Microbial Diversity and Contribution to the Formation of Volatile Compounds during Fine-Flavor Cacao Bean Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Culture Dependant Microbiological Analysis
2.3. DNA Extraction from Isolates
2.4. PCR and Sequencing of Microbial Isolates
2.5. Culture-Independent Analysis
2.6. Metabarcoding Sequencing
2.7. Bioinformatics Analysis
2.8. Laboratory-Scale Fermentation
2.9. Analysis of Volatile Compounds
2.10. Statistical Analysis
3. Results
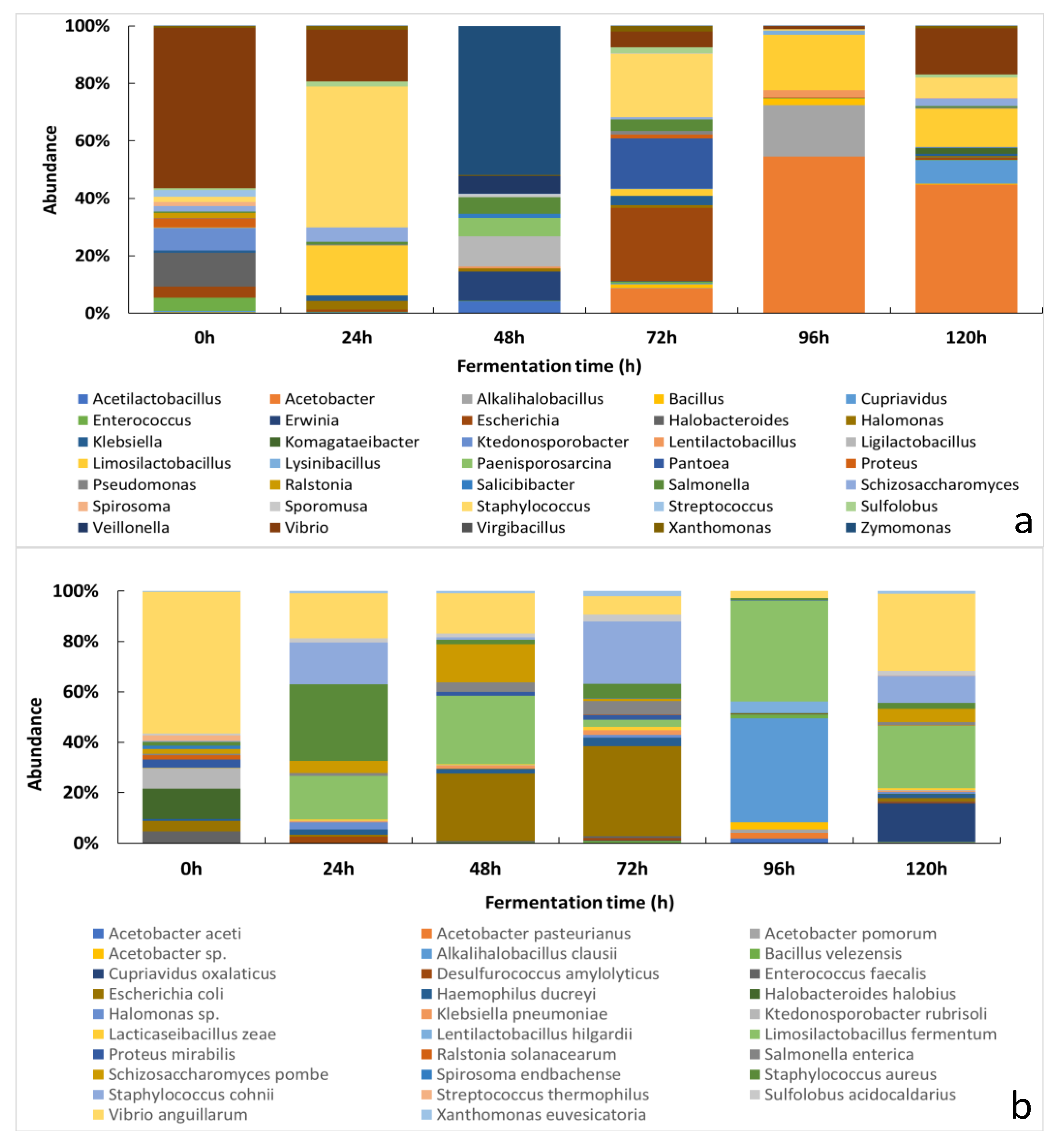
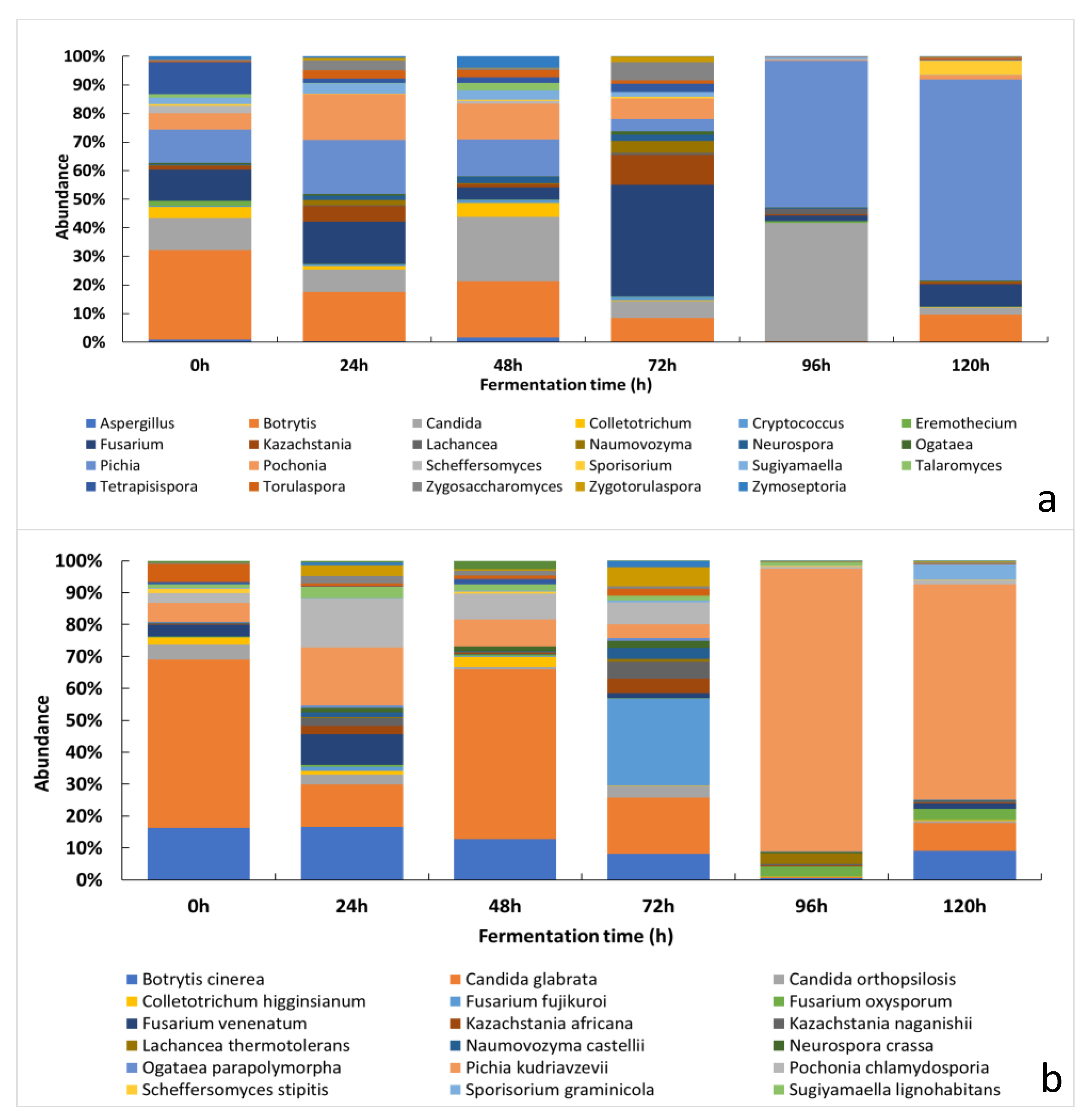
3.1. Culture-Dependent Analysis
3.2. Culture-Independent Analysis
3.2.1. Changes of the Relative Abundance of Bacterial Taxa across the Fermentation Period
3.2.2. Changes of the Relative Abundance of Fungal Taxa across the Fermentation Period
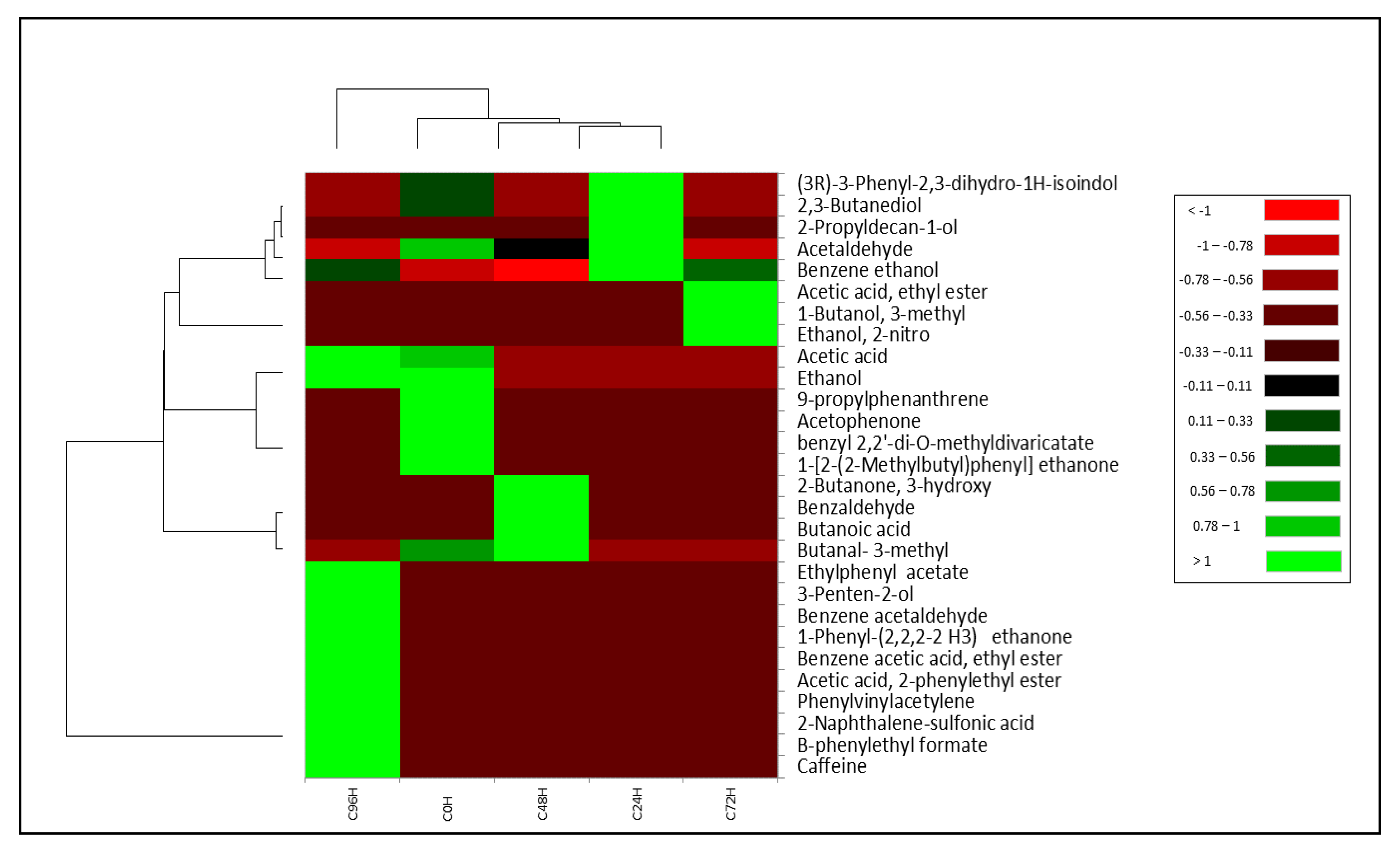
3.3. Volatile Profile
3.4. Laboratory-Scale Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwan, R.F.; De Melo Pereira, G.V.; Fleet, G.H. Microbial Activities during Cocoa Fermentation. In Cocoa and Coffee Fermentation; CRC Press: Boca Raton, FL, USA, 2014; pp. 129–192. [Google Scholar]
- ICCO International Cocoa Organization. Fine or Flavour Cocoa. Available online: https://www.icco.org/fine-or-flavor-cocoa/ (accessed on 29 March 2021).
