Malonyl/Acetyltransferase (MAT) Knockout Decreases Triacylglycerol and Medium-Chain Fatty Acid Contents in Goat Mammary Epithelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
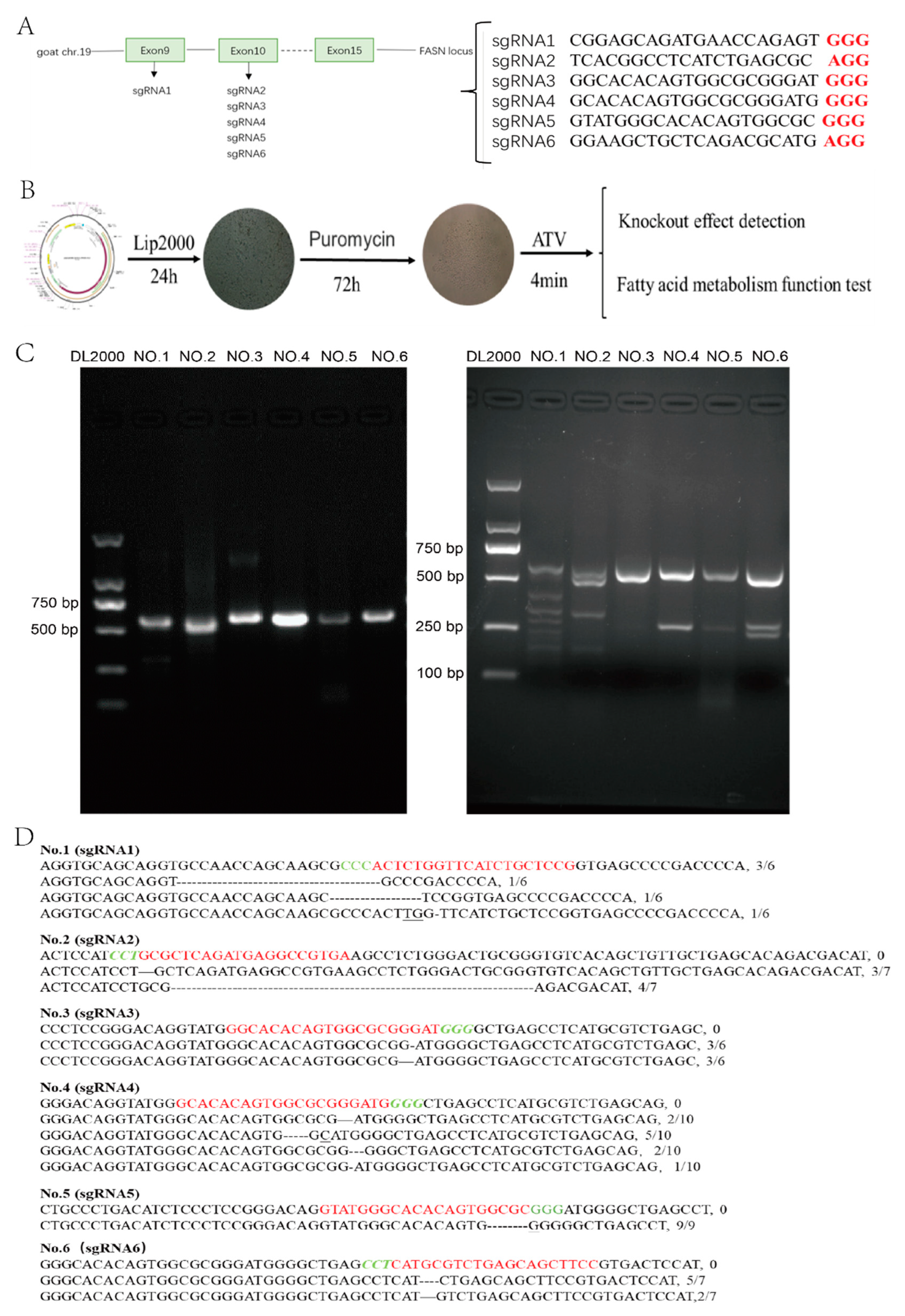
2.2. sgRNA Design and Vector Construction
2.3. Isolation of GMECs
2.4. Cells Culture, Transfection, and Screening
2.5. Cells Culture and T7EN1 Assay
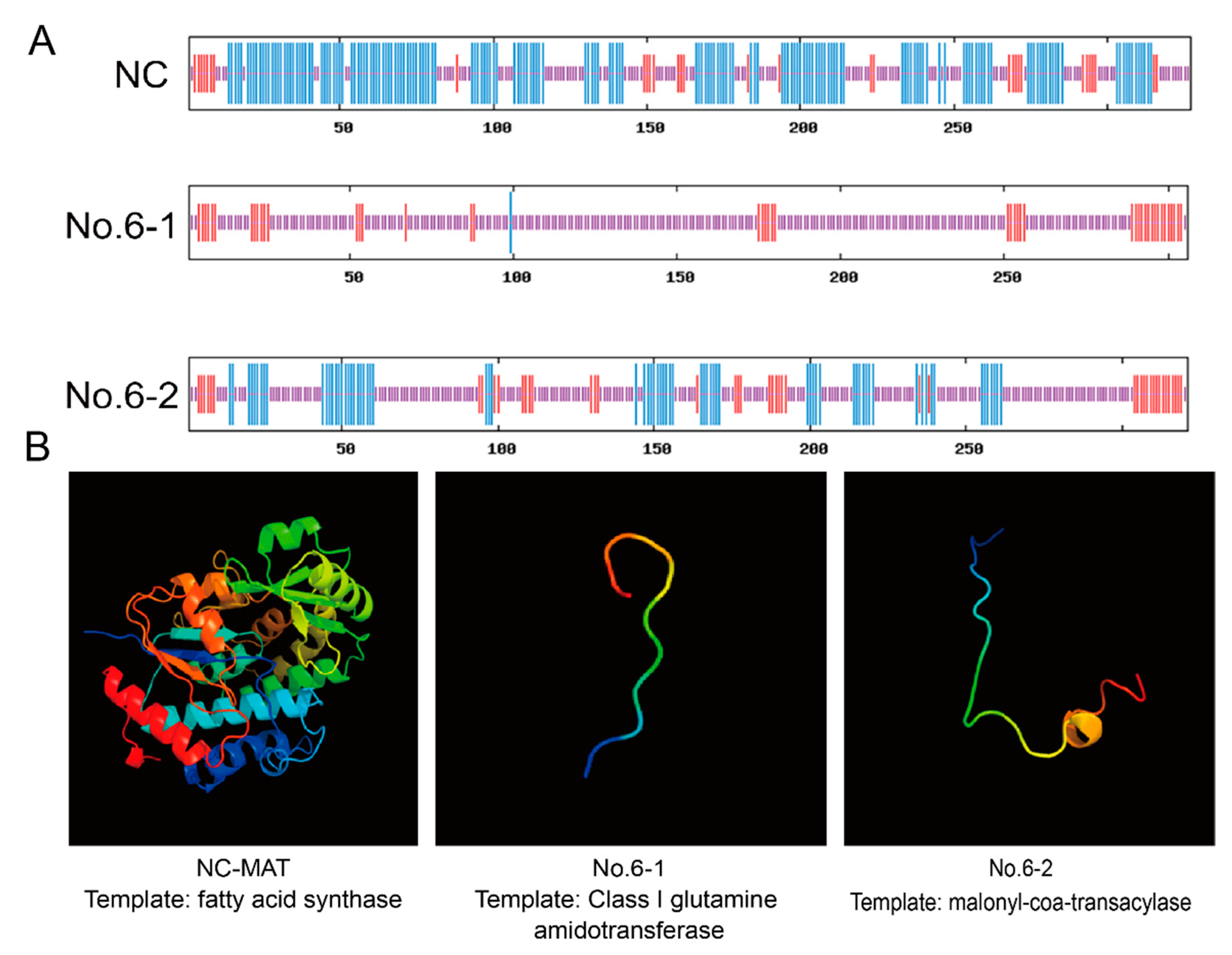
2.6. Genotypic Structure Prediction
2.7. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.8. Protein Extraction and Western Blot Assay
2.9. Off-Target Analysis
2.10. Measurement of Total Cellular Triacylglycerol
2.11. Measurement of Intracellular Fatty Acid Composition
2.12. Measurement of Genes Related to Intracellular Fatty Acid Metabolism
2.13. Measurement of Intracellular Acetyl-CoA
2.14. Measurement of Fatty Acid Metabolism Genes with Acetyl-CoA Addition
2.15. Statistical Analysis
3. Results
3.1. Evaluation of sgRNA for CRISPR-Mediated Repression of MAT
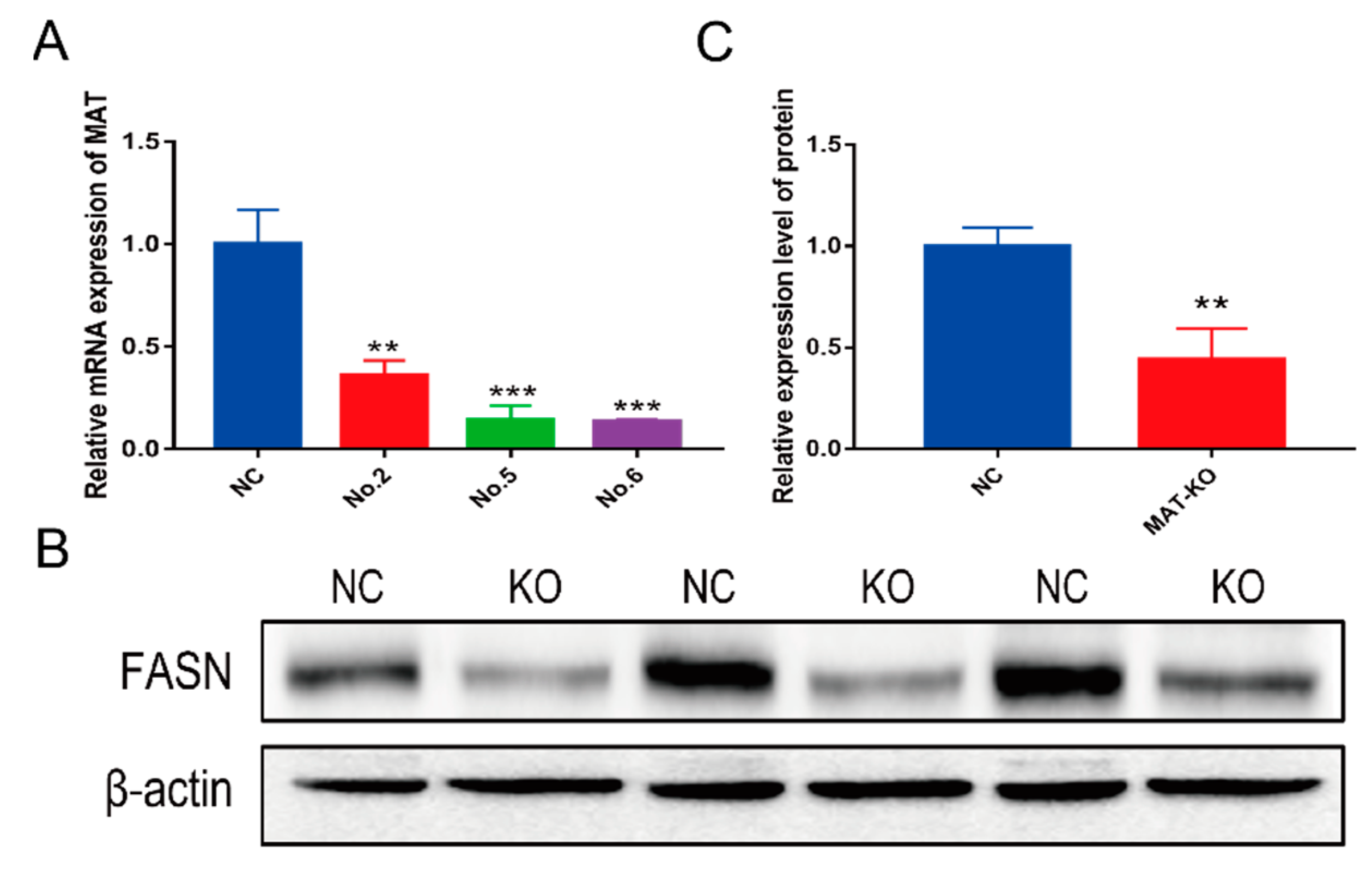
3.2. Decreased Expression of MAT in Genome-Modified GMECs
3.3. The Monoclonal Cells without Off-Target Effects
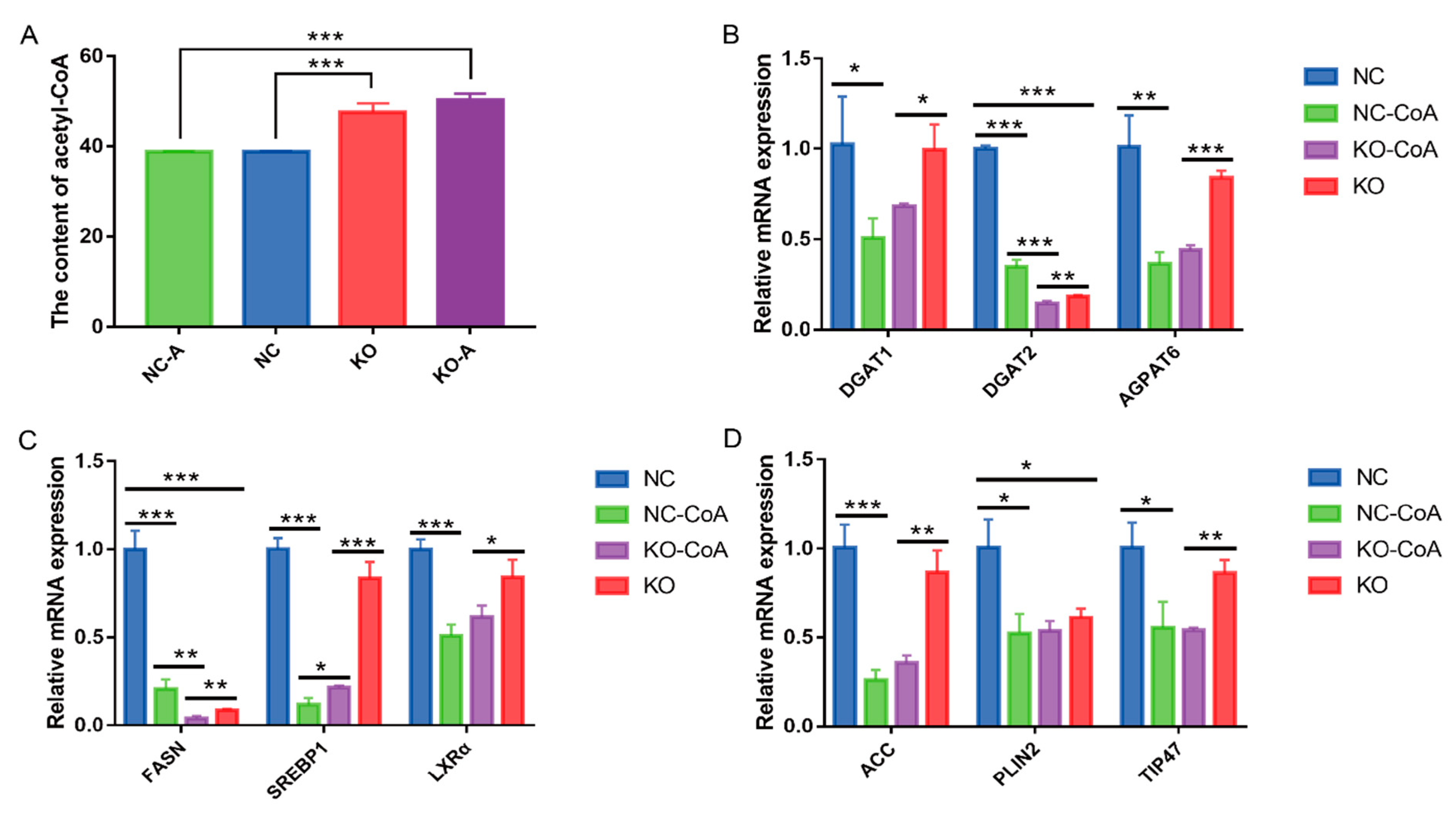
3.4. Knockout of MAT Suppresses Medium-Chain Fatty Acids and Triacylglycerol Synthesis
3.5. Knockout of MAT Decreased the Expression Levels of Genes Related to De Novo Fatty Acid Synthesis
3.6. Acetyl-CoA Suppresses the Fatty Acid Anabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | acetyl-coenzyme A carboxylase alpha |
| AGPAT6 | 1-acylglycerol-3-phosphate O-acyltransferase 6 |
| ATGL | adipose triacylglycerol lipase |
| DGAT1 | diacylglycerol acyl transferase 1 |
| DGAT2 | diacylglycerol acyl transferase 2 |
| FASN | fatty acid synthase |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GPAM | glycerol-3-phosphate acyltransferase 1 |
| LPL | lipoprotein lipase |
| LXRα | liver X receptors alpha |
| MAT | malonyl/acetyltransferase |
| PPARγ | Peroxisome-proliferator-activated receptor gamma |
| SREBP1 | sterol regulatory element-binding transcription protein 1 |
| TIP47 | tail-interacting protein 47 |
| UXT | ubiquitously expressed transcript |
| XDH | Xanthine dehydrogenase |
References
- Zhao, W.; Adjei, M.; Wang, H.; Yangliu, Y.; Zhu, J.; Wu, H. Adipor1 regulates genes involved in milk fat metabolism in goat mammary epithelial cells. Res. Vet. Sci. 2021, 137, 194–200. [Google Scholar] [CrossRef]
