Identification of Marker Peptides for the Whey Protein Quantification in Edam-Type Cheese
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Cheese Samples
2.1.2. Protein and Peptide Standards
2.1.3. Reagents
2.2. Methods
2.2.1. Protein Extraction
2.2.2. In-Solution Protein Digestion
2.2.3. Data-Dependent Acquisition Liquid Chromatography-Tandem Mass Spectrometry (DDA-LC-MS/MS)
2.2.4. In-Silico Peptide Identification
2.2.5. Peptide Selection
2.2.6. Multiple Reaction Monitoring Liquid Chromatography-Tandem Mass Spectrometry (MRM-LC-MS/MS)
2.2.7. Evaluation of the MRM-LC-MS/MS Method
2.2.8. Calculation of Enrichment Factors
2.2.9. Determination of α-Lactalbumin and β-Lactoglobulin Content in Cheese via External Calibration
2.2.10. Degree of Hydrolysis of Potential Marker Peptides during Tryptic In-Vitro Hydrolysis of Milk Protein Standard Mixture
2.2.11. Determination of the Recovery Rate by Adding Milk Protein Standard Mixture to Cheese Phosphate Buffer Suspension
2.2.12. Determination of α-Lactalbumin and β-Lactoglobulin Content in Cheese via Internal Calibration
2.2.13. Two-Sample t-Test (Student’s t-Test)
3. Results and Discussion
3.1. Repeatability, Linearity, and Recovery Rate for the Determination of the Potential Marker Peptides
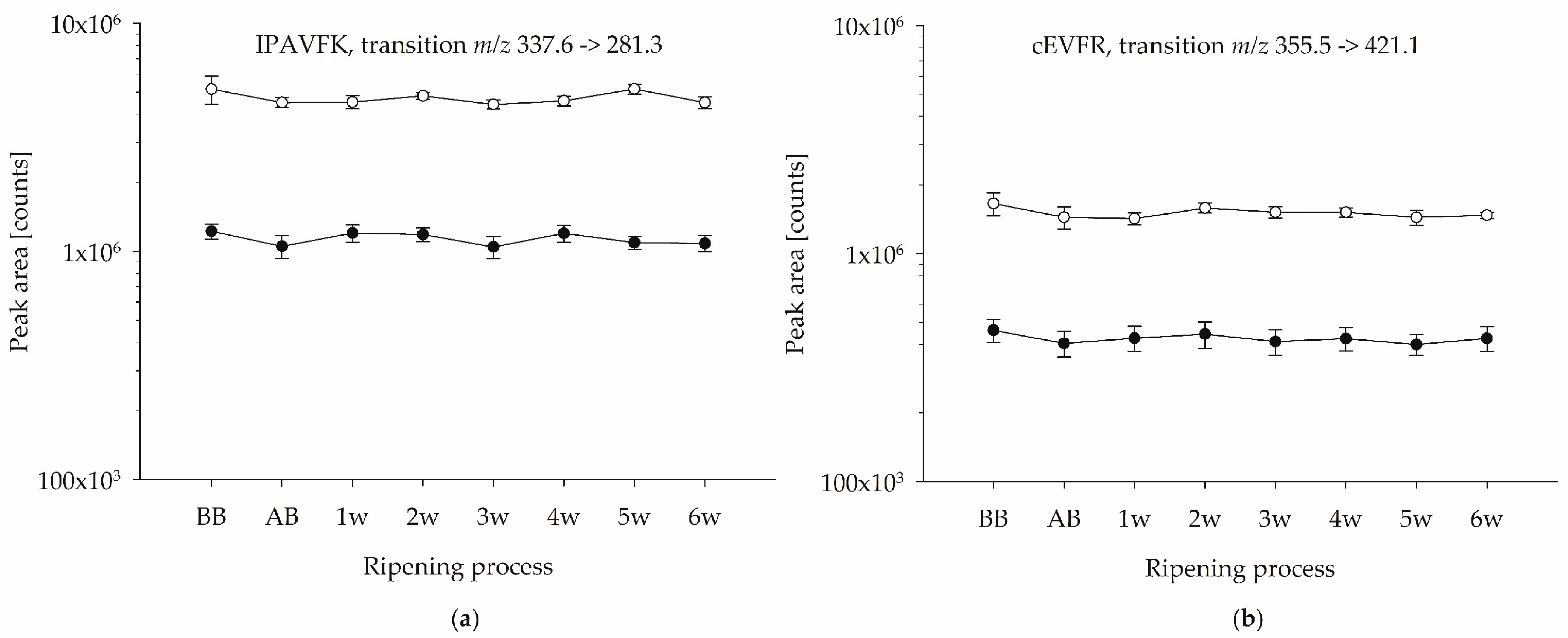
3.2. Stability of Potential Marker Peptides during Cheese Ripening
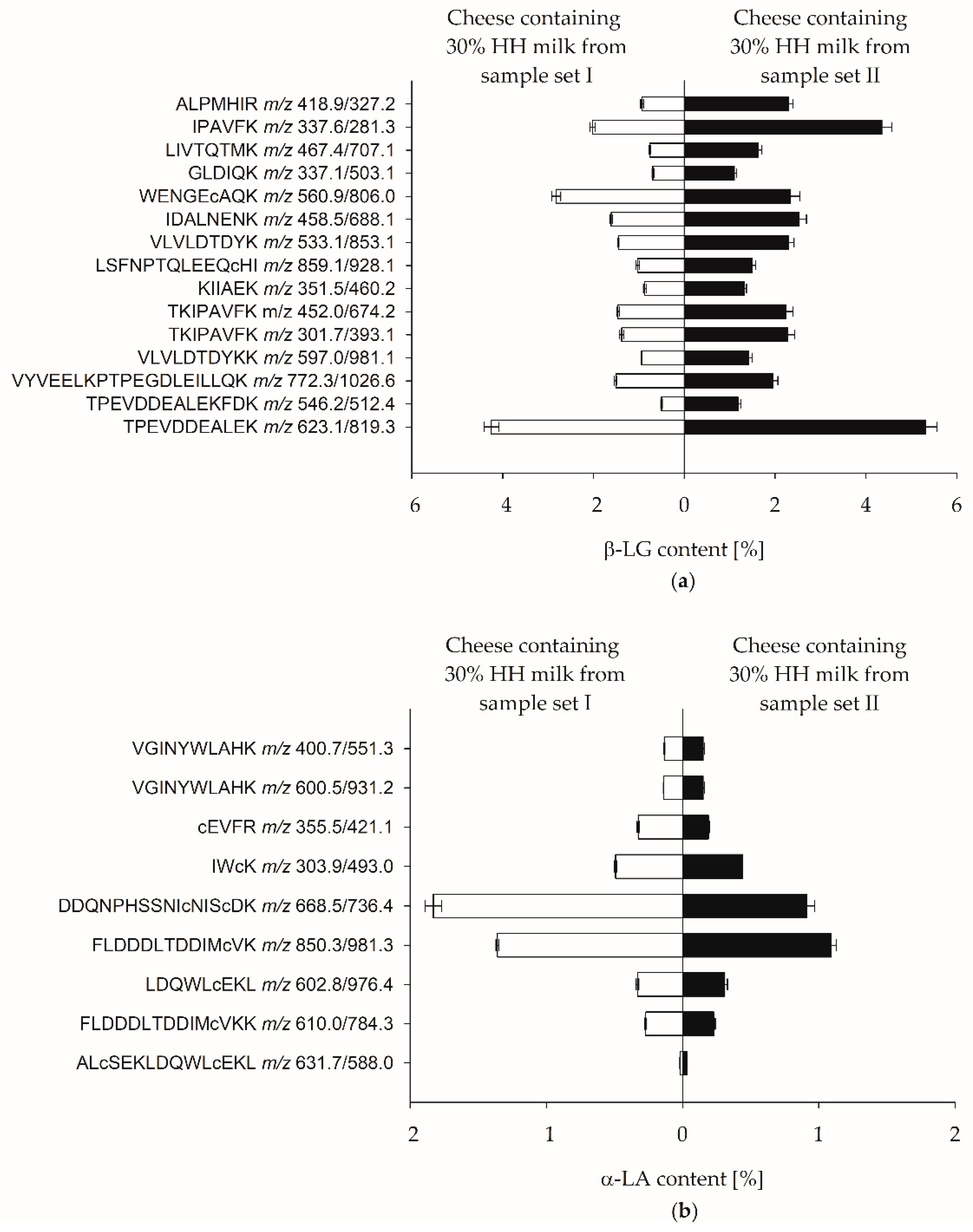
3.3. Enrichment Factors of Individual Whey Proteins
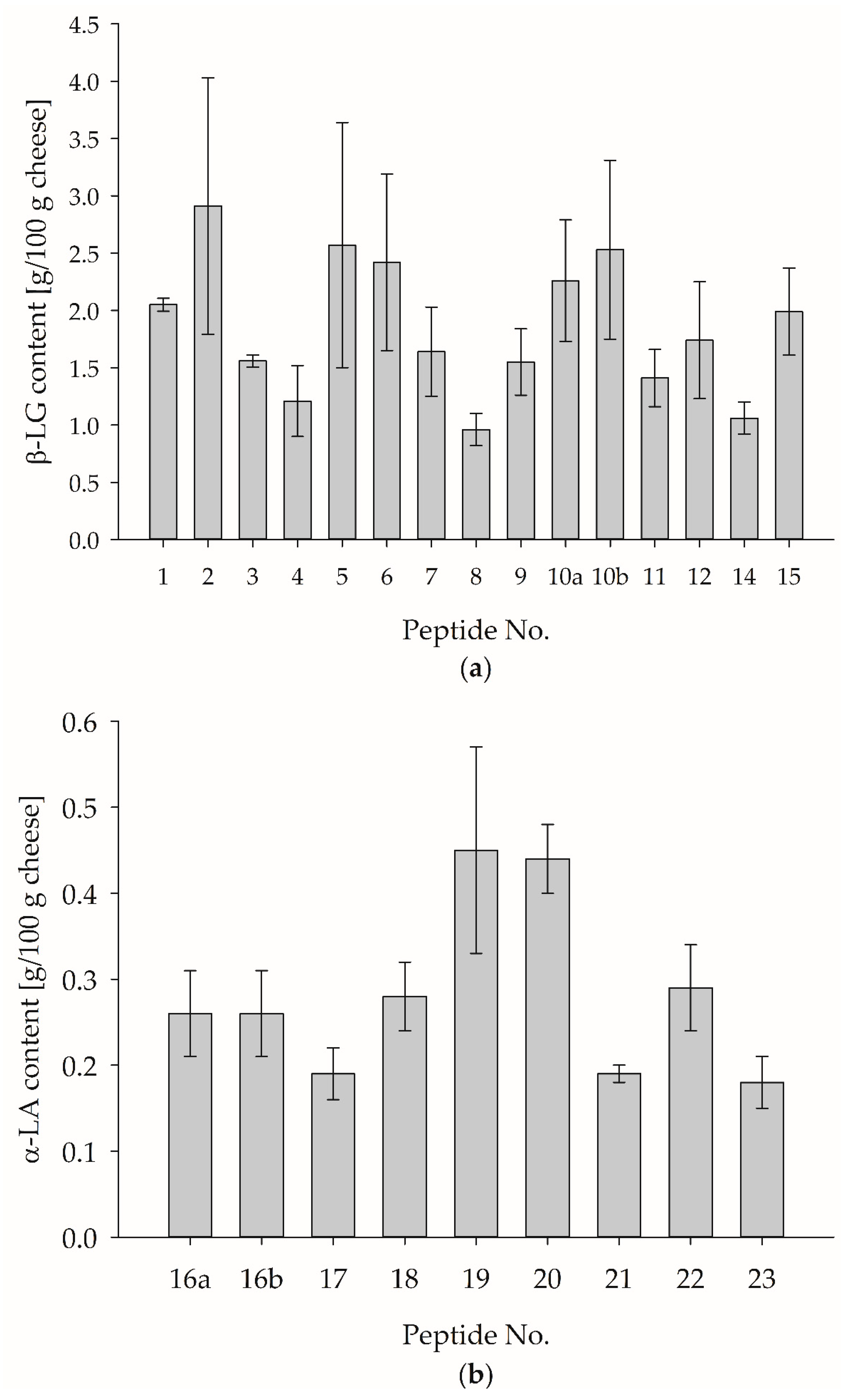
3.4. Quantification of α-Lactalbumin and β-Lactoglobulin in Model Edam-Type Cheese
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sumner, J. Book Review: The World Dairy Situation (2020). Bulletin No. 506/2020 of the International Dairy Federation. Int. J. Dairy Technol. 2021, 74, 778–780. [Google Scholar] [CrossRef]
