Comparison of Chemiluminescence Enzyme Immunoassay (Cl-ELISA) with Colorimetric Enzyme Immunoassay (Co-ELISA) for Imidacloprid Detection in Vegetables
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Optimization Parameters of the Assays
2.3. Sample Preparation
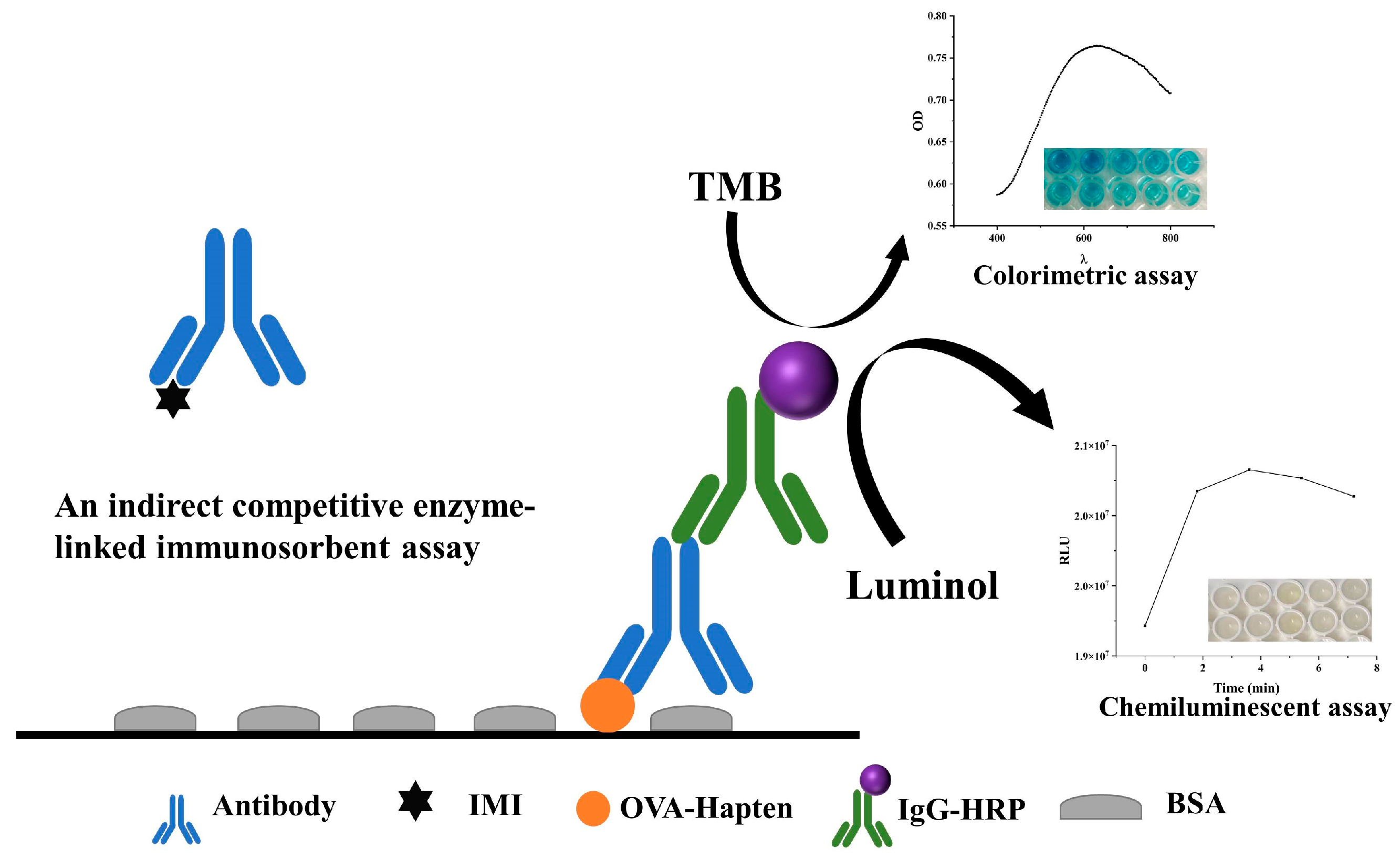
2.4. Co-ELISA Establishment
2.5. Cl-ELISA Establishment
2.6. Calibration Curves
3. Results
3.1. Optimization Parameters of the Assays
3.1.1. Antigen and Antibody Concentrations
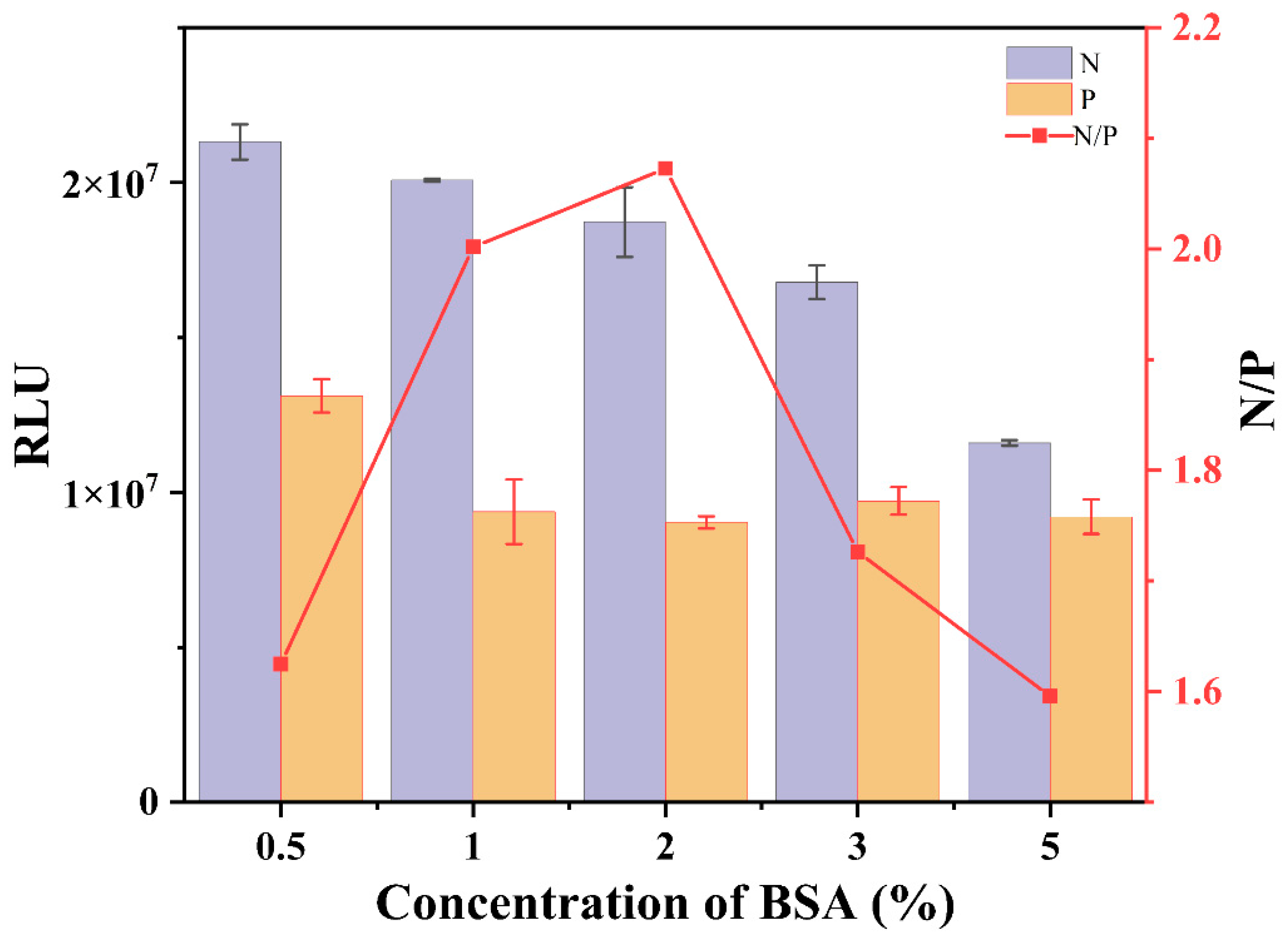
3.1.2. Blocking Agent
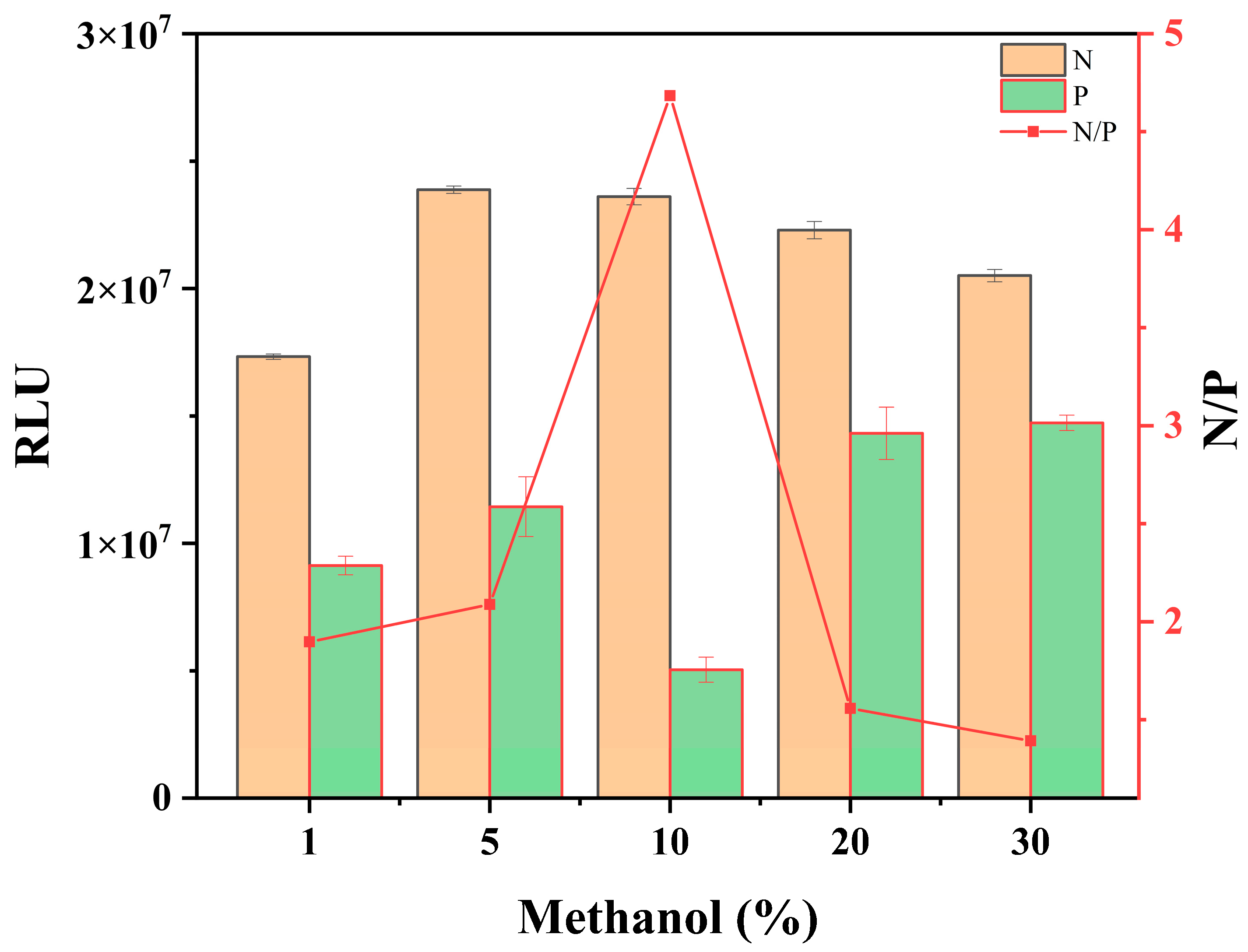
3.1.3. Methanol
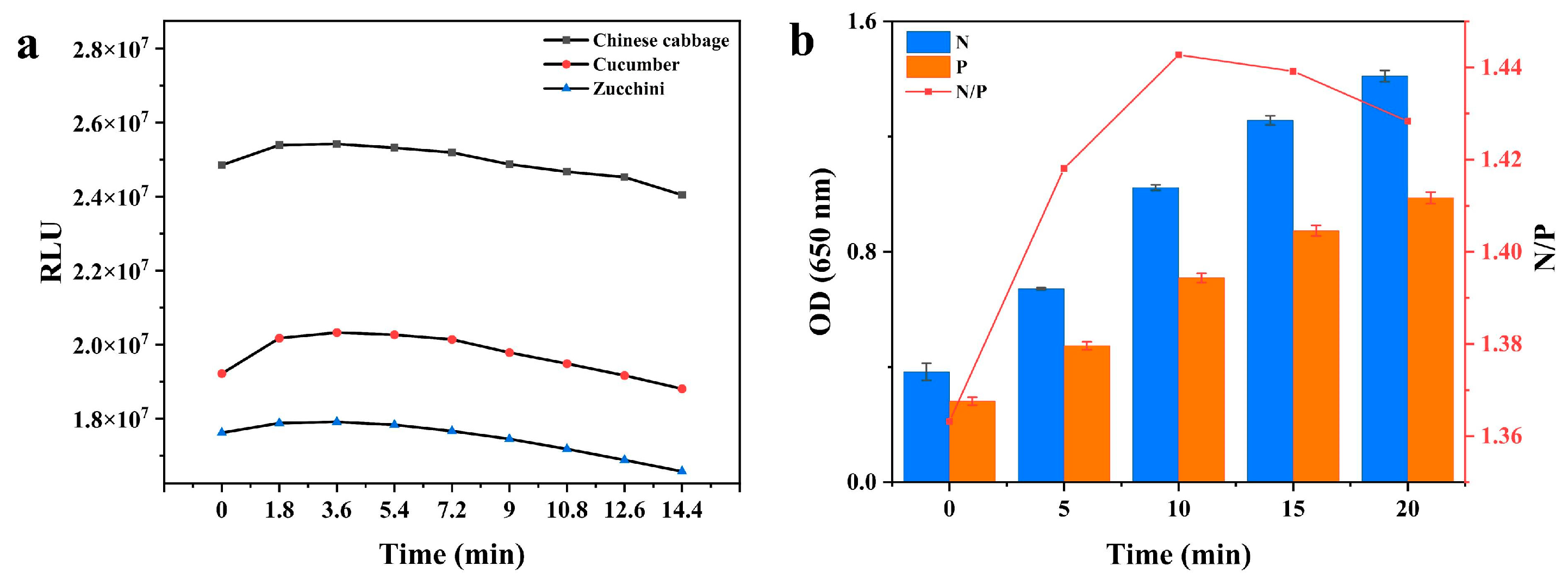
3.1.4. Reaction Time
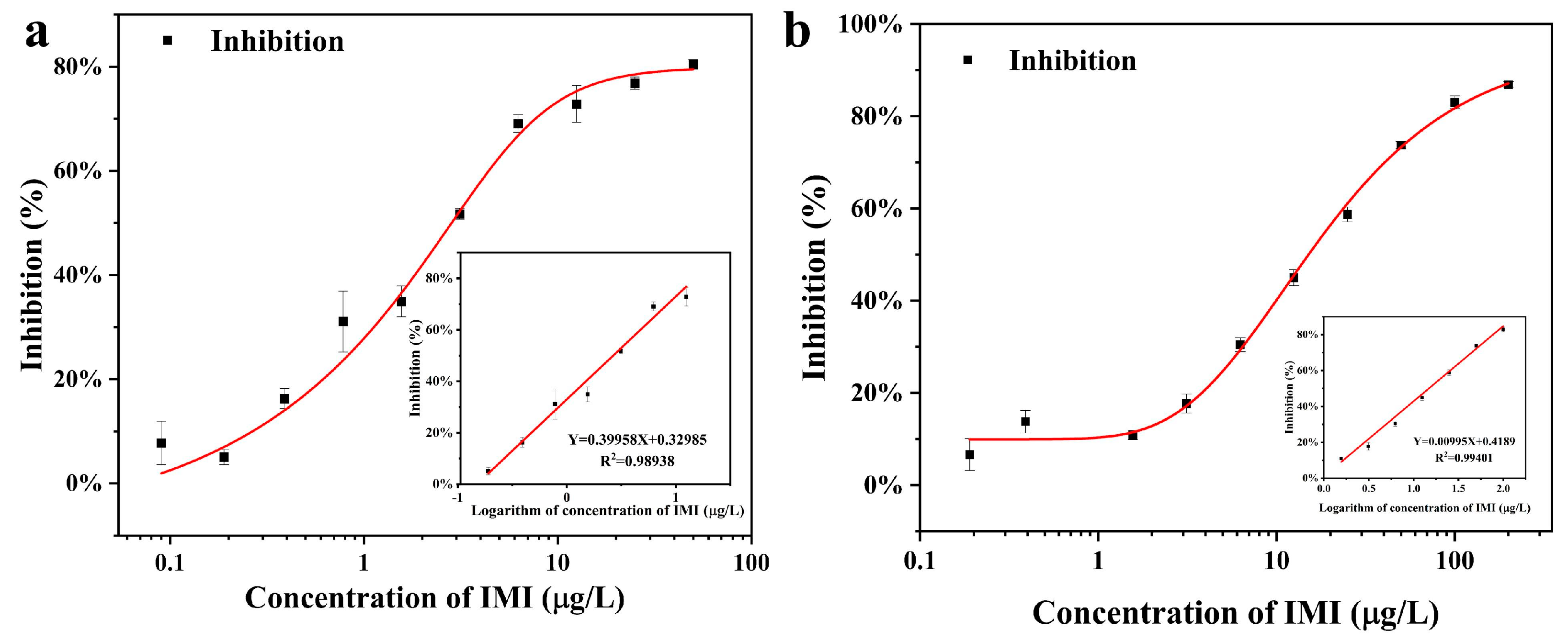
3.2. Calibration Curves
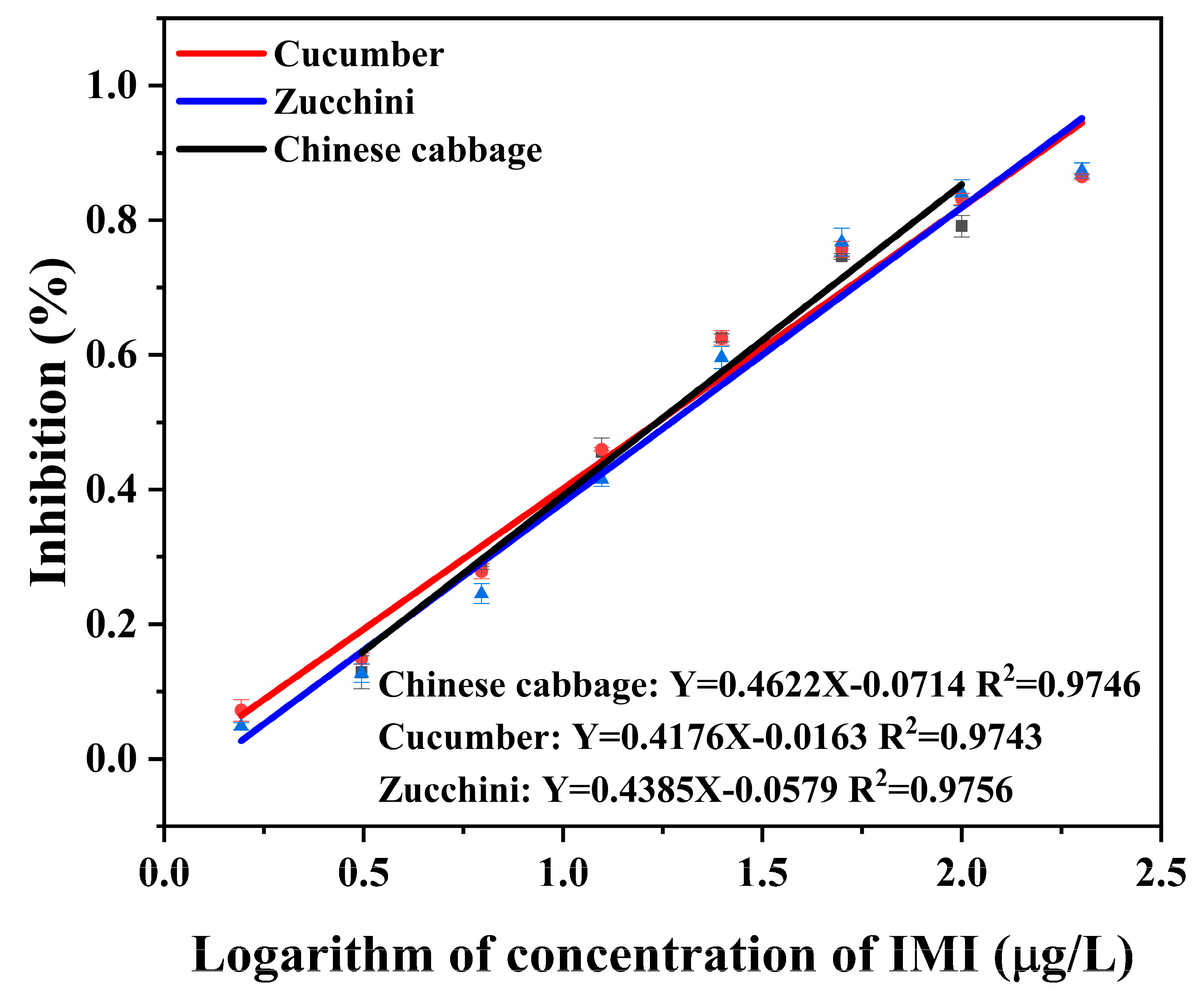
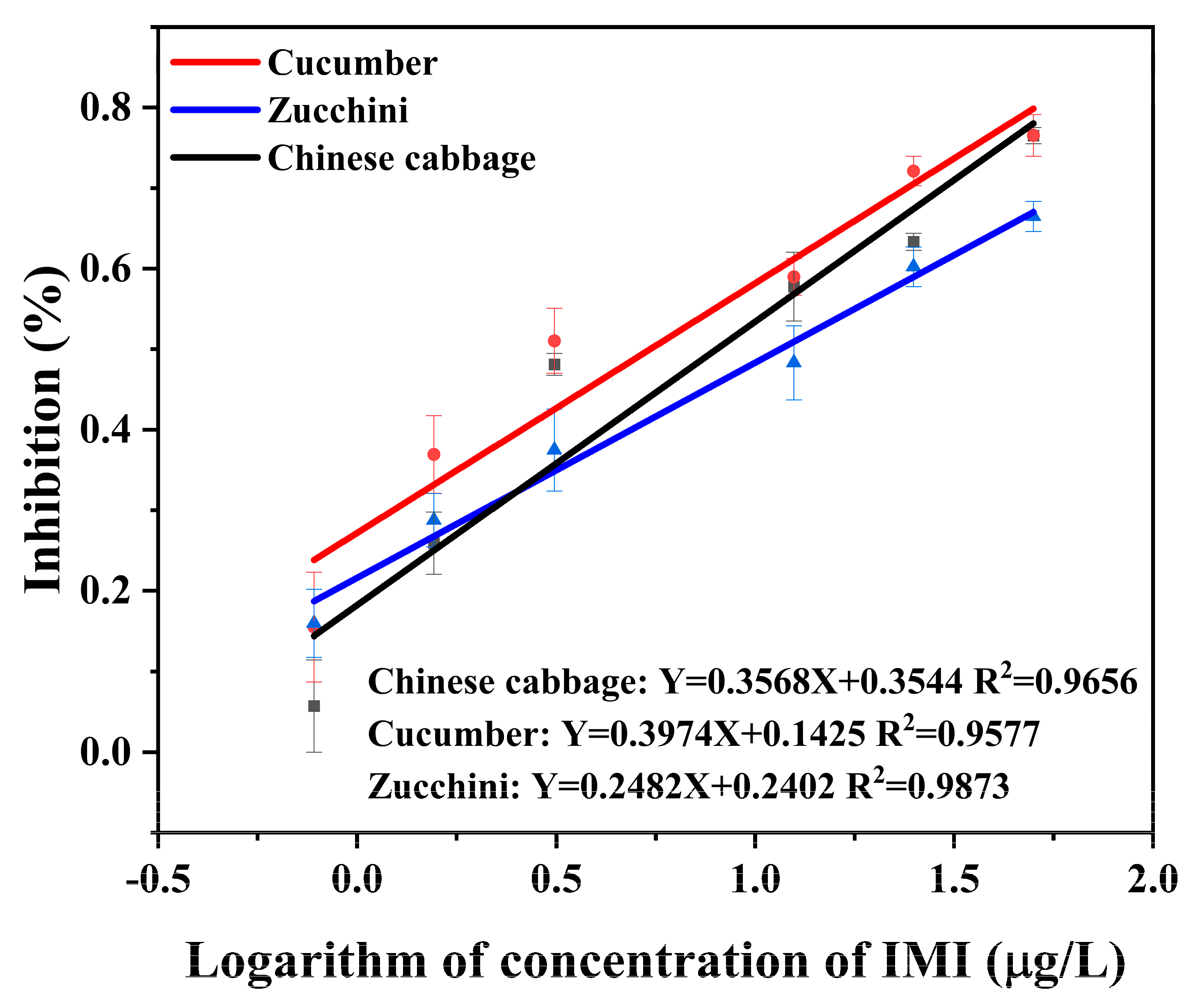
3.3. Optimization of Sample Preparation
3.4. Accuracy and Precision
3.5. Comparison with Other Methods for Detecting IMI
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhai, R.; Zhang, K.; Chen, G.; Liu, G.; Huang, X.; Gao, M.; Zhou, J.; Xu, X.; Li, L.; Zhang, Y.; et al. Residue, Dissipation Pattern, and Dietary Risk Assessment of Imidacloprid in Chinese Chives. Front. Nutr. 2022, 9, 846333. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shen, Q.; Zeng, T.; Li, J.; Li, W.; Wang, Y. Enrichment of imidacloprid and its metabolites in lizards and its toxic effects on gonads. Environ. Pollut. 2020, 258, 113748. [Google Scholar] [CrossRef] [PubMed]
- Seifrtova, M.; Halesova, T.; Sulcova, K.; Riddellova, K.; Erban, T. Distributions of imidacloprid, imidacloprid-olefin and imidacloprid-urea in green plant tissues and roots of rapeseed (Brassica napus) from artificially contaminated potting soil. Pest Manag. Sci. 2017, 73, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, S.; Vicente, V.B.; van Gestel, C.A.M. Long-Term Effects of Imidacloprid, Thiacloprid, and Clothianidin on the Growth and Development of Eisenia andrei. Environ. Toxicol. Chem. 2022, 41, 1686–1695. [Google Scholar] [CrossRef]
