A Nomogram Model for Predicting the Polyphenol Content of Pu-Erh Tea
Abstract
1. Introduction
2. Materials and Methods
2.1. The General Situation of the Research Location
2.2. Experimental Materials
2.3. Statistical Analysis
3. Results
3.1. Single-Factor Analysis
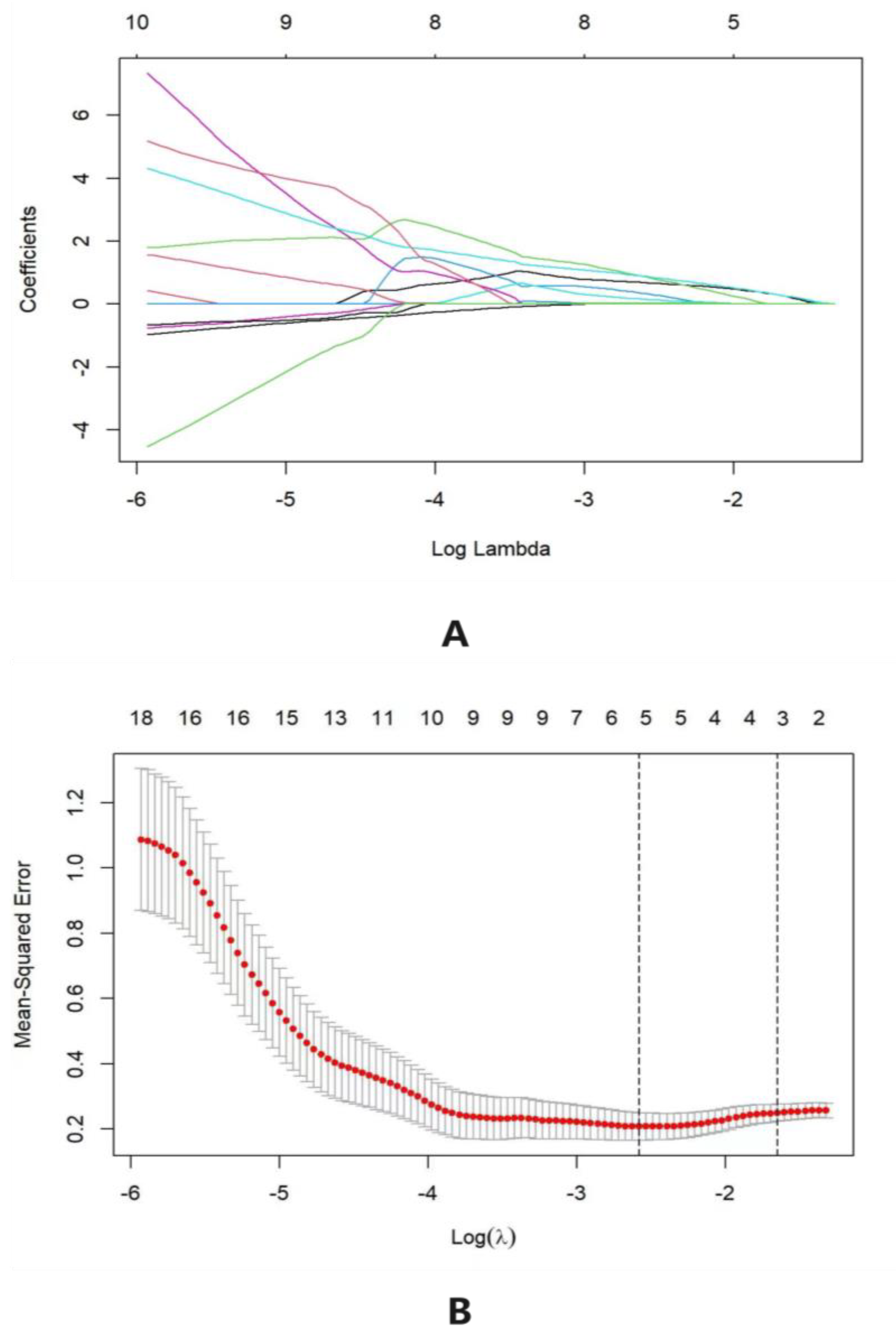
3.2. Model Construction Factor Selection
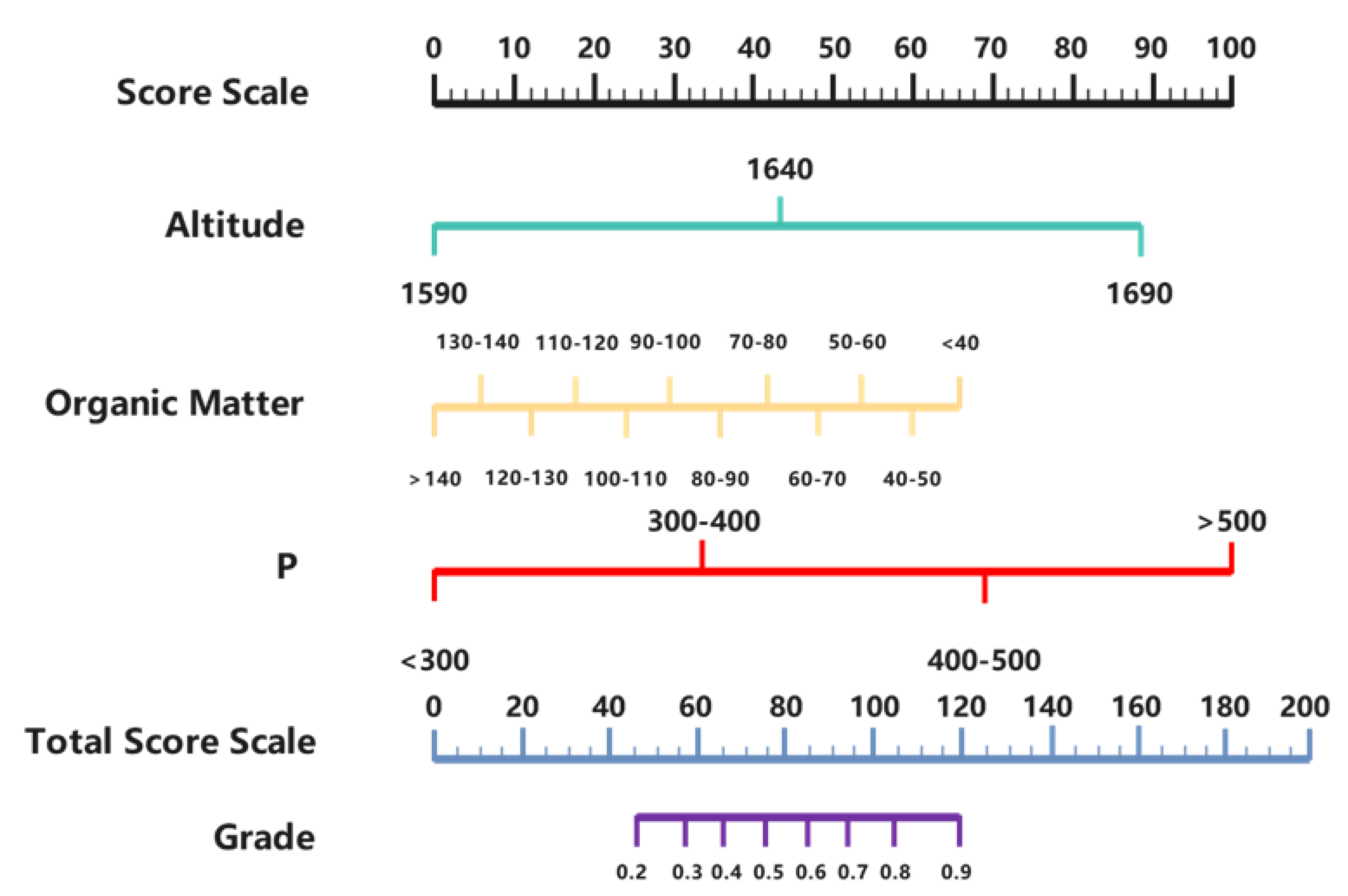
3.3. The Construction of the Nomogram
3.4. The Evaluation of the Accuracy and Stability of the Model
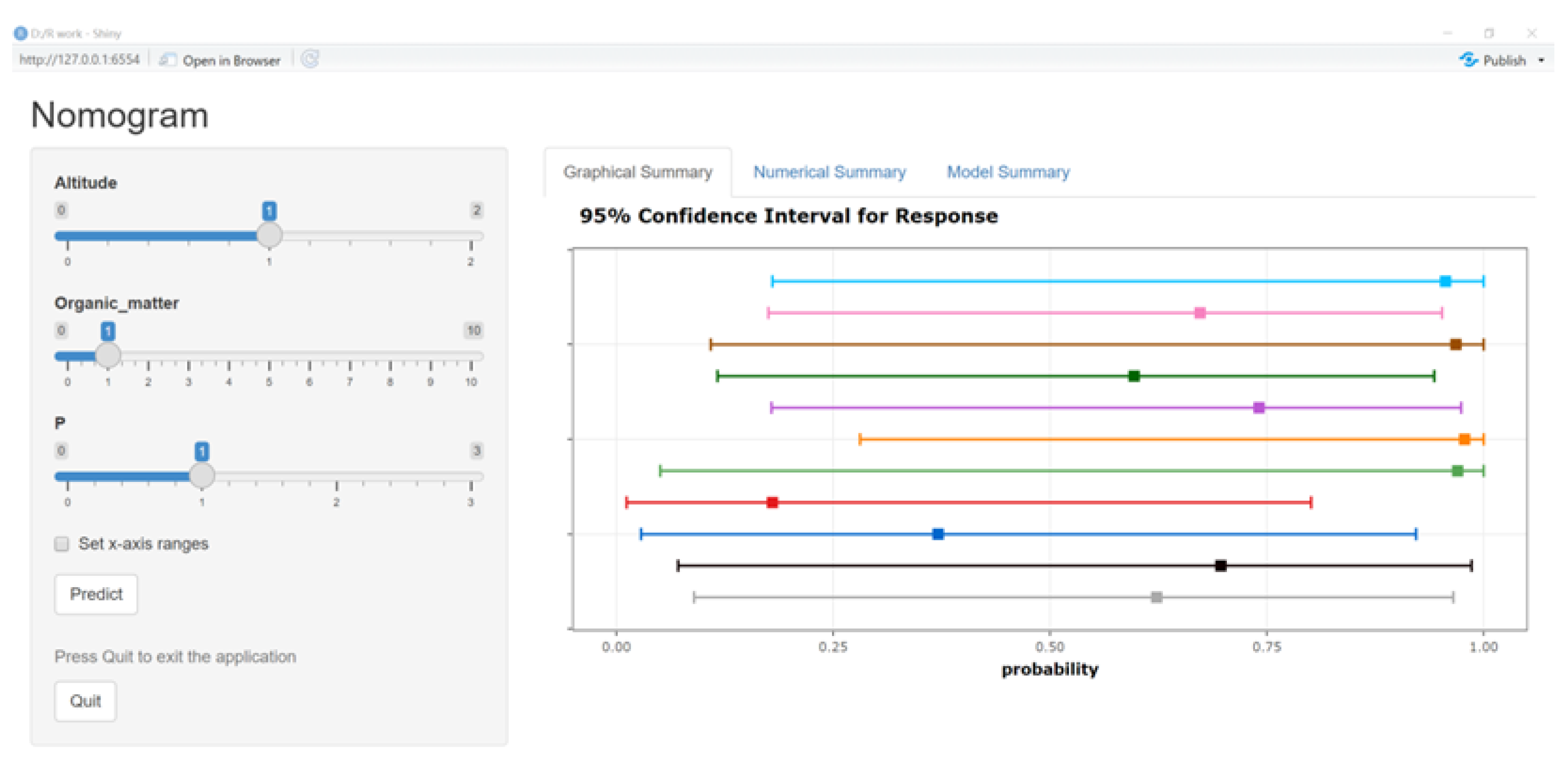
3.5. System Construction and Model Test
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yan, Z.; Zhong, Y.; Duan, Y.; Chen, Q.; Li, F. Antioxidant mechanism of tea polyphenols and its impact on health benefits. Anim. Nutr. 2020, 6, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Q.; Cheng, L.; Zhang, R.; Liu, Y.; Wu, Z.; Zhang, X. Tea Polyphenols Prevent and Intervene in COVID-19 through Intestinal Microbiota. Foods 2022, 11, 506. [Google Scholar] [CrossRef]
- Wang, Y.J.; Jin, G.; Li, L.Q.; Liu, Y.; Kalkhajeh, Y.K.; Ning, J.M.; Zhang, Z.Z. NIR hyperspectral imaging coupled with chemometrics for nondestructive assessment of phosphorus and potassium contents in tea leaves. Infrared Phys. Technol. 2020, 108, 103365. [Google Scholar] [CrossRef]
- Xie, D.; Dai, W.; Lu, M.; Tan, J.; Zhang, Y.; Chen, M.; Lin, Z. Nontargeted metabolomics predicts the storage duration of white teas with 8-C N-ethyl-2-pyrrolidinone-substituted flavan-3-ols as marker compounds. Food Res. Int. 2019, 125, 108635. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Li, H.; Wang, Y.; Fan, K.; Song, Y.; Han, X.; Zhang, J.; Ding, S.; Song, D.; Wang, H.; et al. Prediction of Tea Polyphenols, Free Amino Acids and Caffeine Content in Tea Leaves during Wilting and Fermentation Using Hyperspectral Imaging. Foods 2022, 11, 2537. [Google Scholar] [CrossRef]
- Gaur, N.; Mohanty, B.P. A nomograph to incorporate geophysical heterogeneity in soil moisture downscaling. Water Resour. Res. 2019, 55, 34–54. [Google Scholar] [CrossRef]
- Xie, J.; Shi, D.; Bao, M.; Hu, X.; Wu, W.; Sheng, J.; Xu, K.; Wang, Q.; Wu, J.; Wang, K.; et al. A predictive nomogram for predicting improved clinical outcome probability in patients with COVID-19 in Zhejiang Province, China. Engineering 2022, 8, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, C.T.; Hong, C.Q.; Chu, L.Y.; Huang, X.Y.; Wei, L.-F.; Lin, Y.-R.; Tian, L.-R.; Peng, Y.-H.; Xu, Y.-W. Nomogram based on nutritional and inflammatory indicators for survival prediction of small cell carcinoma of the esophagus. Nutrition 2021, 84, 111086. [Google Scholar] [CrossRef]
- Tan, J.; He, Y.; Li, Z.; Xu, X.; Zhang, Q.; Xu, Q.; Zhang, L.; Xiang, S.; Tang, X.; Zhao, W. Establishment and Validation of a Non-invasive Diagnostic Nomogram to Identify Heart Failure in Patients with Coronary Heart Disease. Front. Cardiovasc. Med. 2022, 9, 826. [Google Scholar] [CrossRef]
- Zhang, K.; Yu, Y.; Dong, J.; Yang, Q.; Xu, X. Adapting & testing use of USLE K factor for agricultural soils in China. Agric. Ecosyst. Environ. 2019, 269, 148–155. [Google Scholar]
- Zhang, S.; Yang, H.; Yang, C.; Yuan, W.; Li, X.; Wang, X.; Zhang, Y.; Cai, X.; Sheng, Y.; Deng, X.; et al. Edge Device Detection of Tea Leaves with One Bud and Two Leaves Based on ShuffleNetv2-YOLOv5-Lite-E. Agronomy 2023, 13, 577. [Google Scholar] [CrossRef]