- McSweeney, P.L.H.; Fox, P.F.; Cotter, P.D.; Everett, D.W. Cheese: Chemistry, Physics and Microbiology, 4th ed.; Elsevier: London, UK, 2017; Volume 1. [Google Scholar]
- De Wit, J.N. Nutritional and Functional Characteristics of Whey Proteins in Food Products. J. Dairy Sci. 1998, 81, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Deeth, H.C.; Bansal, N. Whey Proteins: From Milk to Medicine; Academic Press: London, UK; San Diego, CA, USA; Cambridge, UK; Oxford, UK, 2018. [Google Scholar]
- Hinrichs, J. Incorporation of whey proteins in cheese. Int. Dairy J. 2001, 11, 495–503. [Google Scholar] [CrossRef]
- Guinee, T.P. Effect of high-temperature treatment of milk and whey protein denaturation on the properties of rennet–curd cheese: A review. Int. Dairy J. 2021, 121, 105095. [Google Scholar] [CrossRef]
- Chen, Q.; Ke, X.; Zhang, J.S.; Lai, S.Y.; Fang, F.; Mo, W.M.; Ren, Y.P. Proteomics method to quantify the percentage of cow, goat, and sheep milks in raw materials for dairy products. J. Dairy Sci. 2016, 99, 9483–9492. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.; Harbourne, N.; Oruna-Concha, M.J. Quantification of major camel milk proteins by capillary electrophoresis. Int. Dairy J. 2016, 58, 31–35. [Google Scholar] [CrossRef]
- Schlimme, E.; Clawin-Rädecker, I.; Einhoff, K.; Kiesner, C.; Lorenzen, P.C.; Martin, D.; Meisel, H.; Molkentin, J.; Precht, D. Unterscheidungsmerkmale zur Bewertung der Wärmebehandlung von Milch. Kiel. Milchwirtsch. Forsch. 1996, 48, 5–36. [Google Scholar]
- González de Llano, D.; Santa-María, G. Analysis of whey proteins during ripening of Afuega’l Pitu cheese by reversed phase HPLC Análisis por RP-HPLC de las proteínas del suero del queso Afuega’l Pitu durante la maduración. Food Sci. Technol. Int. 1997, 3, 445–449. [Google Scholar] [CrossRef]