- Cevallos-Cevallos, J.M.; Gysel, L.; Maridueña-Zavala, M.G.; Molina-Miranda, M.J. Time-Related Changes in Volatile Compounds during Fermentation of Bulk and Fine-Flavor Cocoa (Theobroma cacao) Beans. J. Food Qual. 2018, 2018, 1758381. [Google Scholar] [CrossRef] [Green Version]
- Rottiers, H.; Tzompa Sosa, D.A.; De Winne, A.; Ruales, J.; De Clippeleer, J.; De Leersnyder, I.; De Wever, J.; Everaert, H.; Messens, K.; Dewettinck, K. Dynamics of Volatile Compounds and Flavor Precursors during Spontaneous Fermentation of Fine Flavor Trinitario Cocoa Beans. Eur. Food Res. Technol. 2019, 245, 1917–1937. [Google Scholar] [CrossRef]
- Samaniego, I.; Espín, S.; Quiroz, J.; Ortiz, B.; Carrillo, W.; García-Viguera, C.; Mena, P. Effect of the Growing Area on the Methylxanthines and Flavan-3-Ols Content in Cocoa Beans from Ecuador. J. Food Compos. Anal. 2020, 88, 103448. [Google Scholar] [CrossRef]
- Loor, R.G.; Risterucci, A.M.; Courtois, B.; Fouet, O.; Jeanneau, M.; Rosenquist, E.; Amores, F.; Vasco, A.; Medina, M.; Lanaud, C. Tracing the Native Ancestors of the Modern Theobroma cacao L. Population in Ecuador. Tree Genet. Genomes 2009, 5, 421–433. [Google Scholar] [CrossRef]
- Muñoz, M.S.; Rodríguez Cortina, J.; Vaillant, F.E.; Escobar Parra, S. An Overview of the Physical and Biochemical Transformation of Cocoa Seeds to Beans and to Chocolate: Flavor Formation. Crit. Rev. Food Sci. Nutr. 2020, 60, 1593–1613. [Google Scholar] [CrossRef]
- Nieburg, O. Everything You Need to Know about Fine Flavor Cocoa. Available online: https://www.confectionerynews.com/Article/2016/05/10/Everything-you-need-to-know-about-fine-flavor-cocoa (accessed on 12 January 2021).
- De Vuyst, L.; Weckx, S. The Cocoa Bean Fermentation Process: From Ecosystem Analysis to Starter Culture Development. J. Appl. Microbiol. 2016, 121, 5–17. [Google Scholar] [CrossRef]
- Caligiani, A.; Marseglia, A.; Prandi, B.; Palla, G.; Sforza, S. Influence of Fermentation Level and Geographical Origin on Cocoa Bean Oligopeptide Pattern. Food Chem. 2016, 211, 431–439. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; Falony, G.; Romanens, E.; Jimenez, J.C.; Amores, F.; Daniel, H.M.; De Vuyst, L. Species Diversity, Community Dynamics, and Metabolite Kinetics of the Microbiota Associated with Traditional Ecuadorian Spontaneous Cocoa Bean Fermentations. Appl. Environ. Microbiol. 2011, 77, 7698–7714. [Google Scholar] [CrossRef] [Green Version]
- De Vuyst, L.; Leroy, F. Functional Role of Yeasts, Lactic Acid Bacteria and Acetic Acid Bacteria in Cocoa Fermentation Processes. FEMS Microbiol. Rev. 2020, 44, 432–453. [Google Scholar] [CrossRef]
- Geisen, S.; Vaulot, D.; Mahé, F.; Lara, E.; De Vargas, C.; Bass, D. A User Guide to Environmental Protistology: Primers, Metabarcoding, Sequencing, and Analyses. bioRxiv 2019. [Google Scholar] [CrossRef]