- Jorgensen, S.B.; Richter, E.A.; Wojtaszewski, J.F. Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. J. Physiol. 2006, 574 Pt 1, 17–31. [Google Scholar] [CrossRef]
- Prosser, C.G. Compositional and functional characteristics of goat milk and relevance as a base for infant formula. J. Food Sci. 2021, 86, 257–265. [Google Scholar] [CrossRef]
- Li, M.; Lu, X.; Gao, Q.; Wang, M.; Arbab, A.; Sun, Y.; Chen, Z.; Zhang, H.; Karrow, N.A.; Yang, Z.; et al. A Functional 3′ UTR Polymorphism of FADS2 Affects Cow Milk Composition through Modifying Mir-744 Binding. Animals 2019, 9, 1090. [Google Scholar] [CrossRef]
- Yao, D.; Luo, J.; He, Q.; Shi, H.; Li, J.; Wang, H.; Xu, H.; Chen, Z.; Yi, Y.; Loor, J.J. SCD1 Alters Long-Chain Fatty Acid (LCFA) Composition and Its Expression Is Directly Regulated by SREBP-1 and PPARgamma 1 in Dairy Goat Mammary Cells. J. Cell Physiol. 2017, 232, 635–649. [Google Scholar] [CrossRef]
- Wang, H.; Luo, J.; Chen, Z.; Cao, W.T.; Xu, H.F.; Gou, D.M.; Zhu, J.J. MicroRNA-24 can control triacylglycerol synthesis in goat mammary epithelial cells by targeting the fatty acid synthase gene. J. Dairy Sci. 2015, 98, 9001–9014. [Google Scholar] [CrossRef]
- Li, J.; Luo, J.; Xu, H.; Wang, M.; Zhu, J.; Shi, H.; Haile, A.B.; Wang, H.; Sun, Y. Fatty acid synthase promoter: Characterization, and transcriptional regulation by sterol regulatory element binding protein-1 in goat mammary epithelial cells. Gene 2015, 561, 157–164. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Guo, G.; Huo, W.J.; Zhang, S.L.; Pei, C.X.; Zhang, Y.L.; Wang, H. Effects of branched-chain volatile fatty acids on lactation performance and mRNA expression of genes related to fatty acid synthesis in mammary gland of dairy cows. Animals 2018, 12, 2071–2079. [Google Scholar] [CrossRef]
- Mayer, H.K.; Fiechter, G. Physical and chemical characteristics of sheep and goat milk in Austria. Int. Dairy J. 2012, 24, 57–63. [Google Scholar] [CrossRef]
- Schmidely, P.; Andrade, P. Dairy performance and milk fatty acid composition of dairy goats fed high or low concentrate diet in combination with soybeans or canola seed supplementation. Small Rumin. Res. 2011, 99, 135–142. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.; Liu, L.; Da, Z.H.; Zhang, Y.; Hao, C.Y.; Zhu, Q.P. Comparative lipidomics analysis of human, bovine and caprine milk by UHPLC-Q-TOF-MS. Food Chem. 2020, 310, 125865. [Google Scholar] [CrossRef]
