Abstract
We have successfully identified the transcription factor Cmr1 from the fungus Aureobasidium pullulans Hit-lcy3T, which regulates melanin biosynthesis genes. Bioinformatics analysis revealed that the Cmr1 gene encodes a protein of 945 amino acids, containing two Cys2His2 zinc finger domains and a Zn(II)2Cys6 binuclear cluster domain located at the N-terminus of Cmr1. To investigate the function of the Cmr1 gene, we performed gene knockout and overexpression experiments. Our results showed that Cmr1 is a key regulator of melanin synthesis in Hit-lcy3T, and its absence caused developmental defects. Conversely, overexpression of Cmr1 significantly increased the number of chlamydospores in Hit-lcy3T and improved melanin production. RT-qPCR analysis further revealed that overexpression of Cmr1 enhanced the expression of several genes involved in melanin biosynthesis, including Cmr1, PKS, SCD1, and THR1. Melanin extracted from the Hit-lcy3T was characterized using UV and IR spectroscopy. Furthermore, we assessed the antioxidant properties of Hit-lcy3T melanin and found that it possesses strong scavenging activity against DPPH·, ABTS·, and OH·, but weaker activity against O2−·. These findings suggest that Hit-lcy3T melanin holds promise for future development as a functional food additive.
1. Introduction
Melanin is a complex and diverse large molecule named by the Swiss scientist Jöns Berzelius. It is formed by the polymerization of polyhydroxyindole or polyhydroxyphenolic substances and is widely found in animals, plants, and microorganisms [1]. Currently, melanin can be classified into five types based on the precursor substances of its biosynthetic pathway: L-DOPA (L-3,4-dihydroxyphenylalanine) melanin, 1,8-dihydroxynaphthalene (1,8-DHN) melanin, pyomelanin, pheomelanin, and Glutaminyl-4-hydroxybenzene (GHB) melanin. However, most fungi synthesize 1,8-DHN melanin and L-DOPA melanin via the DHN and L-DOPA pathways, respectively. They form complex polyphenol-based polymers through enzyme-catalyzed and chemical reactions between 1,8-DHN and L-Dopaquinone. Melanin’s amorphous quality renders it insoluble in water, most organic solvents, and acidic solutions, but soluble in alkaline solutions. It is tightly bound to macromolecules such as proteins, carbohydrates, and tannins, making it difficult to accurately analyze its precise structure and chemical composition. Melanin provides protection against UV, chemical, and thermal stresses. It also acts as a potent cation chelator and antioxidant, binds and neutralizes antimicrobial peptides and antibiotics drugs, inhibits enzyme activation, and confers dehydration tolerance [2,3].
Daily foods rich in melanin include coffee, cocoa, dark beer, soy sauce, baked foods, mushrooms, and others [4]. These melanins can provide food with antioxidant capacity, inhibit oxidative stress in cells, suppress or kill pathogenic microorganisms, and induce the synthesis of chemopreventive enzymes [5]. In recent years, melanin’s unique biological characteristics have led to its widespread use in a variety of fields, including food, medicine, cosmetics, and material synthesis [6,7]. Lee et al. found that melanin can effectively reduce the weight of high-fat diet mice, improve lipid status, and increase antioxidant enzyme activity to lower blood lipid levels, and water-soluble melanin complexes have been found to improve insulin sensitivity in obese mice fed a high-fat diet and effectively lower blood sugar [8]. Xu et al. verified that melanin could increase mouse inflammatory factors, alleviate inflammatory stress, and protect the liver [9]. In addition, research has shown that 20% of the total antioxidant capacity intake in the Spanish population is provided by melanin in food [10]. All of these indicate the potential application of melanin as a functional food additive [11]. The characteristics of melanin make it highly valuable, but currently, the process of extracting melanin from animals and plants is complex, with low yield and high cost, making it impossible to obtain melanin in large quantities.
Aureobasidium pullulans (A. pullulans), commonly known as black yeast, is a fascinating fungus with a complex lifecycle that exhibits various morphological forms under different environmental conditions or growth stages, including budding spores, mycelia, chlamydospores, and small oval-shaped yeast-like cells [12,13]. One of the most notable characteristics of this species is the production of melanin. Researchers have explored different approaches to optimize the production of melanin by A. pullulans. For example, Mujdeci used carrot peel extract as a substrate to ferment A. pullulans NBRC 100716 and achieved a 100% increase in the total concentration of melanin in the extract by optimizing the fermentation parameters [14]. Mujdeci found that A. pullulans NBRC 100716 can also synthesize natural melanin by fermenting food residues, and characterized it after extraction, with the highest yield reaching 300.0 mM/L [15]. Similarly, Gamal et al. achieved a significant increase in melanin production (up to 550.0 mM/L) by optimizing the fermentation parameters using food waste as a substrate [16]. However, most studies on melanin production by A. pullulans have focused on the selection of fermentation substrates and strain screening, and further research is needed to better understand the synthesis and regulation of melanin by this species.
Cmr1 is a crucial transcriptional activator that regulates the melanin synthesis pathway in fungi. It was first discovered by Gento in 2000 and is typically composed of two Cys2His2 zinc fingers and one Zn(II)2Cys6 binuclear cluster domain [17]. Since then, homologs of Cmr1 have been identified in many fungi, including Helminthosporium maydis, Bipolaris oryzae, Alternaria brassicicola, Alternaria alternate, Grifola frondosa, Botrytis cinerea, and Verticillium dahliae. These homologs play regulatory roles in the DHN melanin synthesis pathway and affect the virulence of pathogenic fungi [18,19,20,21,22,23,24]. Knockout of the Cmr1 homolog or downstream regulated genes typically leads to a reduction or loss of melanin production. In the study of melanin production in A. pullulans, Jiang et al. initially identified the involvement of the Cmr1 gene in melanin synthesis but did not conduct further in-depth research on its function [25].
This paper first isolated and characterized the melanin produced by A. pullulans Hit-lcy3T, and then identified Cmr1 involved in the synthesis of the melanin through bioinformatics analysis of its whole genome. Subsequently, knockout strains of Cmr1 were obtained using homologous recombination, and the functions of Cmr1 were investigated by comparing the morphology, biomass, melanin production, and expression levels of genes involved in melanin biosynthesis between the wild-type (WT), Cmr1 knockout mutant(ΔApCmr1), and Cmr1 overexpression strains (OEX-ApCmr1). Finally, the scavenging activity of the melanin against free radicals including DPPH·, ABTS·, OH·, and O2−· was studied.