- Bordin, G.; Cordeiro Rapaso, F.; de la Calle, B.; Rodriguez, A.R. Identification and quantification of major bovine milk proteins by liquid chromatography. J. Chromatogr. A 2001, 928, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Caçote, H. Detection and quantification of bovine, ovine and caprine milk percentages in protected denomination of origin cheeses by reversed-phase high-performance liquid chromatography of beta-lactoglobulins. J. Chromatogr. A 2003, 1015, 111–118. [Google Scholar] [CrossRef] [PubMed]
- von Oesen, T.; Treblin, M.; Staudacher, A.; Clawin-Rädecker, I.; Martin, D.; Hoffmann, W.; Schrader, K.; Bode, K.; Zink, R.; Rohn, S.; et al. Determination and evaluation of whey protein content in matured cheese via liquid chromatography. LWT 2023, 174, 114347. [Google Scholar] [CrossRef]
- Sinha, A.; Mann, M. A beginner’s guide to mass spectrometry–based proteomics. Biochemist 2020, 42, 64–69. [Google Scholar] [CrossRef]
- Vincent, D.; Elkins, A.; Condina, M.R.; Ezernieks, V.; Rochfort, S. Quantitation and Identification of Intact Major Milk Proteins for High-Throughput LC-ESI-Q-TOF MS Analyses. PLoS ONE 2016, 11, e0163471. [Google Scholar] [CrossRef]
- Kiser, J.Z.; Post, M.; Wang, B.; Miyagi, M. Streptomyces erythraeus Trypsin for Proteomics Applications. J. Proteome Res. 2009, 8, 1810–1817. [Google Scholar] [CrossRef] [PubMed]
- Cristobal, A.; Marino, F.; Post, H.; van den Toorn, H.W.; Mohammed, S.; Heck, A.J. Toward an Optimized Workflow for Middle-Down proteomics. Anal. Chem. 2017, 89, 3318–3325. [Google Scholar] [CrossRef] [PubMed]
- Addona, T.A.; Abbatiello, S.E.; Schilling, B.; Skates, S.J.; Mani, D.; Bunk, D.M.; Spiegelman, C.H.; Zimmerman, L.J.; Ham, A.-J.L.; Keshishian, H. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring–based measurements of proteins in plasma. Nat. Biotechnol. 2009, 27, 633–641. [Google Scholar] [CrossRef]