- He, Q.; Luo, J.; Wu, J.; Yao, W.; Li, Z.; Wang, H.; Xu, H. FoxO1 Knockdown Promotes Fatty Acid Synthesis via Modulating SREBP1 Activities in the Dairy Goat Mammary Epithelial Cells. J. Agric. Food Chem. 2020, 68, 12067–12078. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, J.; Yi, Y.; Zhang, X.; Loor, J.J.; Cao, Y.; Shi, H.; Luo, J. Akt Serine/Threonine Kinase 1 Regulates de Novo Fatty Acid Synthesis through the Mammalian Target of Rapamycin/Sterol Regulatory Element Binding Protein 1 Axis in Dairy Goat Mammary Epithelial Cells. J. Agric. Food Chem. 2018, 66, 1197–1205. [Google Scholar] [CrossRef]
- Maher, T.; Clegg, M.E. A systematic review and meta-analysis of medium-chain triglycerides effects on acute satiety and food intake. Crit. Rev. Food Sci. Nutr. 2021, 61, 636–648. [Google Scholar] [CrossRef]
- Sung, M.H.; Liao, F.H.; Chien, Y.W. Medium-Chain Triglycerides Lower Blood Lipids and Body Weight in Streptozotocin-Induced Type 2 Diabetes Rats. Nutrients 2018, 10, 963. [Google Scholar] [CrossRef]
- Lee, R.; Corley, M.J.; Pang, A.; Arakaki, G.; Abbott, L.; Nishimoto, M.; Miyamoto, R.; Lee, E.; Yamamoto, S.; Maunakea, A.K.; et al. A modified ketogenic gluten-free diet with MCT improves behavior in children with autism spectrum disorder. Physiol. Behav. 2018, 188, 205–211. [Google Scholar] [CrossRef]
- Strzałkowska, N.; Jóźwik, A.; Bagnicka, E.; Krzyżewski, J.; Horbańczuk, K.; Pyzel, B.; Horbańczuk, J.O. Chemical composition, physical traits and fatty acid profile of goat milk as related to the stage of lactation. Anim. Sci. Pap. Rep. 2009, 27, 311–320. [Google Scholar]
- Rial, S.A.; Karelis, A.D.; Bergeron, K.F.; Mounier, C. Gut Microbiota and Metabolic Health: The Potential Beneficial Effects of a Medium Chain Triglyceride Diet in Obese Individuals. Nutrients 2016, 8, 281. [Google Scholar] [CrossRef]
- Lin, X.; Luo, J.; Zhang, L.; Wang, W.; Gou, D. MiR-103 controls milk fat accumulation in goat (Capra hircus) mammary gland during lactation. PLoS ONE. 2013, 8, e79258. [Google Scholar] [CrossRef]