2. Materials and Methods
2.1. Stain, Plasmid, and Culture Medium
A. pullulans Hit-lcy3T and DH5α, as well as the PMD-18T vectors and pBARGPE1-EGFP, were obtained from our laboratory. The culture media used in this study included LB (10.0 g tryptone, 10.0 g NaCl, and 5.0 g yeast extract per liter, pH 7.2), YPD (20.0 g/L glucose, 10.0 g/L yeast extract, and 20.0 g/L peptone), seed medium (50.0 g/L sucrose, 2.0 g/L yeast extract, 1.0 g/L NaCl, 6.0 g/L K2HPO4, 0.2 g/L MgSO4, and 0.6 g/L (NH4)2SO4), and fermentation medium (100.0 g/L sucrose, 2.0 g/L yeast extract, 1.0 g/L NaCl, 6.0 g/L K2HPO4, 0.2 g/L MgSO4, and 0.6 g/L (NH4)2SO4).
2.2. Acquisition and Bioinformatics Analysis of Cmr1 Gene
The Cmr1 gene was found in the genome of A. pullulans Hit-lcy3T, and the primers Cmr1F/Cmr1R were designed to amplify the full-length gene. The amplified gene was sequenced and confirmed by comparison with NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 10 December 2021). SMART (https://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1, accessed on 5 March 2022) was used to predict the conserved domains of Cmr1. BDPG (https://www.fruitfly.org/seq_tools/promoter.html, accessed on 7 March 2022) was used to predict the promoter transcription start site. We then based on Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 25 March 2022) to predict the promoter region of Cmr1. ClustalW (https://www.genome.jp/tools-bin/clustalw, accessed on 25 April 2022) and MEGA Ⅹ software were used to perform multiple strain alignment and construct a phylogenetic tree of the amino acid sequence of Cmr1.
2.3. Melanin Extraction Method
A. pullulans Hit-lcy3T was cultured in a fermentation medium for 6, 7, 8, 9, 10, and 11 days, and the biomass was determined. The melanin was then extracted and purified using an acid–ethanol precipitation method. The fermentation broth was thoroughly mixed, and 50 mL of the mixture was centrifuged at 4000 rpm for 10 min. The supernatant was collected for further processing, and the precipitate was the biomass. The collected supernatant was then mixed with 1 mol/L hydrochloric acid to adjust the pH to 2–3 and shaken vigorously. After that, it was left to stand at 60 °C to induce complete precipitation. The product was then filtered through a 600-mesh filter paper and dried at 60 °C to constant weight to obtain the crude melanin. Finally, the crude product was repeatedly dissolved in a strong base, acid–ethanol precipitated and purified to obtain melanin with higher purity.
2.4. FT-IR Analysis of Melanin
The preparation method of the FT-IR sample was as follows: the isolated melanin was freeze-dried, ground into powder, and then a suitable amount of the powder was placed in a PerkinElmer 100 FT-IR spectrometer (PerkinElmer Inc., Waltham, MA, USA) for analysis under conditions of a resolution of 4 cm−1 and a scanning range of 500–4000 cm−1, to obtain the infrared spectrum of the melanin [26].
2.5. UV–Vis Analysis of Melanin
A suitable amount of melanin extract and standard substance were dissolved in 1 mol/L sodium hydroxide solution and then placed in a UV-6100 type UV–Vis spectrophotometer to scan the absorption spectrum in the wavelength range of 190–800 nm in spectral scan mode [27].
2.6. Plasmid Construction
The primers are listed in Table S1. To construct the Cmr1 transcription factor knockout plasmid, the PMD-18T plasmid was used as the backbone, and the Cmr1 gene was replaced with the hygromycin gene using homologous recombination to achieve the deletion of the Cmr1 gene. First, the Hyg fragment and the upstream and downstream fragments of the Cmr1 gene (Cmr1-up and Cmr1-down) were amplified by PCR. Then, the PMD-18T plasmid was double-digested with EcoRI and BamHI, and the Cmr1-up, Hyg, and Cmr1-down fragments were connected to the PMD-18T knockout plasmid backbone using the ClonExpress II one-step cloning kit, thus resulting in the knockout plasmid PMD-Hyg-△Cmr1.
For the construction of the pBARGPE-EGFP-Cmr1 vector, first, a 3013 bp fragment covering the coding sequence of Cmr1 was amplified from the genome of A. pullulans Hit-lcy3T. Then, EcoRI and KpnI were used to digest the Cmr1 fragment and pBARGPE1-EGFP vector, respectively. Finally, ClonExpress II one-step cloning was used to ligate the recovered Cmr1 fragment into pBARGPE1-EGFP, resulting in the overexpression of plasmid pBARGPE-EGFP-Cmr1.
2.7. Transformation and Validation of Mutant Strains
The successfully constructed overexpression vector pBARGPE-EGFP-Cmr1 and knockout vector PMD-Hyg-△Cmr1 were transformed into A. pullulans protoplasts using the PEG-mediated protoplast transformation method [28]. The genomic DNA of transformants was extracted, and EGFP (egfpF/egfpR) and hygromycin resistance gene (hygR/hygF) were amplified using specific primers and validated by sequencing.
2.8. Effect of Cmr1 on Biomass and Melanin Production
Wild-type, Cmr1 deletion strain, ΔApCmr1, and Cmr1 overexpression strain, OEX-ApCmr1, were cultured in a fermentation medium for 9 days to determine their biomass and melanin production.
2.9. Morphological Observation
The morphological features of A. pullulans Hit-lcy3T and its mutants after 9 days of cultivation on YPD medium were observed using a light microscope (Nikon ECLIPSE E200, Nikon, Tokyo, Japan) and an electron microscope (JSM-6700F, JEOL, Tokyo, Japan). For scanning electron microscopy (SEM) sample preparation, square pieces of 0.5 cm × 0.5 cm were cut from the agar plate, fixed in 2.5% glutaraldehyde at 4 °C for about 12 h, washed twice with phosphate buffer, and treated with graded ethanol (50%, 70%, 90%, 100%). Finally, the samples were placed on conductive tape after treatment with acetone and freeze-dried, and observed under an SEM.
2.10. Analysis of the Transcriptional Levels of Genes Related to the ApCmr1 Biosynthetic Gene Cluster
Total RNA was extracted from the WT and different transformants grown in the fermentation medium. EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) was used for reverse transcription to obtain cDNA. RT-qPCR was performed using Go Taq qPCR Master Mix (Promega, Madison, Wisconsin, USA) in a real-time PCR system (Thermo Fisher Scientific, Wilmington, Massachusetts, USA), with the β-actin gene as the internal control gene. The relative expression levels of Cmr1, PKS, SCD1, and THR1 were quantitatively detected, and expression levels were evaluated using the 2−ΔΔCT method.
2.11. Investigation of Antioxidant Activity of Melanin In Vitro
2.11.1. DPPH· Scavenging Assay
A 0.2 mmol/L solution of DPPH· in absolute ethanol was prepared. Different concentrations of melanin samples and Vc (Vitamin C) solution were mixed with 100 μL of DPPH· solution in absolute ethanol, respectively. The mixtures were thoroughly vortexed, and shielded from light. After 30 min. the OD517 was measured using a multimode plate reader [12].