- Carolina, C.O.; Vaz, A.B.M.; De Castro, G.M.; Lobo, F.; Solar, R.; Rodrigues, C.; Martins Pinto, L.R.; Vandenberghe, L.; Pereira, G.; Miúra da Costa, A.; et al. Integrating Microbial Metagenomics and Physicochemical Parameters and a New Perspective on Starter Culture for Fine Cocoa Fermentation. Food Microbiol. 2020, 93, 103608. [Google Scholar] [CrossRef]
- Maïmouna Kouamé, L. Cocoa Fermentation from Agnéby-Tiassa: Biochemical Study of Microflora. Am. J. BioScience 2015, 3, 203. [Google Scholar] [CrossRef]
- Mota-Gutierrez, J.; Botta, C.; Ferrocino, I.; Giordano, M.; Bertolino, M.; Dolci, P.; Cannoni, M.; Cocolin, L. Dynamics and Biodiversity of Bacterial and Yeast Communities during Fermentation of Cocoa Beans. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, G.; Young, G.M. Food Evolution: The Impact of Society and Science on the Fermentation of Cocoa Beans. Compr. Rev. Food Sci. Food Saf. 2017, 16, 431–455. [Google Scholar] [CrossRef] [PubMed]
- Serra, J.L.; Moura, F.G.; De Melo Pereira, G.V.; Soccol, C.R.; Rogez, H.; Darnet, S. Determination of the Microbial Community in Amazonian Cocoa Bean Fermentation by Illumina-Based Metagenomic Sequencing. LWT 2019, 106, 229–239. [Google Scholar] [CrossRef]
- Verce, M.; Schoonejans, J.; Hernandez Aguirre, C.; Molina-Bravo, R.; De Vuyst, L.; Weckx, S. A Combined Metagenomics and Metatranscriptomics Approach to Unravel Costa Rican Cocoa Box Fermentation Processes Reveals Yet Unreported Microbial Species and Functionalities. Front. Microbiol. 2021, 12, 641185. [Google Scholar] [CrossRef]
- Castro-Alayo, E.M.; Idrogo-Vásquez, G.; Siche, R.; Cardenas-Toro, F.P. Formation of Aromatic Compounds Precursors during Fermentation of Criollo and Forastero Cocoa. Heliyon 2019, 5, 1157. [Google Scholar] [CrossRef] [Green Version]
- Kongor, J.E.; Hinneh, M.; Van de Walle, D.; Afoakwa, E.O.; Boeckx, P.; Dewettinck, K. Factors Influencing Quality Variation in Cocoa (Theobroma cacao) Bean Flavour Profile—A Review. Food Res. Int. 2016, 82, 44–52. [Google Scholar] [CrossRef]
- Santos, D.S.; Rezende, R.P.; Dos Santos, T.F.; De Lima Silva Marques, E.; Ferreira, A.C.R.; De Cerqueira e Silva, A.B.; Romano, C.C.; Da Cruz Santos, D.W.; Dias, J.C.T.; Tavares Bisneto, J.D. Fermentation in Fine Cocoa Type Scavina: Change in Standard Quality as the Effect of Use of Starters Yeast in Fermentation. Food Chem. 2020, 328, 7–12. [Google Scholar] [CrossRef]
- Delgado-Ospina, J.; Acquaticci, L.; Molina-Hernandez, J.B.; Rantsiou, K.; Martuscelli, M.; Kamgang-Nzekoue, A.F.; Vittori, S.; Paparella, A.; Chaves-López, C. Exploring the Capability of Yeasts Isolated from Colombian Fermented Cocoa Beans to Form and Degrade Biogenic Amines in a Lab-Scale Model System for Cocoa Fermentation. Microorganisms 2021, 9, 28. [Google Scholar] [CrossRef]
- Public Health England. UK Standards for Microbiology Investigations Staining Procedures: Staining Procedures Standards for Microbiology Investigations Smi Quality and Consistency in Clinical Laboratories; PHE Publications Gateway Number: 2015075 UK Standards for Microbiology Investigation; PHE: London, UK, 2019.