- Xu, H.; Luo, J.; Tian, H.; Li, J.; Zhang, X.; Chen, Z.; Li, M.; Loor, J.J. Rapid communication: Lipid metabolic gene expression and triacylglycerol accumulation in goat mammary epithelial cells are decreased by inhibition of SREBP-1. J. Anim. Sci. 2018, 96, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.W.; Luo, J.; He, Q.Y.; Wu, M.; Shi, H.B.; Wang, H.; Wang, M.; Xu, H.F.; Loor, J.J. Thyroid hormone responsive (THRSP) promotes the synthesis of medium-chain fatty acids in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 3124–3133. [Google Scholar] [CrossRef] [PubMed]
- Markiewicz-Kęszycka, M.; Czyżak-Runowska, G.; Lipińska, P.; Wójtowski, J. Fatty acid profile of milk-a review. B Vet. I Pulawy. 2013, 57, 135–139. [Google Scholar] [CrossRef]
- Parodi, P.W. Has the association between saturated fatty acids, serum cholesterol and coronary heart disease been over emphasized? Int. Dairy J. 2009, 19, 345–361. [Google Scholar] [CrossRef]
- Fan, Y.; Arbab, A.A.I.; Zhang, H.; Yang, Y.; Lu, X.; Han, Z.; Yang, Z. MicroRNA-193a-5p Regulates the Synthesis of Polyunsaturated Fatty Acids by Targeting Fatty Acid Desaturase 1 (FADS1) in Bovine Mammary Epithelial Cells. Biomolecules 2021, 11, 157. [Google Scholar] [CrossRef]
- Harvatine, K.J.; Boisclair, Y.R.; Bauman, D.E. Liver x receptors stimulate lipogenesis in bovine mammary epithelial cell culture but do not appear to be involved in diet-induced milk fat depression in cows. Physiol. Rep. 2014, 2, e00266. [Google Scholar] [CrossRef]
- Serment, A.; Schmidely, P.; Giger-Reverdin, S.; Chapoutot, P.; Sauvant, D. Effects of the percentage of concentrate on rumen fermentation, nutrient digestibility, plasma metabolites, and milk composition in mid-lactation goats. J. Dairy Sci. 2011, 94, 3960–3972. [Google Scholar] [CrossRef]
- Morris, C.A.; Cullen, N.G.; Glass, B.C.; Hyndman, D.L.; Manley, T.R.; Hickey, S.M.; Lee, M.A. Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm. Genome 2007, 18, 64–74. [Google Scholar] [CrossRef]
- Rudolph, M.C.; Monks, J.; Burns, V.; Phistry, M.; Marians, R.; Foote, M.R.; Bauman, D.E.; Anderson, S.M.; Neville, M.C. Sterol regulatory element binding protein and dietary lipid regulation of fatty acid synthesis in the mammary epithelium. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E918–E927. [Google Scholar] [CrossRef][Green Version]
- Chong, B.M.; Reigan, P.; Mayle-Combs, K.D.; Orlicky, D.J.; Mcmanaman, J.L. Determinants of adipophilin function in milk lipid formation and secretion. Trends Endocrinol. Metab. 2011, 22, 211–217. [Google Scholar] [CrossRef][Green Version]