- Sriapha, C.; Trakulsrichai, S.; Tongpoo, A.; Pradoo, A.; Rittilert, P.; Wananukul, W. Acute Imidacloprid Poisoning in Thailand. Ther. Clin. Risk Manag. 2020, 16, 1081–1088. [Google Scholar] [CrossRef]
- Du, M.; Yang, Q.; Liu, W.; Ding, Y.; Chen, H.; Hua, X.; Wang, M. Development of immunoassays with high sensitivity for detecting imidacloprid in environment and agro-products using phage-borne peptides. Sci. Total Environ. 2020, 723, 137909. [Google Scholar] [CrossRef] [PubMed]
- Rancan, M.; Sabatini, A.G.; Achilli, G.; Galletti, G.C. Determination of Imidacloprid and metabolites by liquid chromatography with an electrochemical detector and post column photochemical reactor. Anal. Chim. Acta 2006, 555, 20–24. [Google Scholar] [CrossRef]
- Iwafune, T.; Ogino, T.; Watanabe, E. Water-based extraction and liquid chromatography-tandem mass spectrometry analysis of neonicotinoid insecticides and their metabolites in green pepper/tomato samples. J. Agric. Food Chem. 2014, 62, 2790–2796. [Google Scholar] [CrossRef]
- Lodevico, R.G.; Li, Q.X. Determination of Total Imidacloprid Residues in Coffee by Gas Chromatography–Mass Spectrometry. Analytical Letters. 2002, 35, 315–326. [Google Scholar] [CrossRef]
- Zhai, R.; Chen, G.; Liu, G.; Huang, X.; Xu, X.; Li, L.; Zhang, Y.; Wang, J.; Jin, M.; Xu, D.; et al. Enzyme inhibition methods based on Au nanomaterials for rapid detection of organophosphorus pesticides in agricultural and environmental samples: A review. J. Adv. Res. 2022, 37, 61–74. [Google Scholar] [CrossRef]
- Li, H.; Jin, R.; Kong, D.; Zhao, X.; Liu, F.; Yan, X.; Lin, Y.; Lu, G. Switchable fluorescence immunoassay using gold nanoclusters anchored cobalt oxyhydroxide composite for sensitive detection of imidacloprid. Sens. Actuators B Chem. 2019, 283, 207–214. [Google Scholar] [CrossRef]
- Perez-Fernandez, B.; Mercader, J.V.; Checa-Orrego, B.I.; de la Escosura-Muniz, A.; Costa-Garcia, A. A monoclonal antibody-based immunosensor for the electrochemical detection of imidacloprid pesticide. Analyst 2019, 144, 2936–2941. [Google Scholar] [CrossRef] [PubMed]