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; John Wiley & Sons: Hoboken, NJ, USA, 2003; ISBN 047150338X. [Google Scholar]
- Zhang, M.; Liu, W.; Nie, X.; Li, C.; Gu, J.; Zhang, C. Molecular Analysis of Bacterial Communities in Biofilms of a Drinking Water Clearwell. Microbes Environ. 2012, 27, 443–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical Innovations for High-Throughput Amplicon Sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.H.; Longley, R.; Bonito, G.; Liao, H.L. A Two-Step PCR Protocol Enabling Flexible Primer Choice and High Sequencing Yield for Illumina Miseq Meta-Barcoding. Agronomy 2021, 11, 1274. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Van den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Read Trimming Tool for Illumina NGS Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Luz Calle, M. Statistical Analysis of Metagenomics Data. Genom. Inform. 2019, 17, e6. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing Large Minimum Evolution Trees with Profiles Instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Ramos, C.L.; Dias, R.; Gabriela Da Cruz, M.; Miguel, P.; Freitas Schwan, R. Impact of Different Cocoa Hybrids (Theobroma cacao L.) and S. Cerevisiae UFLA CA11 Inoculation on Microbial Communities and Volatile Compounds of Cocoa Fermentation. Food Res. Int. 2014, 64, 908–918. [Google Scholar] [CrossRef]
- Frauendorfer, F.; Schieberle, P. Identification of the Key Aroma Compounds in Cocoa Powder Based on Molecular Sensory Correlations. J. Agric. Food Chem. 2006, 54, 5521–5529. [Google Scholar] [CrossRef]
- Frauendorfer, F.; Schieberle, P. Changes in Key Aroma Compounds of Criollo Cocoa Beans during Roasting. J. Agric. Food Chem. 2008, 56, 10244–10251. [Google Scholar] [CrossRef]
- Koné, M.K.; Guéhi, S.T.; Durand, N.; Ban-Koffi, L.; Berthiot, L.; Tachon, A.F.; Brou, K.; Boulanger, R.; Montet, D. Contribution of Predominant Yeasts to the Occurrence of Aroma Compounds during Cocoa Bean Fermentation. Food Res. Int. 2016, 89, 910–917. [Google Scholar] [CrossRef]
- Aculey, P.C.; Snitkjaer, P.; Owusu, M.; Bassompiere, M.; Takrama, J.; Nørgaard, L.; Petersen, M.A.; Nielsen, D.S. Ghanaian Cocoa Bean Fermentation Characterized by Spectroscopic and Chromatographic Methods and Chemometrics. J. Food Sci. 2010, 75, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Teniola, O.D.; Ban-Koffi, L.; Owusu, M.; Andersson, T.S.; Holzapfel, W.H. The Microbiology of Ghanaian Cocoa Fermentations Analysed Using Culture-Dependent and Culture-Independent Methods. Int. J. Food Microbiol. 2007, 114, 168–186. [Google Scholar] [CrossRef] [PubMed]
- Batista, N.N.; Ramos, C.L.; Dias, D.R.; Pinheiro, A.C.M.; Schwan, R.F. The Impact of Yeast Starter Cultures on the Microbial Communities and Volatile Compounds in Cocoa Fermentation and the Resulting Sensory Attributes of Chocolate. J. Food Sci. Technol. 2016, 53, 1101–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arana-Sánchez, A.; Segura-García, L.E.; Kirchmayr, M.; Orozco-Ávila, I.; Lugo-Cervantes, E.; Gschaedler-Mathis, A. Identification of Predominant Yeasts Associated with Artisan Mexican Cocoa Fermentations Using Culture-Dependent and Culture-Independent Approaches. World J. Microbiol. Biotechnol. 2015, 31, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Papalexandratou, Z.; Camu, N.; Falony, G.; De Vuyst, L. Comparison of the Bacterial Species Diversity of Spontaneous Cocoa Bean Fermentations Carried out at Selected Farms in Ivory Coast and Brazil. Food Microbiol. 2011, 28, 964–973. [Google Scholar] [CrossRef]