- Chirala, S.S.; Jayakumar, A.; Gu, Z.W.; Wakil, S.J. Human fatty acid synthase: Role of interdomain in the formation of catalytically active synthase dimer. Proc. Natl. Acad. Sci. USA 2001, 98, 3104–3108. [Google Scholar] [CrossRef]
- Suburu, J.; Shi, L.; Wu, J.; Wang, S.; Samuel, M.; Thomas, M.J.; Kock, N.D.; Yang, G.; Kridel, S.; Chen, Y.Q. Fatty acid synthase is required for mammary gland development and milk production during lactation. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1132–E1143. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.J.; Luo, J.; Sun, Y.T.; Shi, H.B.; Li, J.; Wu, M.; Yu, K.; Haile, A.B.; Loor, J.J. Short communication: Effect of inhibition of fatty acid synthase on triglyceride accumulation and effect on lipid metabolism genes in goat mammary epithelial cells. J. Dairy Sci. 2015, 98, 3485–3491. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Yang, S.; Alim, M.A.; Zhang, Q.; Li, Y.; Liu, L. Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese Holstein cattle population based on a post genome-wide association study. BMC. Genet. 2016, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.B.; Wu, M.; Zhu, J.J.; Zhang, C.H.; Yao, D.W.; Luo, J.; Loor, J.J. Fatty acid elongase 6 plays a role in the synthesis of long-chain fatty acids in goat mammary epithelial cells. J. Dairy Sci. 2017, 100, 4987–4995. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Yoshikawa, T.; Amemiya-Kudo, M.; Hasty, A.H.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 2002, 43, 911–920. [Google Scholar] [CrossRef]
- Lv, Y.; Zhang, S.; Guan, W.; Chen, F.; Zhang, Y.; Chen, J.; Liu, Y. Metabolic transition of milk triacylglycerol synthesis in response to varying levels of palmitate in porcine mammary epithelial cells. Genes Nutr. 2018, 13, 18. [Google Scholar] [CrossRef]
- Zhu, J.J.; Luo, J.; Wang, W.; Yu, K.; Wang, H.B.; Shi, H.B.; Sun, Y.T.; Lin, X.Z.; Li, J. Inhibition of FASN reduces the synthesis of medium-chain fatty acids in goat mammary gland. Animal 2014, 8, 1469–1478. [Google Scholar] [CrossRef]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. Chopchop: A Crispr/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Zhu, J.; Sun, Y.; Luo, J.; Wu, M.; Li, J.; Cao, Y. Specificity protein 1 regulates gene expression related to fatty acid metabolism in goat mammary epithelial cells. Int. J. Mol. Sci. 2015, 16, 1806–1820. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, J.; Wang, W.; Zhao, W.; Lin, X. Characterization and culture of isolated primary dairy goat mammary gland epithelial cells. Sheng Wu Gong Cheng Xue Bao = Chin. J. Biotechnol. 2010, 26, 1123–1127. [Google Scholar]
- Vouillot, L.; Thelie, A.; Pollet, N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 2015, 5, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Cheong, S.A.; Lee, J.G.; Lee, S.W.; Lee, M.S.; Baek, I.J.; Sung, Y.H. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016, 34, 808–810. [Google Scholar] [CrossRef]
- Luo, J.; Zhu, J.J.; Sun, Y.T.; Shi, H.B.; Li, J. Inhibitions of FASN suppress triglyceride synthesis via the control of malonyl-CoA in goat mammary epithelial cells. Anim. Prod. Sci. 2017, 57, 1624–1630. [Google Scholar] [CrossRef]
- Xu, H.F.; Luo, J.; Zhao, W.S.; Yang, Y.C.; Tian, H.B.; Shi, H.B.; Bionaz, M. Overexpression of SREBP1 (sterol regulatory element binding protein 1) promotes de novo fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 783–795. [Google Scholar] [CrossRef]
- Smith, S.; Gagne, H.T.; Pitelka, D.R.; Abraham, S. The effect of dietary fat on lipogenesis in mammary gland and liver from lactating and virgin mice. Biochem. J. 1969, 115, 807–815. [Google Scholar] [CrossRef]
- Strucken, E.M.; Laurenson, Y.C.; Brockmann, G.A. Go with the flow-biology and genetics of the lactation cycle. Front. Genet. 2015, 6, 118. [Google Scholar] [CrossRef]
- Mu, T.; Hu, H.; Ma, Y.; Feng, X.; Zhang, J.; Gu, Y. Regulation of Key Genes for Milk Fat Synthesis in Ruminants. Front. Nutr. 2021, 8, 765147. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Garg, A. Enzymatic activity of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 11: Upregulated in breast and cervical cancers. J. Lipid Res. 2010, 51, 2143–2152. [Google Scholar] [CrossRef]