DPPH· scavenging rate (%) = [1 − (A1 − A2)/A0] × 100%.
A0: 100 μL anhydrous ethanol + 100 μL DPPH· absolute ethanol solution;
A1: 100 μL test solution + 100 μL DPPH· absolute ethanol solution;
A2: 100 μL test solution + 100 μL absolute ethanol solution.
2.11.2. ABTS· Scavenging Assay
First, 4 mmol/L ABTS· reserve solution and 2.6 mmol/L K2S2O8 solution were prepared. Then, the ABTS· reserve solution and the K2S2O8 solution were mixed in a 1:1 ratio and allowed to stand for about 12 h. Finally, it was diluted with distilled water to obtain ABTS· working solution with an absorbance of 0.700 at 734 nm wavelength. Next, 100 μL of different concentrations of melanin solution and Vc solution were mixed into 96-well plates, and then 100 μL ABTS· working solution was added to each well. After 10 min, the absorbance of the sample at 734 nm wavelength was measured [12].
ABTS· scavenging rate (%) = [1 − (A1 − A2)/A0] × 100%
A0: 100 μL distilled water + 100 μL ABTS·;
A1: 100 μL test solution + 100 μL ABTS·;
A2: 100 μL test solution + 100 μL distilled water.
2.11.3. OH· Scavenging Assay
Then, 1 mL melanin solution and Vc solution of different concentrations were added into 1 mL 9 mmol/L FeSO4 and 1 mL H2O2 in turn. The solution was mixed well and incubated at 37 °C for 10 min. Then, 1 mL 9 mmol/L salicylic acid was added and the reaction continued at 37 °C for 30 min. The absorbance of the sample at 536 nm wavelength was measured [12].
OH· scavenging rate (%) = [1 − (A1 − A2)/A0] × 100%.
A0: 1 mL distilled water + 1 mL FeSO4 + 1 mL H2O2 + 1 mL salicylic acid;
A1: 1 mL of the test solution + 1 mL FeSO4 + 1 mL H2O2 + 1 mL salicylic acid;
A2: 1 mL distilled water + 1 mL FeSO4 + 1 mL distilled water + 1 mL salicylic acid.
2.11.4. O2−· Scavenging Assay
In each 6.5 mL reaction mixture, 2 mL L-methionine (39 mmol/L), 2 mL NBT (225 μmol/L), 1 mL disodium EDTA (0.6 mmol/L), 1 mL riboflavin (12 μmol/L), and 0.5 mL of the melanin solution or Vc solution of varying concentrations were added. All solutions were prepared in 0.05 mol/L phosphate buffer (pH 7.8). The reaction mixture containing the sample was incubated at 25 °C for 15 min, and the absorbance was measured at 560 nm. The reaction mixture without the sample served as the control and was kept in the dark. The scavenging activity was calculated as follows [27]:
O2−· scavenging rate (%) = (1 − A1/A0) × 100%
A0: 0.5 mL distilled water + 2 mL L-methionine + 2 mL NBT + 1 mL EDTA + 1 mL riboflavin;
A1: 0.5 mL sample solution + 2 mL L-methionine + 2 mL NBT + 1 mL EDTA + 1 mL riboflavin.
2.12. Statistical Analysis
All data were statistically analyzed using SPSS 19.0 (IBM Corporation). Gene expression analysis was evaluated using one-way analysis of variance (ANOVA). Duncan’s multiple-range test was used to compare data means. p value less than 0.05 was considered statistically significant.
3. Results and Discussion
3.1. Bioinformatics Analysis of Cmr1
The full length of the Cmr1 gene in A. pullulans is 3013 bp, encoding a protein of 945 amino acids. There are two introns at positions 71 bp–122 bp (50 bp) and 986 bp–1015 bp (30 bp), respectively. SMART analysis of the protein sequence revealed that there are two adjacent Cys2His2 zinc finger domains located at positions 2–24 bp and 30–52 bp of the amino acid sequence, respectively, and a Zn(II)2Cys6 binuclear cluster domain located at positions 333–361 bp of the N-terminus of the Cmr1 protein. The upstream 1500 bp sequence of Cmr1 was analyzed using the BDPG tool, revealing that Cmr1 is a member of the GATA family of transcription factors, and its gene promoter region contains three GATA motifs. The cis-acting element analysis of the promoter enhancer region using Plant CARE showed that there are one core promoter element (TATA-box) and three common cis-acting elements (CAAT-box) (Figure S1).
In the amino acid sequence’s alignment with 20 different fungi, a high similarity was observed in the zinc finger structural sequence of the Cmr1 gene (Figure 1A). However, A. pullulans Hit-lcy3T showed a difference at the 9th amino acid position compared to other strains, with histidine instead of glutamine, which is found in other fungi. Due to its distant evolutionary relationship with Candida albicans Ca529L and Saccharomyces cerevisiae 218 S288C, it was not included in the phylogenetic tree (Figure 1B).
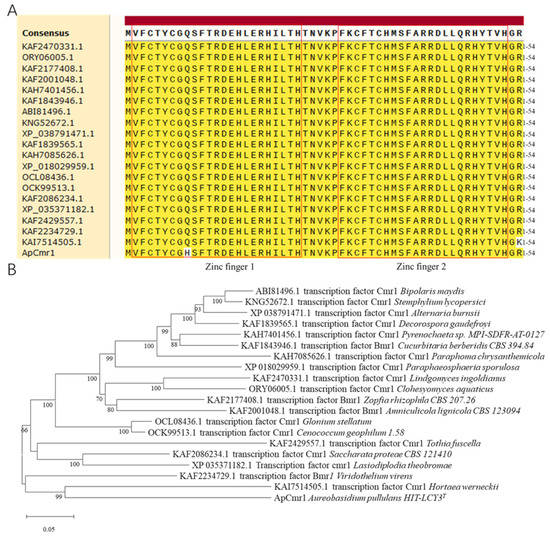
Figure 1.
Bioinformatics analysis of ApCmr1. (A) Comparison of the zinc finger domain of ApCmr1 and the homologous proteins in 20 fungi. (B) Phylogenetic tree of ApCmr1 and its homologous proteins in 20 fungi.
3.2. Fermentation Time of A. pullulans Hit-lcy3T for Melanin Production
The melanin and biomass were measured after different days of fermentation. As shown in Figure 2, both the melanin and biomass increased and then decreased with the increase in fermentation time. At 9 days, the highest melanin yield and biomass were reached at 9.357 g/L and 16.605 g/L, respectively, which were significantly higher than those obtained from Ophiocordyceps sinensis and Streptomyces sp. (1.775 g/L and 1.46 g/L, respectively) [27,29].