- Bor, G.; Man, E.; Ugurlu, O.; Ceylan, A.E.; Balaban, S.; Durmus, C.; Pinar Gumus, Z.; Evran, S.; Timur, S. in vitro Selection of Aptamer for Imidacloprid Recognition as Model Analyte and Construction of a Water Analysis Platform. Electroanalysis 2020, 32, 1922–1929. [Google Scholar] [CrossRef]
- Wang, S.H.; Lo, S.C.; Tung, Y.J.; Kuo, C.W.; Tai, Y.H.; Hsieh, S.Y.; Lee, K.L.; Hsiao, S.R.; Sheen, J.F.; Hsu, J.C.; et al. Multichannel nanoplasmonic platform for imidacloprid and fipronil residues rapid screen detection. Biosens. Bioelectron. 2020, 170, 112677. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, C.; Han, K.; Wei, X.; Xu, Y.; Zou, X.; Zhang, H.; Chen, Z. A signal on-off ratiometric electrochemical sensor coupled with a molecular imprinted polymer for selective and stable determination of imidacloprid. Biosens. Bioelectron. 2020, 154, 112091. [Google Scholar] [CrossRef]
- Li, H.; He, S.; Liu, G.; Li, C.; Ma, Z.; Zhang, X. Residue and dissipation kinetics of toosendanin in cabbage, tobacco and soil using IC-ELISA detection. Food Chem. 2021, 335, 127600. [Google Scholar] [CrossRef]
- Watanabe, E.; Miyake, S.; Yogo, Y. Review of enzyme-linked immunosorbent assays (ELISAs) for analyses of neonicotinoid insecticides in agro-environments. J. Agric. Food Chem. 2013, 61, 12459–12472. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Q.; Huang, H.; Zhang, K.; Bai, L.; Tan, G. Ultrasensitive Immunoassay for the Determination of Imidacloprid in Medicinal Herbs. Anal. Lett. 2022, 1–13. [Google Scholar] [CrossRef]
- Watanabe, E.; Baba, K.; Eun, H.; Miyake, S. Application of a commercial immunoassay to the direct determination of insecticide imidacloprid in fruit juices. Food Chem. 2007, 102, 745–750. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Wen, Y.; Shi, J.; Wang, J. Simultaneous detection of parathion and imidacloprid using broad-specificity polyclonal antibody in enzyme-linked immunosorbent assay. Anal. Methods 2015, 7, 205–210. [Google Scholar] [CrossRef]