- Chen, S.; Fu, Y.; Bian, X.; Zhao, M.; Zuo, Y.; Ge, Y.; Zuo, Y.; Ge, Y.; Xiao, Y.; Xiao, J.; et al. Investigation and dynamic profiling of oligopeptides, free amino acids and derivatives during Pu-erh tea fermentation by ultra-high performance liquid chromatography tandem mass spectrometry. Food Chem. 2022, 371, 131176. [Google Scholar] [CrossRef]
- Cheng, L.; Bai, W.H.; Yang, J.J.; Chou, P.; Ning, W.S.; Cai, Q.; Zhou, C.L. Construction and Validation of Mortality Risk Nomograph Model for Severe/Critical Patients with COVID-19. Diagnostics 2022, 12, 2562. [Google Scholar] [CrossRef] [PubMed]
- König, C.; Grensemann, J.; Czorlich, P.; Schlemm, E.; Kluge, S.; Wicha, S.G. A dosing nomograph for cerebrospinal fluid penetration of meropenem applied by continuous infusion in patients with nosocomial ventriculitis. Clin. Microbiol. Infect. 2022, 28, 1022.e9–1022.e16. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, F.; Pérez-Sánchez, H.; Mehridehnavi, A.; Fassihi, A.; Ghasemi, F. Accelerating big data analysis through LASSO-random forest algorithm in QSAR studies. Bioinformatics 2022, 38, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Z.; Feng, C.; Li, C.; Zhang, H.; Li, Z.; Tu, C.; He, S. Identification of cuproptosis-related lncRNA prognostic signature for osteosarcoma. Front. Endocrinol. 2022, 13, 987942. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Ou, X.; Li, M.F.; Wang, M.D.; Huang, F. Risk stratification system and visualized dynamic nomogram constructed for predicting diagnosis and prognosis in rare male breast cancer patients with bone metastases. Front. Endocrinol. 