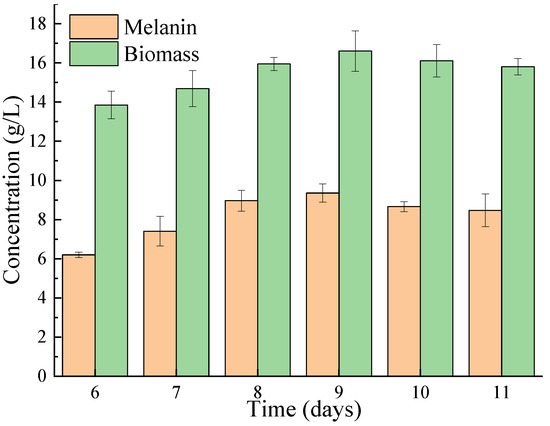
Figure 2.
The effect of fermentation time on the biomass and melanin production of A. pullulans Hit-lcy3T.
3.3. FT-IR Spectroscopy of DHN Melanin
The obtained melanin was characterized using FT-IR spectroscopy. As shown in Figure 3, the infrared spectrum of the DHN melanin extracted from A. pullulans Hit-lcy3T showed similar characteristic peaks as the melanin extracted from A. pullulans NBRC 100716, as reported by Mujdeci. The peak at 3300 cm−1 is attributed to the stretching vibrations of –OH in aromatic ethers. Two obvious peaks at 2978 cm−1 and 2900 cm−1 indicate the presence of C–H stretching vibrations in aromatic hydrocarbons. In comparison with the melanin studied by Centeno and Pacelli, which exhibited two obvious peaks at 2950–2850 cm−1, the two peaks observed in the present study were attributed to stretching vibrations of C–H in aromatic rings caused by lipids or amino acid residues during the extraction process [26,30]. The characteristic peaks of the melanin and synthetic melanin were the same at 1681 cm−1, 1607 cm−1, 1521 cm−1, 1256 cm−1, and 893 cm−1. The strong band at 1681 cm−1 is attributed to the vibration of C=O [27], while the strong band at 1607 cm−1 is attributed to the vibration of the C=C group in aromatic rings [28]. The band at 1521 cm−1 is attributed to the N–H bending vibration in the amide group of the aromatic rings and the peak at 1398 cm−1 is attributed to the bending vibration of C–H in the aromatic hydrocarbons. The peak at 1256 cm−1 is attributed to the stretching vibration of C–N in the amide group of the molecule. The peak observed at 987 cm−1 is related to the out-of-plane bending vibration of C–H in the aromatic amine [31].
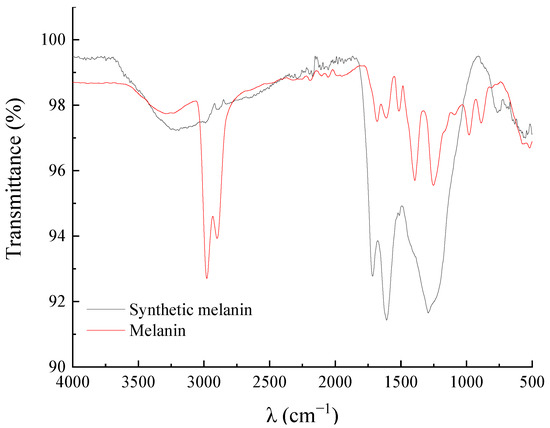
Figure 3.
FT-IR of DHN melanin and synthetic melanin.
3.4. Analysis of UV–Vis Absorption Spectra of DHN Melanin
It has been observed in previous studies that the longer the wavelength, the lower the absorbance of melanin, which is a characteristic feature of melanin. Additionally, the absorption curve of melanin is similar to that of L-DOPA melanin nanoparticles. However, Li et al. found that the absorbance of purified Streptomyces sp. melanin showed almost linear behavior and did not exhibit any maximum or minimum absorption within the range of 200–800 nm [27]. Pacelli et al. compared the absorbance spectra of Cryomyces antarcticus melanin, L-DOPA, and DHN, and found that the highest peak of absorption was in the range of 200–300 nm [30]. Analysis of the UV–Vis absorption spectra of DHN melanin extracted from A. pullulans showed that the maximum absorbance of melanin occurred at 218 nm, which is consistent with the maximum absorbance of synthetic melanin (Figure 4). This is consistent with the results of Thokur’s study on the DHN melanin of Phyllosticta capitalensis [32].
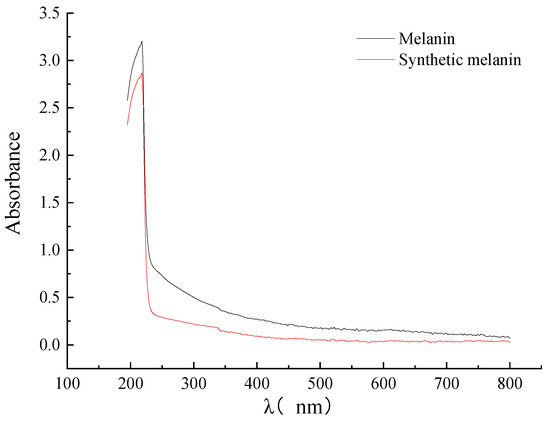
Figure 4.
The UV–Vis of DHN melanin extracted from A. pullulans and the synthetic melanin.
3.5. Schematic and Verification of Gene Knockout
To verify the gene function of Cmr1 in A. pullulans Hit-lcy3T, a knockout vector was constructed using PMD-18T as the vector backbone through homologous recombination, as shown in Figure 5A. Cmr1-up, hyg, Cmr1-down, HindIII, and EcoRI double enzyme digestion PMD-18T vector bands were validated by agarose gel electrophoresis, with band sizes of 1305 bp, 1412 bp, 1289 bp, and 2635 bp, respectively (Figure 5B). As the △Cmr1 vector contains two EcoRI restriction sites while PMD-18T contains only one, EcoRI digestion was performed on both PMD-18T and △Cmr1 vectors separately. The resulting bands for △Cmr1 were 2284 bp and 4610 bp, while PMD-18T only produced a band of 2693 bp (Figure 5C). The amplification with the hygR/hygF primers revealed a 1000 bp hygromycin fragment in ΔAPCmr1, which was not present in the WT strain (Figure 5D).
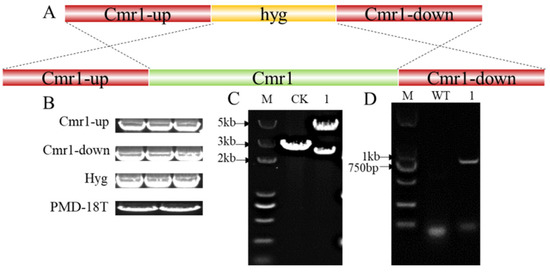
Figure 5.