- Visintin, S.; Alessandria, V.; Valente, A.; Dolci, P.; Cocolin, L. Molecular Identification and Physiological Characterization of Yeasts, Lactic Acid Bacteria and Acetic Acid Bacteria Isolated from Heap and Box Cocoa Bean Fermentations in West Africa. Int. J. Food Microbiol. 2016, 216, 69–78. [Google Scholar] [CrossRef]
- Ho, V.T.T.; Fleet, G.H.; Zhao, J. Unravelling the Contribution of Lactic Acid Bacteria and Acetic Acid Bacteria to Cocoa Fermentation Using Inoculated Organisms. Int. J. Food Microbiol. 2018, 279, 43–56. [Google Scholar] [CrossRef]
- Raspor, P.; Goranovič, D. Biotechnological Applications of Acetic Acid Bacteria. Crit. Rev. Biotechnol. 2008, 28, 101–124. [Google Scholar] [CrossRef]
- Meersman, E.; Steensels, J.; Mathawan, M.; Wittocx, P.J.; Saels, V.; Struyf, N.; Bernaert, H.; Vrancken, G.; Verstrepen, K.J. Detailed Analysis of the Microbial Population in Malaysian Spontaneous Cocoa Pulp Fermentations Reveals a Core and Variable Microbiota. PLoS ONE 2013, 8, e81559. [Google Scholar] [CrossRef] [Green Version]
- Miescher Schwenninger, S.; Freimüller Leischtfeld, S.; Gantenbein-Demarchi, C. High-Throughput Identification of the Microbial Biodiversity of Cocoa Bean Fermentation by MALDI-TOF MS. Lett. Appl. Microbiol. 2016, 63, 347–355. [Google Scholar] [CrossRef]
- Marseglia, A.; Musci, M.; Rinaldi, M.; Palla, G.; Caligiani, A. Volatile Fingerprint of Unroasted and Roasted Cocoa Beans (Theobroma cacao L.) from Different Geographical Origins. Food Res. Int. 2020, 132, 109101. [Google Scholar] [CrossRef] [PubMed]
- Frauendorfer, F.; Schieberle, P. Key Aroma Compounds in Fermented Forastero Cocoa Beans and Changes Induced by Roasting. Eur. Food Res. Technol. 2019, 245, 1907–1915. [Google Scholar] [CrossRef]
- De Melo Pereira, G.V.; Magalhães, K.T.; De Almeida, E.G.; Da Silva Coelho, I.; Schwan, R.F. Spontaneous Cocoa Bean Fermentation Carried out in a Novel-Design Stainless Steel Tank: Influence on the Dynamics of Microbial Populations and Physical-Chemical Properties. Int. J. Food Microbiol. 2013, 161, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Neilson, A.P.; O’Keefe, S.F.; Ogejo, J.A.; Huang, H.; Ponder, M.; Chu, H.S.S.; Jin, Q.; Pilot, G.; Stewart, A.C. A Laboratory-Scale Model Cocoa Fermentation Using Dried, Unfermented Beans and Artificial Pulp Can Simulate the Microbial and Chemical Changes of on-Farm Cocoa Fermentation. Eur. Food Res. Technol. 2019, 245, 511–519. [Google Scholar] [CrossRef]
- Romanens, E.; Näf, R.; Lobmaier, T.; Pedan, V.; Leischtfeld, S.F.; Meile, L.; Schwenninger, S.M. A Lab-Scale Model System for Cocoa Bean Fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 3349–3362. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Weckx, S. The Functional Role of Lactic Acid Bacteria in Cocoa Bean Fermentation. In Biotechnology of Lactic Acid Bacteria: Novel Applications, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 248–278. [Google Scholar] [CrossRef]
- Miguel, M.G.D.C.P.; De Castro Reis, L.V.; Efraim, P.; Santos, C.; Lima, N.; Schwan, R.F. Cocoa Fermentation: Microbial Identification by MALDI-TOF MS, and Sensory Evaluation of Produced Chocolate. LWT Food Sci. Technol. 2017, 77, 362–369. [Google Scholar] [CrossRef] [Green Version]
- Ouattara, H.G.; Koffi, B.L.; Karou, G.T.; Sangaré, A.; Niamke, S.L.; Diopoh, J.K. Implication of Bacillus Sp. in the Production of Pectinolytic Enzymes during Cocoa Fermentation. World J. Microbiol. Biotechnol. 2008, 24, 1753–1760. [Google Scholar] [CrossRef]