- Camerini, S.; Montepeloso, E.; Casella, M.; Crescenzi, M.; Marianella, R.M.; Fuselli, F. Mass spectrometry detection of fraudulent use of cow whey in water buffalo, sheep, or goat Italian ricotta cheese. Food Chem. 2016, 197, 1240–1248. [Google Scholar] [CrossRef]
- Kritikou, A.S.; Aalizadeh, R.; Damalas, D.E.; Barla, I.V.; Baessmann, C.; Thomaidis, N.S. MALDI-TOF-MS integrated workflow for food authenticity investigations: An untargeted protein-based approach for rapid detection of PDO feta cheese adulteration. Food Chem. 2022, 370, 131057. [Google Scholar] [CrossRef]
- Ramachandran, B.; Yang, C.T.; Downs, M.L. Parallel Reaction Monitoring Mass Spectrometry Method for Detection of Both Casein and Whey Milk Allergens from a Baked Food Matrix. J. Proteome Res. 2020, 19, 2964–2976. [Google Scholar] [CrossRef]
- Lutter, P.; Parisod, V.; Weymuth, H. Development and Validation of a Method for the Quantification of Milk Proteins in Food Products Based on Liquid Chromatography with Mass Spectrometric Detection. J. AOAC Int. 2011, 94, 1043–1059. [Google Scholar] [CrossRef]
- Ellingson, D.J.; Shippar, J.J.; Vennard, T.R.; Moloney, C.; O’Connor, D.; O’Regan, J.; McMahon, A.; Affolter, M. Analytical Method for Lactoferrin in Milk-Based Infant Formulas by Signature Peptide Quantification with Ultra-High Performance LC-Tandem Mass Spectrometry. J. AOAC Int. 2019, 102, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liqing, W.; Fei, D.; Bin, Y.; Yi, Y.; Jing, W. Development of an SI-traceable HPLC–isotope dilution mass spectrometry method to quantify β-Lactoglobulin in milk powders. J. Agric. Food Chem. 2014, 62, 3073–3080. [Google Scholar] [CrossRef]
- Le, T.T.; Poulsen, N.A.; Kristiansen, G.H.; Larsen, L.B. Quantitative LC-MS/MS analysis of high-value milk proteins in Danish Holstein cows. Heliyon 2020, 6, e04620. [Google Scholar] [CrossRef]