Construction and validation of the Cmr1 knockout plasmid. (A) Construction strategy of the Cmr1 knockout vector; (B) Cmr1-up, Cmr1-down, hyg, and PMD-18T bands after double enzyme digestion; (C) enzyme digestion (EcoRI) validation of the △Cmr1 vector, where CK represents the PMD-18T vector backbone and 1 represents the △Cmr1 vector; (D) PCR validation of the knockout strain, where WT represents the wild-type strain and 1 represents the ΔAPCmr1 knockout strain.
3.6. Construction and Verification of Cmr1 Overexpression Plasmid
Due to the presence of a strong gpdA promoter in pBARGPE-EGFP, it was selected as the backbone for the overexpression vector. The construction process of pBARGPE-EGFP-Cmr1 overexpression plasmid involved double digestion with EcoRI and KpnI, followed by ligation with the amplified Cmr1 gene from the genome, as illustrated in Figure 6A. With the pBARGPE-EGFP and pBARGPE-EGFP-Cmr1 as templates, the amplification, using primers epc1-F and epc1-R, showed that a band of 1700 bp was obtained from pBARGPE-EGFP-Cmr1, whereas no band was detected from pBARGPE-EGFP (Figure 6B). Since pBARGPE-EGFP-Cmr1 contains two BamHI restriction sites, while pBARGPE-EGFP only has one BamHI restriction site, BamHI digestion was performed to verify both pBARGPE-EGFP and pBARGPE-EGFP-Cmr1. The results showed that pBARGPE-EGFP-Cmr1 produced two bands of 2672 bp and 6999 bp, respectively, after digestion, while pBARGPE-EGFP produced only a single band of 6700 bp. Furthermore, amplification of the green fluorescent protein-encoding gene using egfpF/egfpR primers revealed a band of 720 bp corresponding to the EGFP gene in the OEX-ApCmr1 strain but not in the WT strain (Figure 6C). Fluorescence inverted microscope analysis (Figure 6E) confirmed the successful expression of the green fluorescent protein in the OEX-ApCmr1 strain.
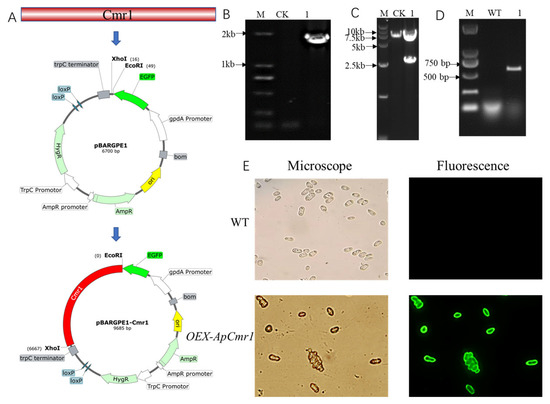
Figure 6.
Constructing the Cmr1 overexpression plasmid and validating the mutant strain. (A) Construction strategy for Cmr1 overexpression vector; (B) PCR validation of pBARGPE-EGFP-Cmr1 plasmid, with CK representing the empty vector as negative control and 1 representing pBARGPE-EGFP-Cmr1 plasmid; (C) enzyme digestion validation (BamHI) of pBARGPE-EGFP vector, with CK representing the empty vector as negative control and 1 representing pBARGPE-EGFP-Cmr1 plasmid; (D) PCR validation of the overexpression strain, with WT representing wild-type strain, and 1 representing the OEX-ApCmr1 overexpression strain; (E) green fluorescence validation of the overexpression strain, with WT representing wild-type strain and OEX-ApCmr1 as the overexpression strain.
3.7. Effect of Cmr1 Mutant Strain on Morphology and Melanin Production
As shown in Figure 7, after 9 days of cultivation in a YPD medium, the surface of the ΔApCmr1 mutant strain was overall white, smooth, and moist, without aerial mycelia, while both the WT and OEX-ApCmr1 strains had abundant aerial mycelia on the YPD medium plate. The OEX-ApCmr1 mutant strain exhibited significantly higher DHN melanin secretion than the WT strain. Under the microscope, there were significant differences in melanin secretion among the two mutant strains and the WT strain, with no melanin generation in the ΔApCmr1 strain, while the OEX-ApCmr1 strain had extremely high melanin content, staining the entire cell black. SEM observation revealed that the morphology of A. pullulans after knocking out the ApCmr1 gene was mostly yeast-like, and the yeast cells of the mutant strain had obvious defects in their edges. Overexpression of ApCmr1 led to chlamydospores becoming the main form, which was consistent with Mujdeci finding that the main form of melanin synthesis is chlamydospores [15]. It is speculated that the GATA-type transcription factor Cmr1, as a key transcription activation factor, not only regulates the synthesis of melanin but may also have an effect on the cell morphology of A. pullulans.
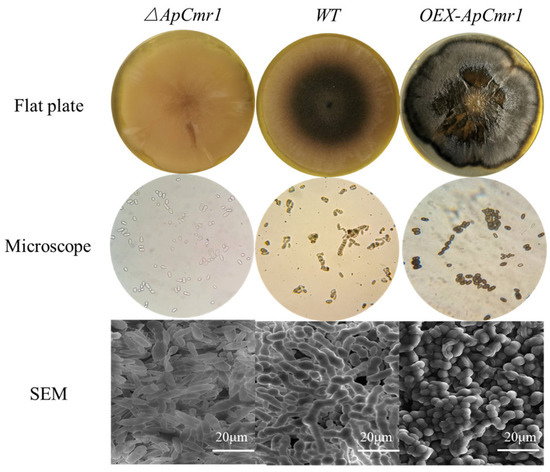
Figure 7.
Morphological observation of the mutant strain.
WT and each mutant strain were cultured in a fermentation medium for 9 days, and the production of melanin and biomass is shown in Figure 8. The melanin of the ΔAPCmr1 strain disappeared, and its biomass was reduced compared to the WT and OEX-APCmr1. This is partly due to the decrease in the expression level of melanin-related genes and the possible decrease in biomass due to the developmental defects of the strain caused by the knockout of the Cmr1 gene. However, the melanin production in the OEX-ApCmr1 strain was 1.436 times that of the WT, and its biomass also slightly increased, which was one of the reasons for the increase in melanin production.
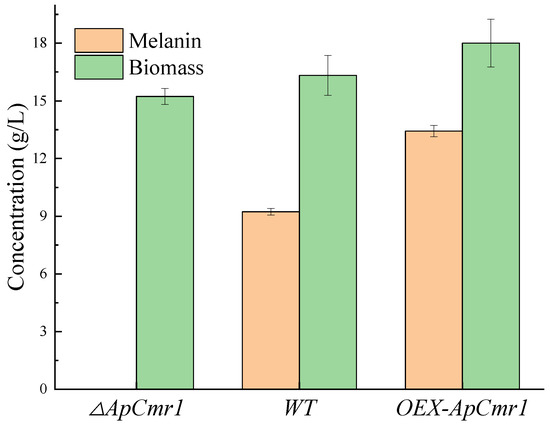
Figure 8.