- Yue, Y.; Chen, J.; Zhang, M.; Yin, Y.; Dong, Y. Determination of Organophosphorus Pesticides in Vegetables and Fruit by an Indirect Competitive Enzyme-Linked Immunosorbent Assay (ic-ELISA) and a Lateral-Flow Immunochromatographic (LFIC) Strip Assay. Anal. Lett. 2022, 55, 1701–1718. [Google Scholar] [CrossRef]
- Girotti, S.; Maiolini, E.; Ghini, S.; Eremin, S.; Mañes, J. Quantification of Imidacloprid in Honeybees: Development of a Chemiluminescent ELISA. Anal. Lett. 2010, 43, 466–475. [Google Scholar] [CrossRef]
- Hu, Y.M.; Feng, H.W.; Liu, S.; Liu, C.; Zhao, P.Y.; Zhang, M.; Zhang, L.; Zhao, J.; Li, J.Z.; Yu, X.M.; et al. The preparation of polyclonal antibody against chlordimeform and establishment of detection by indirect competitive ELISA. J. Environ. Sci. Health B 2022, 57, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.M.; He, S.P.; Zhang, L.; Zhao, H.W.; Zhao, J.; Li, Z.; Li, X.F.; Wang, B.M. Systematic optimization of antibody and coating antigen concentrations in enzyme linked immunosorbent assay checkerboard assay. Chin. J. Anal. Chem. 2008, 36, 1191–1195. [Google Scholar]
- Mukherjee, M.; Nandhini, C.; Bhatt, P. Colorimetric and chemiluminescence based enzyme linked apta-sorbent assay (ELASA) for ochratoxin A detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 244, 118875. [Google Scholar] [CrossRef]
- Zou, R.B.; Liu, Y.; Wang, S.J.; Zhang, Y.; Guo, Y.R.; Zhu, G.N. Development and evaluation of chemiluminescence enzyme-linked immunoassay for residue detection of three organophosphorus pesticides. Chin. J. Pestic. Sci. 2017, 19, 37–45. [Google Scholar]
- Ding, Y.; Hua, X.; Sun, N.; Yang, J.; Deng, J.; Shi, H.; Wang, M. Development of a phage chemiluminescent enzyme immunoassay with high sensitivity for the determination of imidaclothiz in agricultural and environmental samples. Sci. Total Environ. 2017, 609, 854–860. [Google Scholar] [CrossRef]
