawat, R.; Kong, Y. Oat proteins: A perspective on functional properties. LWT-Food Sci. Technol. 2021, 152, 112307. [Google Scholar] [CrossRef]
- Esfandi, R.; Willmore, W.G.; Tsopmo, A. Peptidomic analysis of hydrolyzed oat bran proteins, and their in vitro antioxidant and metal chelating properties. Food Chem. 2019, 279, 49–57. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Pliszka, M.; Vegarud, G.E.; Iwaniak, A.; Minkiewicz, P. Angiotensin I-converting enzyme inhibitory peptides in oat (Avena sativa L.) proteins-derived digests—In silico and in vitro study. New Biotechnol. 2016, 33, S173. [Google Scholar] [CrossRef]
- Fuentes, L.R.; Richard, C.; Chen, L. Sequential alcalase and flavourzyme treatment for preparation of α-amylase, α-glucosidase, and dipeptidyl peptidase (DPP)-IV inhibitory peptides from oat protein. J. Funct. Foods 2021, 87, 104829. [Google Scholar] [CrossRef]
- Wei, M.; Tang, M.; Wang, L.; Cheng, X.; Wu, Y.; Ouyang, J. Endogenous bioactive compounds of naked oats (Avena nuda L.) inhibit α-amylase and α-glucosidase activity. LWT-Food Sci. Technol. 2021, 149, 111902. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Zhuang, Y.; Li, Y.; Shi, P.Q.; Tian, H.; Li, X.T.; Chen, X. Isolation of novel ACE-inhibitory peptide from naked oat globulin hydrolysates in silico approach: Molecular docking, in vivo antihypertension and effects on renin and intracellular endothelin-1. J. Food Sci. 2020, 85, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Jodayree, S.; Smith, J.C.; Tsopmo, A. Use of carbohydrase to enhance protein extraction efficiency and antioxidative properties of oat bran protein hydrolysates. Food Res. Int. 2012, 46, 69–75. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonic acid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, P.; Zheng, Y.; Guo, M.; Zhuang, Y.; Huo, X. Millet bran protein hydrolysates derived peptides-zinc chelate: Structural characterization, security prediction in silico, zinc transport capacity and stability against different food processing conditions. J. Food Sci. 2023, 88, 477–490. [Google Scholar] [CrossRef]
- Zaharuddin, N.D.; Barkia, I.; Ibadullah, W.Z.W.; Zarei, M.; Saari, N. Identification, molecular docking, and kinetic studies of six novel angiotensin-I-converting enzyme (ACE) inhibitory peptides derived from Kenaf (Hibiscus cannabinus L.) seed. Int. J. Biol. Macromol. 2022, 220, 1512–1522. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Sharma, M.; Nagpal, G.; Chauhan, J.S.; Singh, S.; Gautam, A.; Raghava, G.P. AHTPDB: A comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res. 2015, 43, D956–D962. [Google Scholar] [CrossRef]
- Zarei, M.; Ghanbari, R.; Zainal, N.; Ovissipour, R.; Saari, N. Inhibition kinetics, molecular docking, and stability studies of the effect of papain-generated peptides from palm kernel cake proteins on angiotensin-converting enzyme (ACE). Food Chem. Mol. Sci. 2022, 5, 100147. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S.; Open Source Drug Discovery Consortium. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, B.; Ananthanarayana, L.; Jamdar, S. Purification, identification, and characterization of novel angiotensin Iconverting enzyme (ACE) inhibitory peptides from alcalase digested horse gram flour. LWT-Food Sci. Technol. 2019, 103, 155–161. [Google Scholar] [CrossRef]
- Fadimu, G.J.; Gan, C.Y.; Olalere, O.A.; Farahnaky, A.; Gill, H.; Truong, T. Novel antihypertensive peptides from lupin protein hydrolysate: An in-silico identification and molecular docking studies. Food Chem. 2023, 407, 135082. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, S.; Zhu, X.; Li, Q.; Fan, Y.; Cheng, D.; Li, B. A novel calcium-binding peptide from Antarctic krill protein hydrolysates and identification of binding sites of calcium-peptide complex. Food Chem. 2018, 243, 389–395. [Google Scholar] [CrossRef]
- Wei, G.; Zhao, Q.; Wang, D.; Fan, Y.; Shi, Y.; Huang, A. Novel ACE inhibitory, antioxidant and α-glucosidase inhibitory peptides identified from fermented rubing cheese through peptidomic and molecular docking. LWT 2022, 159, 113196. [Google Scholar] [CrossRef]
- Chen, J.; Yu, X.; Chen, Q.; Wu, Q.; He, Q. Screening and mechanisms of novel angiotensin-I-converting enzyme inhibitory peptides from rabbit meat proteins: A combined in silico and in vitro study. Food Chem. 2022, 370, 131070. [Google Scholar] [CrossRef]