Biomass and melanin production of mutant strains.
3.8. Expression of Melanin Synthesis-Related Gene
After 9 days of fermentation in the culture medium, RNA was extracted from WT and various mutant strains. Reverse transcription was performed to obtain cDNA, and RT-qPCR analysis was conducted on WT, ∆ApCmr1, and OEX-ApCmr1 strains (Figure 9). The results revealed that the expression levels of Cmr1, PKS, SCD1, and THR1 genes were nearly undetectable after knocking out the Cmr1 gene, providing further evidence that Cmr1 acts as a transcriptional regulator of melanin synthesis in A. pullulans, which is consistent with the findings of Chi’s study [33]. Overexpression of the Cmr1 gene resulted in a 5.488-fold increase in the relative expression of Cmr1, and 7.324-fold, 8.535-fold, and 8.434-fold increases in the relative expression of PKS, SCD1, and THR1 genes, respectively. However, the expression levels were still lower than the relative expression levels of Bmr1, PKS, SCD1, and THR1 genes in the Bipolaris oryzae strain after overexpression of the Bmr1 gene [19].
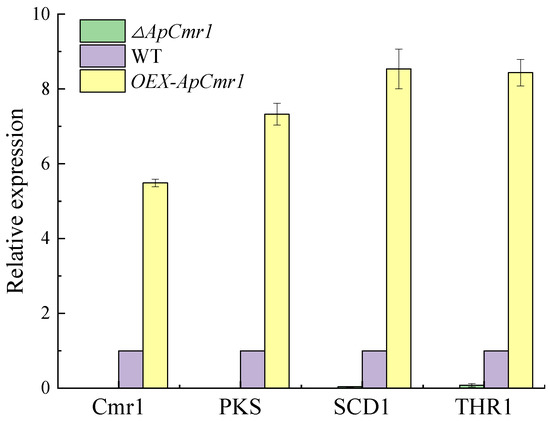
Figure 9.
The effect of Cmr1 on the expression of melanin synthesis genes.
3.9. In Vitro Antioxidant Activity of Melanin
Studies have shown that melanin from microorganisms has certain antioxidant activity. To explore the antioxidant activity of melanin, Vc was selected as a control to evaluate its ability to scavenge DPPH·, ABTS·, OH·, and O2−·. The results of melanin scavenging DPPH· are shown in Figure 10A. As the concentration of Vc and melanin increased, the scavenging rate of DPPH· also increased, reaching a maximum of 84.96%, which is slightly lower than the ability of melanin extracted from Cryptococcus rajasthanensis KY627764 to scavenge DPPH· [34]. The melanin scavenging activity against ABTS· is shown in Figure 10B. With the increase in melanin concentration, the scavenging rate of ABTS· also increased, reaching a maximum of 80.44%, but it was still lower than that of Vc. The melanin scavenging activity against OH· is shown in Figure 10C. With the increase in melanin concentration, the scavenging rate of OH· also increased, reaching a maximum of 81.57%. The ability of melanin to scavenge O2−· increased with the concentration (Figure 10D), reaching a scavenging rate of 17.83% at 100 μg/mL, but was weaker compared to the ability of melanin extracted from Streptomyces sp. by Li et al. to scavenge O2−· [27]. These results suggest that the melanin produced by A. pullulans has a strong ability to scavenge DPPH·, ABTS·, and OH·, but a weaker ability to scavenge O2−·.
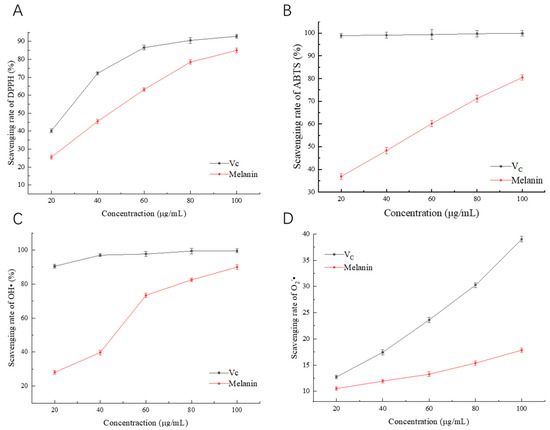
Figure 10.
Antioxidant activity of melanin. (A) Scavenging rate of DPPH·. (B) Scavenging rate of ABTS·. (C) Scavenging rate of OH·. (D) Scavenging rate of O2−·.
Currently, research on the synthesis and application of melanin is increasing, and the potential of melanin as a new type of functional active substance has been gradually explored. Most fungi synthesize melanin through two pathways: tyrosine (L-DOPA pathway) or malonyl CoA (DHN pathway) [35]. In the DOPA pathway, tyrosinase first catalyzes L-tyrosine (monophenol) and L-DOPA (Bisphenol) to generate L-dopaquinone under the catalysis of the enzyme. Then, L-dopaquinone undergoes enzyme-catalyzed and chemical reactions to form a complex chemical structure, a non-homogeneous polymeric material known as L-DOPA melanin. In the DHN pathway, the precursor of malonyl CoA is endogenously produced. Under the catalysis of polyketide synthase, five malonyl CoA are consecutively decarboxylated and condensed to generate 1,3,6,8-tetrahydroxynaphthalene (THN). THN then undergoes reduction and dehydration to generate 1,8- DHN, which is finally polymerized to obtain DHN melanin. Cmr1 is a key transcriptional regulatory factor of DHN melanin that regulates the genes involved in the DHN melanin synthesis pathway in most fungi. It specifically binds to the upstream of the polyketide synthase gene to initiate the synthesis of melanin [36]. Studies have shown that in melanin within A. pullulans, DHN melanin is the primary form, which is synthesized by the polymerization of 1,8-DHN, and is important for maintaining cell wall integrity and morphology.
Knocking out the Cmr1 gene caused the disappearance of melanin in A. pullulans, as the expression levels of genes related to melanin synthesis were almost zero after knocking out Cmr1. Moreover, the biomass of the mutant strains decreased, and they mainly showed a yeast-like morphology. Since fungal melanin is a part of the cell wall composition, the strains that underwent knockout displayed developmental defects around the cell surface, which is consistent with Liang’s study. The reason for this was the loss of melanin, which led to the loss of the originally filled areas of melanin in the cell wall, resulting in irregular developmental defects in the cell wall, demonstrating that melanin plays an important role in maintaining the fungal cell wall morphology [37]. The OEX-ApCmr1 strain showed increased expression levels of Cmr1, PKS, SCD1, and THR1 genes related to melanin synthesis by 5.488-fold, 7.324-fold, 8.535-fold, and 8.434-fold, respectively, and melanin production was 1.436-times higher than that of the WT strain. This may be due to the overexpression of melanin in the OEX-ApCmr1 strain, which bound more to the cell wall and was not released into the fermentation broth. Morphological observations showed that the cells of the OEX-ApCmr1 strain were predominantly chlamydospores and hyphae, consistent with previous research that suggested that chlamydospores and hyphae were the primary forms of melanin production [38].