- Kleinnijenhuis, A.J.; van Gool, M.P.; van Holthoon, F.L.; van den Noort, M.; Huppertz, T. Quantification of bovine α-lactalbumin in infant milk formula using LC-MS. Int. Dairy J. 2021, 113, 104899. [Google Scholar] [CrossRef]
- De Oliveira, L.V.A.; Kleemann, C.R.; Molognoni, L.; Daguer, H.; Hoff, R.B.; Prudencio, E.S. Reference LC-MS/MS method to detect fresh cheeses adulteration with whey. Food Res. Int. 2022, 156, 111140. [Google Scholar] [CrossRef]
- Bär, C.; Mathis, D.; Neuhaus, P.; Dürr, D.; Bisig, W.; Egger, L.; Portmann, R. Protein profile of dairy products: Simultaneous quantification of twenty bovine milk proteins. Int. Dairy J. 2019, 97, 167–175. [Google Scholar] [CrossRef]
- Hao, X.; Fu, L.; Shao, L.; Chen, Q.; Dorus, B.; Cao, X.; Fang, F. Quantification of major milk proteins using ultra-performance liquid chromatography tandem triple quadrupole mass spectrometry and its application in milk authenticity analysis. Food Control 2022, 131, 108455. [Google Scholar] [CrossRef]
- Treblin, M.; von Oesen, T.; Lüneburg, J.; Clawin-Rädecker, I.; Martin, D.; Schrader, K.; Zink, R.; Hoffmann, W.; Fritsche, J.; Rohn, S. High-Performance Thin-Layer Chromatography-Immunostaining as a Technique for the Characterization of Whey Protein Enrichment in Edam Cheese. Foods 2022, 11, 534. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.M., Jr.; Jimenez-Flores, R.; Bleck, G.T.; Brown, E.M.; Butler, J.E.; Creamer, L.K.; Hicks, C.L.; Hollar, C.M.; Ng-Kwai-Hang, K.F.; Swaisgood, H.E. Nomenclature of the Proteins of Cows’ Milk—Sixth Revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef]
- Pierce Biotechnology. Instructions Pierce Trypsin Protease, MS Grade. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0011821_Pierce_Trypsin_Protease_MS_Grade_UG.pdf (accessed on 6 December 2022).