- Harris, C.A.; Haas, J.T.; Streeper, R.S.; Stone, S.J.; Kumari, M.; Yang, K.; Han, X.; Brownell, N.; Gross, R.W.; Zechner, R.; et al. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 2011, 52, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhang, M.; Xing, Z.; Coleman, D.N.; Liang, Y.; Loor, J.J.; Yang, G. Knockout of butyrophilin subfamily 1 member A1 (BTN1A1) alters lipid droplet formation and phospholipid composition in bovine mammary epithelial cells. J. Anim. Sci. Biotechnol. 2020, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Padhi, E.; Hasegawa, Y.; Larke, J.; Parenti, M.; Wang, A.; Hernell, O.; Lonnerdal, B.; Slupsky, C. Compositional Dynamics of the Milk Fat Globule and Its Role in Infant Development. Front. Pediatr. 2018, 6, 313. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.B.; Yu, K.; Luo, J.; Li, J.; Tian, H.B.; Zhu, J.J.; Sun, Y.T.; Yao, D.W.; Xu, H.F.; Shi, H.P.; et al. Adipocyte differentiation-related protein promotes lipid accumulation in goat mammary epithelial cells. J. Dairy Sci. 2015, 98, 6954–6964. [Google Scholar] [CrossRef]
- Li, J.; Luo, J.; Wang, H.; Shi, H.; Zhu, J.; Sun, Y.; Yu, K.; Yao, D. Adipose triglyceride lipase regulates lipid metabolism in dairy goat mammary epithelial cells. Gene 2015, 554, 125–130. [Google Scholar] [CrossRef]
- Xu, H.F.; Luo, J.; Zhang, X.Y.; Li, J.; Bionaz, M. Activation of liver X receptor promotes fatty acid synthesis in goat mammary epithelial cells via modulation of SREBP1 expression. J. Dairy Sci. 2019, 102, 3544–3555. [Google Scholar] [CrossRef]
- Yao, D.; Yang, C.; Ma, J.; Chen, L.; Luo, J.; Ma, Y.; Loor, J.J. cAMP Response Element Binding Protein 1 (CREB1) Promotes Monounsaturated Fatty Acid Synthesis and Triacylglycerol Accumulation in Goat Mammary Epithelial Cells. Animals 2020, 10, 1871. [Google Scholar] [CrossRef]
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef]
- Wente, S.R.; Rout, M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010, 2, a000562. [Google Scholar] [CrossRef]
- Zaidi, N.; Swinnen, J.V.; Smans, K. ATP-citrate lyase: A key player in cancer metabolism. Cancer Res. 2012, 72, 3709–3714. [Google Scholar]
- Menzies, K.J.; Zhang, H.; Katsyuba, E.; Auwerx, J. Protein acetylation in metabolism—Metabolites and cofactors. Nat. Rev. Endocrinol. 2016, 12, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Sivanand, S.; Viney, I.; Wellen, K.E. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem. Sci. 2018, 43, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Busato, S.; Bionaz, M. The interplay between non-esterified fatty acids and bovine peroxisome proliferator-activated receptors: Results of an in vitro hybrid approach. J. Anim. Sci. Biotechnol. 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Chilliard, Y.; Ferlay, A.; Rouel, J.; Lamberet, G. A review of nutritional and physiological factors affecting goat milk lipid synthesis and lipolysis. J. Dairy Sci. 2003, 86, 1751–1770. [Google Scholar] [CrossRef]