2022, 13, 1013338. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, L.; Pu, S.; Liu, Y.; He, J.; Wang, K. Can we reliably identify the pathological outcomes of neoadjuvant chemotherapy in patients with breast cancer? Development and validation of a logistic regression nomogram based on preoperative factors. Ann. Surg. Oncol. 2021, 28, 2632–2645. [Google Scholar] [CrossRef]
- Zou, J.; Chen, H.; Liu, C.; Cai, Z.; Yang, J.; Zhang, Y.; Li, S.; Lin, H.; Tan, M. Development and validation of a nomogram to predict the 30-day mortality risk of patients with intracerebral hemorrhage. Front. Neurosci. 2022, 16, 942100. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, X.; Liang, S. Abstract WP123: Risk Factors and Nomogram to Predict Intracranial Hemorrhage in Stroke Patients Undergoing Thrombolysis. Stroke 2020, 51 (Suppl. 1), AWP123. [Google Scholar] [CrossRef]
- Sun, X.; Zheng, Z.; Liang, J.; Chen, R.L.; Huang, H.; Yao, X.; Lei, W.; Peng, M.; Cheng, J.; Zhang, N. Development and validation of a simple clinical nomogram for predicting obstructive sleep apnea. J. Sleep Res. 2022, 31, e13546. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.; Cao, Y.; Gui, M.; Lin, J.; Zhang, Q.; Li, Y.; Ji, S.; Bu, B. Dynamic nomogram for predicting generalized conversion in adult-onset ocular myasthenia gravis. Neurol. Sci. 2023, 44, 1383–1391. [Google Scholar] [CrossRef]