- Wang, T.; Lin, S.; Cui, P.; Bao, Z.; Liu, K.; Jiang, P.; Zhu, B.; Sun, N. Antarctic krill derived peptide as a nanocarrier of iron through the gastrointestinal tract. Food Biosci. 2020, 36, 100657. [Google Scholar] [CrossRef]
- Pina, A.; Roque, A. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J. Mol. Recognit. 2009, 22, 162–168. [Google Scholar] [CrossRef]
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Montone, C.M.; Chiozzi, R.Z.; Laganà, A. Recent trends and analytical challenges in plant bioactive peptide separation, identification and validation. Anal. Bioanal. Chem. 2018, 410, 3425–3444. [Google Scholar] [CrossRef]
- Lin, Z.; Lai, J.; He, P.; Pan, L.; Zhang, Y.; Zhang, M.; Wu, H. Screening, ACE-inhibitory mechanism and structure-activity relationship of a novel ACE-inhibitory peptide from Lepidium meyenii (Maca) protein hydrolysate. Food Biosci. 2023, 52, 102374. [Google Scholar] [CrossRef]

| Peptide Sequence | GTTGGMGT | GYHGH | GLRAAAAAAEGG | QYVPF | GAAAALV | PSSPPS |
|---|---|---|---|---|---|---|
| Mass (Da) | 579.73 | 569.65 | 1014.25 | 652.81 | 571.75 | 570.66 |
| Matched sequence in Avena nuda a | H.GTTGGMG.T | G.GYHGH.G | G.GLRAAAAAAEGG.M | Q.QYVPF.A | L.GAAAALV.F | -.PSSPPS.V |
| SVMS b | −0.24 | 0.90 | −0.50 | 1.21 | −0.94 | −0.31 |
| Antihypertension prediction | Non-AHT | AHT | Non-AHT | AHT | Non-AHT | Non-AHT |
| ACE-inhibitory activity (IC50: μmol/L) | ND | 321.94 | ND | 243.36 | ND | ND |
| Zinc chelating capacity (mg/g) | 3.36 ± 0.16 e | 14.85 ± 0.39 d | 4.95 ± 0.07 e | 0.32 ± 0.09 g | 1.27 ± 0.22 ef | 0.42 ± 0.06 g |
| Basic or acidic amino acid content (%) | 0.00% | 40.00% | 16.67% | 0.00% | 0.00% | 0.00% |
| Hydrophobic amino acid content (%) | 12.50% | 0.00% | 50.00% | 60.00% | 85.71% | 50.00% |
| Physicochemical properties | ||||||
| Hydrophobicity | 0.08 | −0.09 | 0.01 | 0.08 | 0.32 | −0.17 |
| Amphiphilicity | 0.00 | 1.59 | 0.31 | 1.26 | 0.00 | 0.00 |
| Hydrophilicity | −0.30 | −0.66 | 0.10 | −1.22 | −0.76 | 0.15 |
| Isoelectric point | 5.88 | 7.25 | 6.36 | 5.88 | 5.88 | 5.88 |
| Security c | ||||||
| Toxicity | Non-Toxin | Non-Toxin | Non-Toxin | Non-Toxin | Non-Toxin | Non-Toxin |
| Allergenicity | No | No | No | No | No | No |
| Ligand | T-Score | C-Score | Interaction Mode | ACE Residues and the Length of Hydrogen Bonds Formed between ACE and Ligand |
|---|---|---|---|---|
| QYVPF | 8.95 | 4 | Hydrogen bond | Thr372: 2.60Å; Asp377: 2.85Å; Lys454: 2.93Å |
| Hydrophobic interaction | Glu162, Pro163, Gln369, Cys370, Trp279, Cys352, His353, Val380, Ala354, Lys511, Tyr520, Asp415, Val379, Phe527, Tyr523, Phe457, Gln281, Thr166, Glu376, Asn374, Ala170, Asn167 | |||
| GYHGH | 8.04 | 3 | Hydrogen bond | Arg468: 2.02Å; Ser526: 2.14Å; Arg522: 1.98Å; Val518: 2.07Å; Ser461: 2.08Å |
| Hydrophobic interaction | Gln369, Asp377, Ser355, His383, Tyr520, Phe457, Val380, Val379, Trp279, Asn277, Thr166 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Li, J.; Cheng, C.; Zheng, Y.; Li, H.; Zhu, Z.; Yan, Y.; Hao, W.; Qin, N. Study on the In Silico Screening and Characterization, Inhibition Mechanisms, Zinc-Chelate Activity, and Stability of ACE-Inhibitory Peptides Identified in Naked Oat Bran Albumin Hydrolysates. Foods 2023, 12, 2268. https://doi.org/10.3390/foods12112268
Li Y, Li J, Cheng C, Zheng Y, Li H, Zhu Z, Yan Y, Hao W, Qin N. Study on the In Silico Screening and Characterization, Inhibition Mechanisms, Zinc-Chelate Activity, and Stability of ACE-Inhibitory Peptides Identified in Naked Oat Bran Albumin Hydrolysates. Foods. 2023; 12(11):2268. https://doi.org/10.3390/foods12112268
Chicago/Turabian StyleLi, Yan, Junru Li, Chaoxia Cheng, Yajun Zheng, Hanxu Li, Zilin Zhu, Yuxiang Yan, Wenhui Hao, and Nan Qin. 2023. "Study on the In Silico Screening and Characterization, Inhibition Mechanisms, Zinc-Chelate Activity, and Stability of ACE-Inhibitory Peptides Identified in Naked Oat Bran Albumin Hydrolysates" Foods 12, no. 11: 2268. https://doi.org/10.3390/foods12112268
APA StyleLi, Y., Li, J., Cheng, C., Zheng, Y., Li, H., Zhu, Z., Yan, Y., Hao, W., & Qin, N. (2023). Study on the In Silico Screening and Characterization, Inhibition Mechanisms, Zinc-Chelate Activity, and Stability of ACE-Inhibitory Peptides Identified in Naked Oat Bran Albumin Hydrolysates. Foods, 12(11), 2268. https://doi.org/10.3390/foods12112268