| Fatty Acid | NC | MAT-KO | Fatty Acid | NC | MAT-KO |
|---|---|---|---|---|---|
| C10:0 (%) | 2.73 ± 0.44 | 1.56 ± 0.12 | C18:1 (%) | 10.09 ± 0.40 | 11.75 ± 0.17 * |
| C12:0 (%) | 3.05 ± 0.28 | 1.81 ± 0.04 * | C18:0 (%) | 24.18 ± 0.99 | 24.94 ± 0.12 |
| C14:0 (%) | 10.40 ± 0.47 | 8.48 ± 0.07 * | C20:4 (%) | 0.21 ± 0.02 | 0.29 ± 0.02 |
| C15:0 (%) | 0.88 ± 0.04 | 0.90 ± 0.02 | C22:6 (%) | 0.10 ± 0.02 | 0.10 ± 0.02 |
| C16:1 (%) | 0.24 ± 0.03 | 0.32 ± 0.00 | MCFA (%) | 5.78 ± 0.68 | 3.37 ± 0.13 * |
| C16:0 (%) | 46.19 ± 1.23 | 47.30 ± 0.18 | PUFA (%) | 1.13 ± 0.10 | 1.67 ± 0.82 * |
| C17:1 (%) | 0.15 ± 0.02 | 0.20 ± 0.02 | UFA (%) | 11.61 ± 0.44 | 13.93 ± 0.21 ** |
| C17:0 (%) | 0.96 ± 0.02 | 1.08 ± 0.02 * | SFA/UFA | 7.64 ± 0.32 | 6.18 ± 0.11 * |
| C18:2 (%) | 0.82 ± 0.07 | 1.28 ± 0.05 ** | TAG (mg/g protein) | 285.64 ± 23.30 | 182.97 ± 12.12 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Luo, J.; Tian, H.; Niu, H.; An, X.; Wang, X.; Zang, S. Malonyl/Acetyltransferase (MAT) Knockout Decreases Triacylglycerol and Medium-Chain Fatty Acid Contents in Goat Mammary Epithelial Cells. Foods 2022, 11, 1291. https://doi.org/10.3390/foods11091291
Yao W, Luo J, Tian H, Niu H, An X, Wang X, Zang S. Malonyl/Acetyltransferase (MAT) Knockout Decreases Triacylglycerol and Medium-Chain Fatty Acid Contents in Goat Mammary Epithelial Cells. Foods. 2022; 11(9):1291. https://doi.org/10.3390/foods11091291
Chicago/Turabian StyleYao, Weiwei, Jun Luo, Huibin Tian, Huimin Niu, Xuetong An, Xinpei Wang, and Saige Zang. 2022. "Malonyl/Acetyltransferase (MAT) Knockout Decreases Triacylglycerol and Medium-Chain Fatty Acid Contents in Goat Mammary Epithelial Cells" Foods 11, no. 9: 1291. https://doi.org/10.3390/foods11091291
APA StyleYao, W., Luo, J., Tian, H., Niu, H., An, X., Wang, X., & Zang, S. (2022). Malonyl/Acetyltransferase (MAT) Knockout Decreases Triacylglycerol and Medium-Chain Fatty Acid Contents in Goat Mammary Epithelial Cells. Foods, 11(9), 1291. https://doi.org/10.3390/foods11091291