- Chen, G.; Lu, M.; Shi, Z.; Xia, S.; Ren, Y.; Liu, Z.; Ren, Y.; Liu, Z.; Liu, X.; Li, Z.; et al. Development and validation of machine learning prediction model based on computed tomography angiography–derived hemodynamics for rupture status of intracranial aneurysms: A Chinese multicenter study. Eur. Radiol. 2020, 30, 5170–5182. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Yang, M.; Han, Z.; Que, Z.; Luo, T.; Tian, J. Establishment of a Nomogram-Based Prognostic Model (LASSO-COX Regression) for Predicting Progression-Free Survival of Primary Non-Small Cell Lung Cancer Patients Treated with Adjuvant Chinese Herbal Medicines Therapy: A Retrospective Study of Case Series. Front. Oncol. 2022, 12, 882278. [Google Scholar] [CrossRef]
- Luo, N.; Li, Y.; Yang, B.; Liu, B.; Dai, Q. Prediction Model for Tea Polyphenol Content with Deep Features Extracted Using 1D and 2D Convolutional Neural Network. Agriculture 2022, 12, 1299. [Google Scholar] [CrossRef]




| Factor Name | B | S.E | Wald | P | Exp(B) | 95% CI for EXP(B) | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| Altitude | 1.673 | 0.680 | 6.050 | 0.014 | 5.326 | 1.405 | 20.191 |
| Arsenic | 0.267 | 0.305 | 0.769 | 0.381 | 1.306 | 0.719 | 2.373 |
| Chrome | −0.082 | 0.235 | 0.123 | 0.726 | 0.921 | 0.582 | 1.459 |
| Lead | −0.281 | 0.169 | 2.779 | 0.096 | 0.755 | 0.542 | 1.051 |
| Nickel | 1.526 | 0.746 | 4.184 | 0.041 | 4.600 | 1.066 | 19.852 |
| Mercury | 1.977 | 1.178 | 2.816 | 0.093 | 7.222 | 0.718 | 72.696 |
| Available Cadmium | 1.884 | 0.898 | 4.398 | 0.036 | 6.579 | 1.131 | 38.265 |
| Available Chromium | 0.509 | 0.461 | 1.215 | 0.270 | 1.663 | 0.673 | 4.109 |
| Available Nickel | 0.056 | 0.138 | 0.165 | 0.684 | 1.058 | 0.807 | 1.386 |