- Shin, S.; Choi, M.; Shim, J.; Park, S. Hook effect detection and detection-range-controllable one-step immunosensor for inflammation monitoring. Sens. Actuators B Chem. 2020, 304, 127408. [Google Scholar] [CrossRef]
- Song, G.; Huang, L.; Huang, Y.; Liu, W.; Wang, M.; Hua, X. Electrofusion preparation of anti-triazophos monoclonal antibodies for development of an indirect competitive enzyme-linked immunosorbent assay. J. Immunol. Methods 2022, 500, 113184. [Google Scholar] [CrossRef]
- Parra, J.; Mercader, J.V.; Agullo, C.; Abad-Somovilla, A.; Abad-Fuentes, A. Generation of anti-azoxystrobin monoclonal antibodies from regioisomeric haptens functionalized at selected sites and development of indirect competitive immunoassays. Anal. Chim. Acta 2012, 715, 105–112. [Google Scholar] [CrossRef]
- Ma, H.; Xu, Y.; Li, Q.X.; Xu, T.; Wang, X.; Li, J. Application of enzyme-linked immunosorbent assay for quantification of the insecticides imidacloprid and thiamethoxam in honey samples. Food Addit. Contam: Part A. 2009, 26, 713–718. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Wang, K.; Li, W.; Shelver, W.L.; Li, Q.X.; Li, J.; Xu, T. Selection of phage-displayed peptides for the detection of imidacloprid in water and soil. Anal. Biochem. 2015, 485, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.Z.; Shi, X.H.; Cao, Y.C.; Zhou, C.R. Solubility of Imidacloprid in Different Solvents. J. Chem. Eng. Data 2008, 53, 615–618. [Google Scholar] [CrossRef]
- Gui, W.J.; Liu, Y.H.; Wang, C.M.; Liang, X.; Zhu, G.N. Development of a direct competitive enzyme-linked immunosorbent assay for parathion residue in food samples. Anal. Biochem. 2009, 393, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, Q.; Zhang, L.; Ling, S.; Jia, H.; Wu, Y. Dissipation, occurrence, and risk assessment of 12 pesticides in Dendrobium officinale Kimura et Migo. Ecotoxicol. Environ. Saf. 2021, 222, 112487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, M.; Bu, T.; Bai, F.; Zhao, S.; Cao, Y.; He, K.; Wu, H.; Xi, J.; Wang, L. Immunochromatographic Assay based on Sc-TCPP 3D MOF for the rapid detection of imidacloprid in food samples. Food Chem. 2023, 401, 134131. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fernandez, B.; Mercader, J.V.; Abad-Fuentes, A.; Checa-Orrego, B.I.; Costa-Garcia, A.; Escosura-Muniz, A. Direct competitive immunosensor for Imidacloprid pesticide detection on gold nanoparticle-modified electrodes. Talanta 2020, 209, 120465. [Google Scholar] [CrossRef]
- Guo, Y.; Zou, R.; Si, F.; Liang, W.; Zhang, T.; Chang, Y.; Qiao, X.; Zhao, J. A sensitive immunoassay based on fluorescence resonance energy transfer from up-converting nanoparticles and graphene oxide for one-step detection of imidacloprid. Food Chem. 2021, 335, 127609. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, W.Y.; Zhang, W. Simultaneous Determination of Residues of 38 Pesticides in Fruits by QuEChERS Combined with High Performance Liquid Chromatography-Tandem Mass Spectrometry. Food Sci. 2020, 41, 288–296. [Google Scholar]
- Tursen, ·J.; Yang, T.; Bai, L.; Tan, R. Determination of imidacloprid and acetamiprid in grenadine juice by vortex-assisted dispersive liquid-liquid microextraction coupled with HPLC. Chin. J. Anal. Lab. 2019, 38, 815–818. [Google Scholar]
- Xue, Q.H.; Huang, Q.; Sun, X.H. Effects of Spent Mushroom Compost on Degradation of Imidacloprid in Soil and Plant. IOP Conf. Ser. Earth Environ. Sci. 2019, 252, 052042. [Google Scholar] [CrossRef]
- Liu, J.G.; Sun, H.; Cheng, X.; Yang, J.; Hou, Z.G. Determination of Paclobutrazol and Other Four Pesticides in the Vegetable by HPLC-MS/MS. Agrochemicals 2012, 51, 358–363. [Google Scholar]
- Badawy, M.E.I.; Ismail, A.M.E.; Ibrahim, A.I.H. Quantitative analysis of acetamiprid and imidacloprid residues in tomato fruits under greenhouse conditions. J. Environ. Sci. Health B 2019, 54, 898–905. [Google Scholar] [CrossRef] [PubMed]
| Antibody Concentration (mg/L) | OVA-Hapten Concentration (mg/L) | |||
|---|---|---|---|---|
| 10 | 5 | |||
| RLUmax/IC50 | R2 | RLUmax/IC50 | R2 | |
| 4 | 2.15 × 106 | 0.9676 | - | - |
| 2 | 7.56 × 105 | 0.9491 | - | - |