- Rasch, B.; Friese, M.; Hofmann, W.; Naumann, E. Quantitative Methoden 1: Einführung in die Statistik für Psychologen und Sozialwissenschaftler; Springer: Heidelberg, Germany, 2014; Volume 4. [Google Scholar]


| Sample Set | HH Milk in Cheese Milk [%] (w/w) | Replication of Production (n) | Production Stages | |
|---|---|---|---|---|
| Cheese Curd before Ripening | Cheese during Ripening [Weeks] | |||
| I | 0 | 9 | BB, AB | 1, 2, 3, 4, 5, 6 |
| 10 | 3 | – | 6 | |
| 20 | 3 | – | 6 | |
| 30 | 3 | BB, AB | 1, 2, 3, 4, 5, 6 | |
| II | 0 | 1 | BB, AB | 3, 6 |
| 10 | 1 | – | 6 | |
| 20 | 1 | – | 6 | |
| 30 | 1 | BB, AB | 3, 6 | |
| Protein | Concentration in MPSM [mg mL−1] |
|---|---|
| α-LA | 1.08 |
| β-LG (A + B) | 2.89 |
| BSA | 0.32 |
| LF | 0.07 |
| IgG | 0.42 |
| αs-CN | 16.97 |
| β-CN | 9.95 |
| κ-CN | 2.86 |
| Protein | No. | Peptide Sequence of PMP | Rt. [min] | Prec. Mass m/z | Prec. Charge z | Frag. 1 m/z | Frag. 2 m/z | Frag. 3 m/z |
|---|---|---|---|---|---|---|---|---|
| β-LG | 1 | ALPMHIR | 13.3 | 418.9 | 2 | 327.2 | 653.2 | 425.1 |
| 2 | IPAVFK | 16.0 | 337.6 | 2 | 281.3 | 561.2 | 464.2 | |
| 3 | LIVTQTMK | 13.2 | 467.4 | 2 | 707.1 | 608.1 | 227.0 | |
| 4 | GLDIQK | 9.7 | 337.1 | 2 | 503.1 | 170.9 | 388.1 | |
| 5 | WENGEcAQK | 7.6 | 560.9 | 2 | 806.0 | 692.2 | 158.5 | |
| 6 | IDALNENK | 8.9 | 458.5 | 2 | 688.1 | 504.1 | 229.0 | |
| 7 | VLVLDTDYK | 18.0 | 533.1 | 2 | 853.1 | 754.1 | 641.0 | |
| 8 | LSFNPTQLEEQcHI | 23.1 | 859.1 | 2 | 928.1 | 1254.1 | 815.0 | |
| 9 | KIIAEK | 7.0 | 351.5 | 2 | 460.2 | 573.1 | 128.9 | |
| 10a | TKIPAVFK [M + 2H]2+ | 15.7 | 452.0 | 2 | 674.2 | 561.1 | 343.3 | |
| 10b | TKIPAVFK [M + 3H]3+ | 15.7 | 301.7 | 3 | 393.1 | 256.1 | 511.0 | |
| 11 | VLVLDTDYKK | 15.3 | 597.0 | 2 | 981.1 | 882.0 | 491.2 | |
| 12 | VYVEELKPTPEGDLEILLQK | 26.7 | 772.3 | 3 | 1026.6 | 791.1 | 627.9 | |
| 13 | PMHIR | 7.8 | 327.0 | 2 | 425.3 | 229.1 | 556.3 | |
| 14 | TPEVDDEALEKFDK | 17.5 | 546.2 | 3 | 512.4 | 655.0 | 719.7 | |
| 15 | TPEVDDEALEK | 11.6 | 623.1 | 2 | 819.3 | 573.1 | 328.2 | |
| α-LA | 16a | VGINYWLAHK [M + 3H]3+ | 21.2 | 400.7 | 3 | 551.3 | 522.6 | 466.3 |
| 16b | VGINYWLAHK [M + 2H]2+ | 21.2 | 600.5 | 2 | 931.2 | 654.1 | 817.1 | |
| 17 | cEVFR | 9.6 | 355.5 | 2 | 421.1 | 322.0 | 290.0 | |
| 18 | IWcK | 8.6 | 303.9 | 2 | 493.0 | 307.1 | 460.9 | |
| 19 | DDQNPHSSNIcNIScDK | 9.6 | 668.5 | 3 | 736.4 | 896.5 | 766.2 | |
| 20 | FLDDDLTDDIMcVK | 24.8 | 850.3 | 2 | 981.3 | 1439.7 | 1324.5 | |
| 21 | LDQWLcEKL | 22.5 | 602.8 | 2 | 976.4 | 848.4 | 537.2 | |