The mechanism by which melanin acts as an antioxidant is still unclear. However, recent research has found that its antioxidant activity is related to the monomer 1,8-DHN. This is due to the unique pattern of hydroxylation of the hydrogen-bonding ring in 1,8-DHN, which facilitates the rapid transfer of H atoms [39]. Our research has found that melanin has good scavenging activity on OH·, DPPH·, and ABTS·, but a weak effect on O2−·. The reason for this is that when O2−· extracts H atoms to form OOH· or H2O2, the transfer of the two hydrogen atoms only occurs at the adjacent carbon position. However, during the polymerization of 1,8-DHN to form melanin, one or two of the adjacent hydroxy groups are replaced, resulting in a weaker scavenging activity on O2−· [40]. In the food industry, natural pigments are valued for their safety, nutrition, and certain bioactivity. Therefore, melanin can be used as a natural coloring agent and additive in food production. Melanin has already been used to prepare yogurt to enhance its antioxidant and free radical scavenging properties, as well as to develop various new types of food, such as black bean vermicelli, black tofu skin, and black caviar [7]. It can also be used as a functional food to better exert its role in reducing blood cholesterol, lowering blood sugar, and anti-tumor activity. There are many microorganisms in nature that can produce pigments. By using genetic engineering methods to obtain genes encoding pigment synthesis and constructing microorganism pigment production engineering bacteria, and effectively regulating their metabolic pathways, it is possible to achieve industrial production of pigments. With the rapid development of biotechnology, microbial pigments are expected to replace synthetic pigments as mainstream food coloring agents.
4. Conclusions
In this study, we investigated the regulatory gene Cmr1 in melanin synthesis through the construction of gene knockout and overexpression strains. Our results showed that Cmr1 is a key regulator of melanin synthesis in Hit-lcy3T, and its absence caused developmental defects. Furthermore, the expression levels of Cmr1, PKS, SCD1, and THR1 genes related to melanin synthesis were regulated by Cmr1. Finally, we found that melanin possesses strong scavenging activity against DPPH·, ABTS·, and OH·, but weaker activity against O2−·. These findings suggest that Hit-lcy3T melanin has the potential to be used as a functional food additive. This study provides a theoretical basis and guidance for the efficient production of melanin strains of A. pullulans and research on the antioxidant activity of melanin.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods12112135/s1, Figure S1: promoter prediction analysis of ApCmr1; Table S1: primers used in this study.
Author Contributions
Conceptualization, W.W. and K.Z.; methodology, C.L.; software, S.Z.; validation, J.G.; formal analysis, W.Z.; investigation, X.R.; resources, H.C.; supervision, Q.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Butler, M.J.; Day, A.W. Fungal melanins: A review. Can. J. Microbiol. 1998, 44, 1115–1136. [Google Scholar] [CrossRef]
- Dadachova, E.; Casadevall, A. Ionizing radiation: How fungi cope, adapt, and exploit with the help of melanin. Curr. Opin. Microbiol. 2008, 11, 525–531. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Echavarria, A.P.; Pagan, J.; Ibarz, A. Melanoidins Formed by Maillard Reaction in Food and Their Biological Activity. Food Eng. Rev. 2012, 4, 203–223. [Google Scholar] [CrossRef]
- Fogliano, V.; Morales, F.J. Estimation of dietary intake of melanoidins from coffee and bread. Food Funct. 2011, 2, 117–123. [Google Scholar] [CrossRef]
- Huang, L.; Liu, M.Y.; Huang, H.Y.; Wen, Y.Q.; Zhang, X.Y.; Wei, Y. Recent Advances and Progress on Melanin-like Materials and Their Biomedical Applications. Biomacromolecules 2018, 19, 1858–1868. [Google Scholar] [CrossRef]
- Tran-Ly, A.N.; Reyes, C.; Schwarze, F.W.M.R.; Ribera, J. Microbial production of melanin and its various applications. World J. Microbiol. Biotechnol. 2020, 36, 170. [Google Scholar] [CrossRef]
- Lee, J.H.; Hyun, C.K. Insulin-Sensitizing and Beneficial Lipid-Metabolic Effects of the Water-Soluble Melanin Complex Extracted from Inonotus obliquus. Phytother. Res. 2014, 28, 1320–1328. [Google Scholar] [CrossRef]
- Xu, C.; Chen, T.T.; Li, J.L.; Jin, M.Z.; Ye, M. The structural analysis and its hepatoprotective activity of melanin isolated from Lachnum sp. Process. Biochem. 2020, 90, 249–256. [Google Scholar] [CrossRef]
- Mesias, M.; Delgado-Andrade, C. Melanoidins as a potential functional food ingredient. Curr. Opin. Food Sci. 2017, 14, 37–42. [Google Scholar] [CrossRef]
- Dufosse, L. Microbial production of food grade pigments. Food Technol. Biotechnol. 2006, 44, 313–321. [Google Scholar]
- Zhang, K.; Zhou, W.; Wang, W.; Zhao, S.S.; Lin, C.Y.; Ru, X.; Guan, J.Q.; Cong, H.; Yang, Q. Area Gene Regulates the Synthesis of beta-Glucan with Antioxidant Activity in the Aureobasidium pullulans. Foods 2023, 12, 660. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhang, K.; Zhao, S.; Wang, W.; Ru, X.; Song, J.; Cong, H.; Yang, Q. Screening and identification of a strain of Aureobasidium pullulans and its application in potato starch industrial waste. Environ. Res. 2022, 214, 113947. [Google Scholar] [CrossRef] [PubMed]
- Mujdeci, G.N. Experimental modeling and optimization of melanin production by Aureobasidium pullulans NBRC 100716 in carrot peel extract. Environ. Prog. Sustain. Energy 2022, 41, e13919. [Google Scholar] [CrossRef]
- Mujdeci, G.N. Natural Melanin Synthesized by Aureobasidium pullulans Using Food Wastes and its Characterization. Appl. Food Biotechnol. 2021, 8, 307–318. [Google Scholar]
- Bialy, H.E.; Elsayed, M.; Khalifa, M.A. Isolation and Characterization of melanized yeast form of Aureobasidium pullulans and physiological studies on the melanization process. J. Nucl. Technol. Appl. Sci. 2017, 5, 57–72. [Google Scholar]