| 1 | 8.47 × 106 | 0.9266 | 1.49 × 106 | 0.9307 |
| 0.5 | 4.19 × 106 | 0.9582 | 6.24 × 106 | 0.9960 |
| 0.1 | - | - | 2.31 × 106 | 0.8743 |
| Antibody Dilution Times | OVA-Hapten Concentration (mg/L) | |||
|---|---|---|---|---|
| 20 | 10 | 5 | 2.5 | |
| 2 | 1.8459 | 1.1196 | 0.7011 | 0.5497 |
| 4 | 1.1879 | 0.8834 | 0.6110 | 0.5007 |
| 8 | 0.7198 | 0.6742 | 0.6787 | 0.5687 |
| 16 | 0.6613 | 0.6390 | 0.5700 | 0.4688 |
| 32 | 0.4950 | 0.6464 | 0.6006 | 0.5197 |
| 64 | 0.5268 | 0.4675 | 0.5115 | 0.5437 |
| 128 | 0.3867 | 0.4048 | 0.4691 | 0.4861 |
| 256 | 0.4395 | 0.4617 | 0.5157 | 0.3846 |
| Parameters | Cl-ELISA | Co-ELISA |
|---|---|---|
| Antigen (mg/L) | 5 | 10 |
| Antibody (mg/L) | 0.5 | 2 |
| Linearity (μg/L) | 0.19–25 | 1.56–200 |
| R2 | 0.9940 | 0.9893 |
| Reaction time (min) | 3.6 | 10 |
| IC50 (μg/L) | 1.56 | 8.15 |
| Method 1 | Method 1 | Method 2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Spiked (μg/L) | Co-ELISA (μg/L) | Recovery (%) | RSD (%) | Spiked (μg/L) | Cl-ELISA (μg/L) | Recovery (%) | RSD (%) | Spiked (μg/L) | Cl-ELISA (μg/L) | Recovery (%) | RSD (%) |
| Chinese cabbage | 10 | 11.35 ± 1.87 | 113.5 | 5.4 | 5 | 1.34 ± 0.46 | 26.8 | 7.5 | 5 | 5.47 ± 0.14 | 109.3 | 4.7 |
| 50 | 58.74 ± 1.94 | 117.5 | 6.2 | 10 | 5.85 ± 1.13 | 58.5 | 5.6 | 10 | 7.03 ± 0.11 | 70.3 | 1.5 | |
| 100 | 81.71 ± 2.02 | 81.7 | 6.9 | 20 | 18.19 ± 0.56 | 91.0 | 1.4 | 20 | 13.94 ± 0.17 | 69.7 | 8.1 | |
| cucumber | 10 | 8.21 ± 1.14 | 82.1 | 0.7 | 5 | 5.06 ± 0.04 | 101.2 | 8.1 | 5 | 4.49 ± 0.69 | 89.7 | 7.9 |
| 50 | 49.11 ± 1.20 | 98.2 | 1.7 | 10 | 7.23 ± 0.09 | 72.3 | 24.0 | 10 | 11.41 ± 0.47 | 114.1 | 1.0 | |
| 100 | 79.38 ± 1.31 | 79.4 | 3.2 | 20 | 9.30 ± 0.60 | 46.5 | 19.5 | 20 | 14.82 ± 0.60 | 74.1 | 5.5 | |
| zucchini | 10 | 11.11 ± 2.05 | 111.1 | 7.9 | 5 | 9.35 ± 0.30 | 187.0 | 43.8 | 5 | 6.07 ± 0.17 | 120.6 | 5.0 |
| 50 | 58.81 ± 2.42 | 117.6 | 11.1 | 10 | 23.74 ± 0.18 | 237.5 | 45.7 | 10 | 6.99 ± 0.43 | 69.9 | 15.0 | |
| 100 | 82.55 ± 2.50 | 82.5 | 11.6 | 20 | 27.10 ± 0.19 | 135.5 | 52.7 | 20 | 19.94 ± 0.24 | 104.9 | 8.8 | |