| 22 | FLDDDLTDDIMcVKK | 23.4 | 610.0 | 3 | 784.3 | 841.0 | 555.4 | |
| 23 | ALcSEKLDQWLcEKL | 24.9 | 631.7 | 3 | 588.0 | 774.8 | 855.1 |
| Peptide No. | Potential Marker Peptide | CV Repeatability of Multiple Tryptic Hydrolyses [%] | CV Repeatability of Multiple Measurements [%] | |||
|---|---|---|---|---|---|---|
| Cheese a | MPSM b | Cheese c | MPSM c | Pepmix d | ||
| 1 | ALPMHIR | 1.97 | 4.49 | 0.43 | 1.09 | 1.24 |
| 2 | IPAVFK | 0.78 | 5.12 | 0.20 | 1.15 | 1.02 |
| 3 | LIVTQTMK | 1.48 | 6.67 | 0.53 | 1.07 | 1.07 |
| 4 | GLDIQK | 2.18 | 5.68 | 0.47 | 1.61 | 1.32 |
| 5 | WENGEcAQK | 2.01 | 8.58 | 1.34 | 2.36 | 2.11 |
| 6 | IDALNENK | 1.70 | 5.70 | 0.40 | 1.62 | 2.07 |
| 7 | VLVLDTDYK | 1.59 | 3.91 | 0.62 | 0.94 | 0.92 |
| 8 | LSFNPTQLEEQcHI | 3.17 | 5.94 | 1.82 | 0.98 | 6.53 |
| 9 | KIIAEK | 3.57 | 2.22 | 1.02 | 1.31 | 1.36 |
| 10a | TKIPAVFK [M + 2H]2+ | 2.67 | 5.77 | 0.57 | 1.07 | 1.70 |
| 10b | TKIPAVFK [M + 3H]3+ | 2.92 | 6.46 | 0.89 | 0.96 | 1.31 |
| 11 | VLVLDTDYKK | 2.54 | 2.99 | 1.20 | 1.50 | 2.58 |
| 12 | VYVEELKPTPEGDLEILLQK | 1.85 | 5.68 | 0.41 | 1.03 | 8.65 |
| 13 | PMHIR | 17.45 | 48.19 | 17.76 | 10.41 | 1.90 |
| 14 | TPEVDDEALEKFDK | 3.40 | 1.62 | 0.43 | 1.03 | 1.71 |
| 15 | TPEVDDEALEK | 2.41 | 2.85 | 0.51 | 0.87 | 1.34 |
| 16a | VGINYWLAHK [M + 3H]3+ | 13.68 | 3.55 | 0.75 | 1.06 | 1.06 |
| 16b | VGINYWLAHK [M + 2H]2+ | 11.08 | 4.53 | 3.28 | 0.80 | 1.77 |
| 17 | cEVFR | 1.94 | 3.16 | 0.80 | 0.92 | 1.75 |
| 18 | IWcK | 2.61 | 5.53 | 0.69 | 1.36 | 1.30 |
| 19 | DDQNPHSSNIcNIScDK | 3.98 | 8.94 | 1.38 | 2.54 | 3.29 |
| 20 | FLDDDLTDDIMcVK | 2.77 | 7.75 | 0.84 | 0.92 | 3.85 |
| 21 | LDQWLcEKL | 3.51 | 4.80 | 0.95 | 1.12 | 1.66 |
| 22 | FLDDDLTDDIMcVKK | 7.25 | 5.69 | 1.21 | 1.37 | 6.56 |
| 23 | ALcSEKLDQWLcEKL | 12.15 | 5.37 | 2.82 | 1.18 | 9.54 |
| Peptide No. | Potential Marker Peptide | RCPeptide Spiked CPBS Hydrolysates [%] | RCPeptide Spiked MPSM Hydrolysates [%] |
|---|---|---|---|
| 1 | ALPMHIR | 99.8 ± 6.5 | 106.6 ± 10.1 |
| 2 | IPAVFK | 103.1 ± 1.4 | 114.5 ± 0.8 |
| 3 | LIVTQTMK | 79.3 ± 1.3 | 78.1 ± 5.0 |
| 4 | GLDIQK | 99.4 ± 3.5 | 119.6 ± 3.4 |
| 5 | WENGEcAQK | 94.5 ± 2.1 | 92.5 ± 1.2 |
| 6 | IDALNENK | 97.6 ± 2.5 | 107.1 ± 2.4 |
| 7 | VLVLDTDYK | 105.2 ± 1.5 | 110.9 ± 3.2 |
| 8 | LSFNPTQLEEQcHI | 131.4 ± 22.1 | 117.2 ± 19.7 |
| 9 | KIIAEK | 97.1 ± 6.1 | 105.2 ± 3.6 |
| 10a | TKIPAVFK [M + 2H]2+ | 95.0 ± 2.0 | 104.6 ± 3.1 |
| 10b | TKIPAVFK [M + 3H]3+ | 92.3 ± 5.2 | 115.9 ± 1.9 |
| 11 | VLVLDTDYKK | 94.7 ± 3.5 | 92.5 ± 9.1 |
| 12 | VYVEELKPTPEGDLEILLQK | n.d. | n.d. |
| 14 | TPEVDDEALEKFDK | 145.5 ± 5.4 | 169.1 ± 7.2 |