| PH | 0.204 | 0.454 | 0.202 | 0.653 | 1.226 | 0.503 | 2.988 |
| Zn | −0.155 | 0.558 | 0.077 | 0.781 | 0.857 | 0.287 | 2.555 |
| Cu | 0.123 | 0.141 | 0.753 | 0.386 | 1.131 | 0.857 | 1.492 |
| Organic Matter | 0.482 | 0.203 | 5.604 | 0.018 | 1.619 | 1.086 | 2.411 |
| N | 1.146 | 0.483 | 5.638 | 0.018 | 3.147 | 1.222 | 8.107 |
| P | 1.452 | 0.632 | 5.279 | 0.022 | 4.272 | 1.238 | 14.744 |
| K | −0.523 | 0.249 | 4.417 | 0.036 | 0.593 | 0.364 | 0.965 |
| Available Potassium | 1.248 | 0.719 | 3.016 | 0.082 | 3.484 | 0.852 | 14.250 |
| Available Phosphorus | 0.525 | 0.534 | 0.970 | 0.325 | 1.691 | 0.594 | 4.813 |
| Alkaline Hydrolysis Nitrogen | 1.466 | 0.708 | 4.293 | 0.038 | 4.332 | 1.082 | 17.335 |
| Mg | −0.507 | 0.399 | 1.613 | 0.204 | 0.602 | 0.275 | 1.317 |
| Fluoride | −0.134 | 0.519 | 0.067 | 0.796 | 0.875 | 0.316 | 2.419 |
| Cation | 0.111 | 0.471 | 0.055 | 0.814 | 1.117 | 0.443 | 2.814 |
| Factor Name | B | S.E | Wald | P | Exp(B) | 95% CI for EXP(B) | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| Organic Matter | 0.155 | 0.373 | 1.17 | 0.677 | 1.17 | 0.56 | 2.43 |
| P | 1.722 | 1.126 | 5.60 | 0.126 | 5.60 | 0.62 | 50.86 |
| Altitude | 1.888 | 1.428 | 6.61 | 0.186 | 6.61 | 0.4 | 108.52 |
| Altitude (m) | Organic Matter (g/kg) | P (mg/kg) | Tea Polyphenol (mg/kg) | Grade | Correct |
|---|---|---|---|---|---|
| 1690 | 88.9 | 300 | 39.53 | 0.909 | √ |
| 1690 | 104 | 499 | 38.87 | 0.993 | √ |
| 1690 | 97.4 | 411 | 40.06 | 0.995 | √ |
| 1690 | 113 | 490 | 39.42 | 0.991 | √ |
| 1690 | 163 | 429 | 38.28 | 0.983 | √ |
| 1690 | 112 | 499 | 38.4 | 0.991 | √ |
| 1690 | 41.4 | 359 | 42.99 | 0.995 | √ |