- Tsuji, G.; Kenmochi, Y.; Takano, Y.; Sweigard, J.; Farrall, L.; Furusawa, I.; Horino, O.; Kubo, Y. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 2000, 38, 940–954. [Google Scholar]
- Eliahu, N.; Igbaria, A.; Rose, M.S.; Horwitz, B.A.; Lev, S. Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot. Cell 2007, 6, 421–429. [Google Scholar] [CrossRef]
- Kihara, J.; Moriwaki, A.; Tanaka, N.; Tanaka, C.; Ueno, M.; Arase, S. Characterization of the BMR1 gene encoding a transcription factor for melanin biosynthesis genes in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 2008, 281, 221–227. [Google Scholar] [CrossRef]
- Cho, Y. The transcription factor Amr1 induces melanin biosynthesis and conidium production but differentially suppresses virulence in Alternaria brassicicola. Phytopathology 2011, 101, S36. [Google Scholar]
- Valiante, V.; Baldin, C.; Hortschansky, P.; Jain, R.; Thywissen, A.; Strassburger, M.; Shelest, E.; Heinekamp, T.; Brakhage, A.A. The Aspergillus fumigatus conidial melanin production is regulated by the bifunctional bHLH DevR and MADS-box RlmA transcription factors. Mol. Microbiol. 2016, 102, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, A.; Shimoda, T.; Sato, M.; Fujimori, F.; Hirama, J.; Nishibori, K. A putative transcription factor Gf.BMR1 in Grifola frondosa, the homolog of BMR1 in Bipolaris oryzae, was strongly induced by near-ultraviolet light and blue light. Mycoscience 2015, 56, 177–182. [Google Scholar] [CrossRef]
- Schumacher, J. DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 2016, 99, 729–748. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Hu, X.P.; Fang, Y.L.; Anchieta, A.; Goldman, P.H.; Hernandez, G.; Klosterman, S.J. Transcription factor VdCmr1 is required for pigment production, protection from UV irradiation, and regulates expression of melanin biosynthetic genes in Verticillium dahliae. Microbiology 2018, 164, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chi, Z.; Liu, G.L.; Hu, Z.; Zhao, S.Z.; Chi, Z.M. Melanin biosynthesis in the desert-derived Aureobasidium melanogenum XJ5-1 is controlled mainly by the CWI signal pathway via a transcriptional activator Cmr1. Curr. Genet. 2020, 66, 173–185. [Google Scholar] [CrossRef]
- Centeno, S.A.; Shamir, J. Surface enhanced Raman scattering (SERS) and FTIR characterization of the sepia melanin pigment used in works of art. J. Mol. Struct. 2008, 873, 149–159. [Google Scholar] [CrossRef]
- Li, C.; Ji, C.; Tang, B. Purification, characterisation and biological activity of melanin from Streptomyces sp. FEMS Microbiol. Lett. 2018, 365, fny077. [Google Scholar] [CrossRef]
- Zhang, K.; Lin, C.Y.; Zhao, S.S.; Wang, W.; Zhou, W.; Ru, X.; Cong, H.; Yang, Q. The role of pH transcription factor Appacc in upregulation of pullulan biosynthesis in Aureobasidium pullulans using potato waste as a substrate. Int. J. Biol. Macromol. 2023, 242, 124794. [Google Scholar] [CrossRef]
- Dong, C.H.; Yao, Y.J. Isolation, characterization of melanin derived from Ophiocordyceps sinensis, an entomogenous fungus endemic to the Tibetan Plateau. J. Biosci. Bioeng. 2012, 113, 474–479. [Google Scholar] [CrossRef]
- Pacelli, C.; Cassaro, A.; Maturilli, A.; Timperio, A.M.; Gevi, F.; Cavalazzi, B.; Stefan, M.; Ghica, D.; Onofri, S. Multidisciplinary characterization of melanin pigments from the black fungus Cryomyces antarcticus. Appl. Microbiol. Biotechnol. 2020, 104, 6385–6395. [Google Scholar] [CrossRef]
- Pal, A.K.; Gajjar, D.U.; Vasavada, A.R. DOPA and DHN pathway orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2014, 52, 10–18. [Google Scholar] [PubMed]
- Suryanarayanan, T.S.; Ravishankar, J.P.; Venkatesan, G.; Murali, T.S. Characterization of the melanin pigment of a cosmopolitan fungal endophyte. Mycol. Res. 2004, 108, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Kong, C.C.; Wang, Z.Z.; Wang, Z.; Liu, G.L.; Hu, Z.; Chi, Z.M. The signaling pathways involved in metabolic regulation and stress responses of the yeast-like fungi Aureobasidium spp. Biotechnol. Adv. 2022, 55, 107898. [Google Scholar] [CrossRef] [PubMed]
- Barretto, D.A.; Vootla, S.K. Biological activities of melanin pigment extracted from Bombyx mori gut-associated yeast Cryptococcus rajasthanensis KY627764. World J. Microbiol. Biotechnol. 2020, 36, 159. [Google Scholar] [CrossRef]
- El-Bialy, H.A.; El-Gamal, M.S.; Elsayed, M.A.; Saudi, H.A.; Khalifa, M.A. Microbial melanin physiology under stress conditions and gamma radiation protection studies. Radiat. Phys. Chem. 2019, 162, 178–186. [Google Scholar] [CrossRef]
- Jia, S.L.; Chi, Z.; Chen, L.; Liu, G.L.; Hu, Z.; Chi, Z.M. Molecular evolution and regulation of DHN melanin-related gene clusters are closely related to adaptation of different melanin-producing fungi. Genomics 2021, 113, 1962–1975. [Google Scholar] [CrossRef]
- Liang, X.; Li, B.; Zhao, X.; Yao, L.; Kong, Y.; Liu, W.; Zhang, R.; Sun, G. 1,8-Dihydroxynaphthalene Melanin Biosynthesis in Colletotrichum fructicola Is Developmentally Regulated and Requires the Cooperative Function of Two Putative Zinc Finger Transcription Factors. Phytopathology 2022, 112, 2174–2186. [Google Scholar] [CrossRef]
- Dehoog, G.S.; Yurlova, N.A. Conidiogenesis, Nutritional Physiology and Taxonomy of Aureobasidium and Hormonema. Anton. Leeuw. Int. J. G. 1994, 65, 41–54. [Google Scholar] [CrossRef]
- Lino, V.; Manini, P.; Galeotti, M.; Salamone, M.; Bietti, M.; Crescenzi, O.; Napolitano, A.; d’Ischia, M. Antioxidant Activities of Hydroxylated Naphthalenes: The Role of Aryloxyl Radicals. Chempluschem 2023, 88, e202200449. [Google Scholar] [CrossRef]
- Tiwari, M.K.; Jena, N.R.; Mishra, P.C. Mechanisms of scavenging superoxide, hydroxyl, nitrogen dioxide and methoxy radicals by allicin: Catalytic role of superoxide dismutase in scavenging superoxide radical. J. Chem. Sci. 2018, 130, 105. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).