| 15 | TPEVDDEALEK | 102.0 ± 2.2 | 112.3 ± 1.9 |
| 16a | VGINYWLAHK [M + 3H]3+ | 127.6 ± 10.0 | 141.9 ± 10.0 |
| 16b | VGINYWLAHK [M + 2H]2+ | 125.2 ± 13.2 | 117.3 ± 12.2 |
| 17 | cEVFR | 70.8 ± 3.5 | 89.5 ± 5.3 |
| 18 | IWcK | 97.6 ± 3.0 | 104.7 ± 2.3 |
| 19 | DDQNPHSSNIcNIScDK | 48.0 ± 2.5 | 53.9 ± 4.9 |
| 20 | FLDDDLTDDIMcVK | 9.5 ± 2.0 | 3.9 ± 2.5 |
| 21 | LDQWLcEKL | 116.0 ± 14.0 | 150.2 ± 12.1 |
| 22 | FLDDDLTDDIMcVKK | n.d. | n.d. |
| 23 | ALcSEKLDQWLcEKL | n.d. | n.d. |
| Cheese | Sample Set | EF of β-LG Based on Potential Marker Peptides a | EF of β-LG Based on Estimated β-LG Content b |
|---|---|---|---|
| Cheese containing 10% HH milk | I | 2.3 ± 0.2 | 1.9 |
| II | 2.0 ± 0.2 | 1.9 | |
| Cheese containing 20% HH milk | I | 3.5 ± 0.4 | 3.0 |
| II | 3.1 ± 0.4 | 2.8 | |
| Cheese containing 30% HH milk | I | 4.8 ± 0.7 | 4.0 |
| II | 4.1 ± 0.6 | 3.7 |
| Cheese | Sample Set | EF of α-LA Based on Potential Marker Peptides a | EF α-LA Based on Estimated α-LA Content b |
|---|---|---|---|
| Cheese containing 10% HH milk | I | 1.8 ± 0.3 | 1.5 |
| II | 1.6 ± 0.1 | 1.4 | |
| Cheese containing 20% HH milk | I | 2.5 ± 0.6 | 2.6 |
| II | 2.1 ± 0.1 | 1.9 | |
| Cheese containing 30% HH milk | I | 3.3 ± 0.9 | 2.7 |
| II | 2.7 ± 0.2 | 2.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Oesen, T.; Treblin, M.; Clawin-Rädecker, I.; Martin, D.; Maul, R.; Hoffmann, W.; Schrader, K.; Wegner, B.; Bode, K.; Zink, R.; et al. Identification of Marker Peptides for the Whey Protein Quantification in Edam-Type Cheese. Foods 2023, 12, 2002. https://doi.org/10.3390/foods12102002
von Oesen T, Treblin M, Clawin-Rädecker I, Martin D, Maul R, Hoffmann W, Schrader K, Wegner B, Bode K, Zink R, et al. Identification of Marker Peptides for the Whey Protein Quantification in Edam-Type Cheese. Foods. 2023; 12(10):2002. https://doi.org/10.3390/foods12102002
Chicago/Turabian Stylevon Oesen, Tobias, Mascha Treblin, Ingrid Clawin-Rädecker, Dierk Martin, Ronald Maul, Wolfgang Hoffmann, Katrin Schrader, Benjamin Wegner, Katja Bode, Ralf Zink, and et al. 2023. "Identification of Marker Peptides for the Whey Protein Quantification in Edam-Type Cheese" Foods 12, no. 10: 2002. https://doi.org/10.3390/foods12102002
APA Stylevon Oesen, T., Treblin, M., Clawin-Rädecker, I., Martin, D., Maul, R., Hoffmann, W., Schrader, K., Wegner, B., Bode, K., Zink, R., Rohn, S., & Fritsche, J. (2023). Identification of Marker Peptides for the Whey Protein Quantification in Edam-Type Cheese. Foods, 12(10), 2002. https://doi.org/10.3390/foods12102002