| Methods | LOD (μg/L) | Recovery (%) | Sample | RSD (%) | Reference |
|---|---|---|---|---|---|
| HPLC—MS/MS | 5.9 | 70.1–119.3 | fruits | 2.7–12.4 | [39] |
| HPLC | 2.7 | 88.9–93.9 | juice | 3.2–4.1 | [40] |
| HPLC | 10 | 83.3–90.8 | soil | 2.4–4.4 | [41] |
| HPLC—MS/MS | 2.1 | 85.9–103.8 | vegetable | 2.6–12.1 | [42] |
| HPLC | 8.53 | 88.9–90.2 | vegetable | 5.5–6.8 | [43] |
| ITS | 0.04 | 80.0–124.0 | vegetable | 0.1–2.7 | [36] |
| Immunoassay (UCNPs/GO) | 0.08 | 76.8–101.8 | Water, vegetable, tea, honey | - | [38] |
| Co-ELISA | 1.56 | 79.4–117.6 | vegetable | 0.7–11.6 | This work |
| Cl-ELISA | 0.19 | 69.7–120.6 | vegetable | 1.0–15.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, R.; Chen, G.; Liu, G.; Huang, X.; Xu, X.; Li, L.; Zhang, Y.; Xu, D.; Abd El-Aty, A.M. Comparison of Chemiluminescence Enzyme Immunoassay (Cl-ELISA) with Colorimetric Enzyme Immunoassay (Co-ELISA) for Imidacloprid Detection in Vegetables. Foods 2023, 12, 196. https://doi.org/10.3390/foods12010196
Zhai R, Chen G, Liu G, Huang X, Xu X, Li L, Zhang Y, Xu D, Abd El-Aty AM. Comparison of Chemiluminescence Enzyme Immunoassay (Cl-ELISA) with Colorimetric Enzyme Immunoassay (Co-ELISA) for Imidacloprid Detection in Vegetables. Foods. 2023; 12(1):196. https://doi.org/10.3390/foods12010196
Chicago/Turabian StyleZhai, Rongqi, Ge Chen, Guangyang Liu, Xiaodong Huang, Xiaomin Xu, Lingyun Li, Yanguo Zhang, Donghui Xu, and A. M. Abd El-Aty. 2023. "Comparison of Chemiluminescence Enzyme Immunoassay (Cl-ELISA) with Colorimetric Enzyme Immunoassay (Co-ELISA) for Imidacloprid Detection in Vegetables" Foods 12, no. 1: 196. https://doi.org/10.3390/foods12010196
APA StyleZhai, R., Chen, G., Liu, G., Huang, X., Xu, X., Li, L., Zhang, Y., Xu, D., & Abd El-Aty, A. M. (2023). Comparison of Chemiluminescence Enzyme Immunoassay (Cl-ELISA) with Colorimetric Enzyme Immunoassay (Co-ELISA) for Imidacloprid Detection in Vegetables. Foods, 12(1), 196. https://doi.org/10.3390/foods12010196