| 1590 | 82.6 | 315 | 27.85 | 0.381 | √ |
| 1590 | 43.8 | 353 | 34.91 | 0.697 | √ |
| 1590 | 42.7 | 417 | 38.14 | 0.926 | √ |
| 1590 | 51.6 | 336 | 32.08 | 0.623 | √ |
| 1590 | 42.5 | 394 | 30.42 | 0.697 | √ |
| 1590 | 21 | 42.4 | 24.53 | 0.371 | √ |
| 1590 | 65.5 | 113 | 40.54 | 0.18 | |
| 1640 | 144 | 587 | 38.15 | 0.97 | √ |
| 1640 | 76.2 | 420 | 35.09 | 0.978 | √ |
| 1640 | 55.7 | 214 | 37.42 | 0.741 | √ |
| 1640 | 74.3 | 34.8 | 25.99 | 0.597 | |
| 1640 | 23.5 | 367 | 32.77 | 0.968 | |
| 1640 | 64.6 | 274 | 34.17 | 0.673 | √ |
| 1640 | 49.6 | 366 | 34.18 | 0.956 | √ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Yang, C.; Sheng, Y.; Liu, X.; Yuan, W.; Deng, X.; Li, X.; Huang, W.; Zhang, Y.; Li, L.; et al. A Nomogram Model for Predicting the Polyphenol Content of Pu-Erh Tea. Foods 2023, 12, 2128. https://doi.org/10.3390/foods12112128
Zhang S, Yang C, Sheng Y, Liu X, Yuan W, Deng X, Li X, Huang W, Zhang Y, Li L, et al. A Nomogram Model for Predicting the Polyphenol Content of Pu-Erh Tea. Foods. 2023; 12(11):2128. https://doi.org/10.3390/foods12112128
Chicago/Turabian StyleZhang, Shihao, Chunhua Yang, Yubo Sheng, Xiaohui Liu, Wenxia Yuan, Xiujuan Deng, Xinghui Li, Wei Huang, Yinsong Zhang, Lei Li, and et al. 2023. "A Nomogram Model for Predicting the Polyphenol Content of Pu-Erh Tea" Foods 12, no. 11: 2128. https://doi.org/10.3390/foods12112128
APA StyleZhang, S., Yang, C., Sheng, Y., Liu, X., Yuan, W., Deng, X., Li, X., Huang, W., Zhang, Y., Li, L., Lv, Y., Wang, Y., & Wang, B. (2023). A Nomogram Model for Predicting the Polyphenol Content of Pu-Erh Tea. Foods, 12(11), 2128. https://doi.org/10.3390/foods12112128