- Adler, P.; Bolten, C.J.; Dohnt, K.; Hansen, C.E.; Wittmann, C. Core Fluxome and Metafluxome of Lactic Acid Bacteria under Simulated Cocoa Pulp Fermentation Conditions. Appl. Environ. Microbiol. 2013, 79, 5670–5681. [Google Scholar] [CrossRef] [Green Version]
- Bastos, V.S.; Santos, M.F.S.; Gomes, L.P.; Leite, A.M.O.; Flosi Paschoalin, V.M.; Del Aguila, E.M. Analysis of the Cocobiota and Metabolites of Moniliophthora Perniciosa-Resistant Theobroma cacao Beans during Spontaneous Fermentation in Southern Brazil. J. Sci. Food Agric. 2018, 98, 4963–4970. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Montealegre, M.E.; Dávila-Mora, L.L.; Botero-Rute, L.M.; Reyes, A.; Caro-Quintero, A. Fine Resolution Analysis of Microbial Communities Provides Insights into the Variability of Cocoa Bean Fermentation. Front. Microbiol. 2020, 11, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Z.; Luo, J.; Niu, Y.; Wang, P.; Wang, R.; Sun, X. Olfactory Impact of Esters on Rose Essential Oil Floral Alcohol Aroma Expression in Model Solution. Food Res. Int. 2019, 116, 211–222. [Google Scholar] [CrossRef]
- Rodriguez-Campos, J.; Escalona-Buendía, H.B.; Contreras-Ramos, S.M.; Orozco-Avila, I.; Jaramillo-Flores, E.; Lugo-Cervantes, E. Effect of Fermentation Time and Drying Temperature on Volatile Compounds in Cocoa. Food Chem. 2012, 132, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s Microbes: Tools for Distinguishing the Living from the Dead in Microbial Ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Dyrhovden, R.; Øvrebø, K.K.; Nordahl, M.V.; Nygaard, R.M.; Ulvestad, E.; Kommedal, Ø. Bacteria and Fungi in Acute Cholecystitis. A Prospective Study Comparing next Generation Sequencing to Culture. J. Infect. 2020, 80, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Aldrete-Tapia, J.; Martínez-Peniche, R.; Hernández-Iturriaga, M. Yeast and Bacterial Diversity, Dynamics and Fermentative Kinetics during Small-Scale Tequila Spontaneous Fermentation. Food Microbiol. 2019, 86, 103339. [Google Scholar] [CrossRef]
| Primer | Sequence | PCR Step |
|---|---|---|
| ITS4 | 5′TCCTCCGCTTATTGATATGC 3′ | 1 |
| ITS1F | 5′CTTGGTCATTTAGAGGAAGTAA 3′ | |
| LROR | 5′ACCCGCTGAACTTAAGC 3′ | |
| LR3 | 5′CCGTGTTTCAAGACGGG 3′ | |
| 806R | 5′GGACTACHVGGGTWTCTAAT 3′ | |
| 341F | 5′CCT ACG GGN GGC WGC AG 3′ | |
| ITS4 f1 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNAATCCTCCGCTTATTGATATGC 3′ | 2 |
| ITS4 f2 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNTNNNAATCCTCCGCTTATTGATATGC 3′ | |
| ITS4 f3 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNCTNNNAATCCTCCGCTTATTGATATGC 3′ | |
| ITS4 f4 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNACTNNNAATCCTCCGCTTATTGATATGC 3′ | |
| ITS4 f5 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNGACTNNNAATCCTCCGCTTATTGATATGC 3′ | |
| ITS4 f6 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNTGACTNNNAATCCTCCGCTTATTGATATGC 3′ | |
| ITS1F f1 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNNNNNTTCTTGGTCATTTAGAGGAAGTAA 3′ | |
| ITS1F f2 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNTNNNNTTCTTGGTCATTTAGAGGAAGTAA 3′ | |
| ITS1F f3 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNCTNNNNTTCTTGGTCATTTAGAGGAAGTAA 3′ | |
| ITS1F f4 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNACTNNNNTTCTTGGTCATTTAGAGGAAGTAA 3′ | |
| ITS1F f5 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNGACTNNNNTTCTTGGTCATTTAGAGGAAGTAA 3′ | |
| ITS1F f6 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNTGACTNNNNTTCTTGGTCATTTAGAGGAAGTAA 3′ | |
| LROR f1 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNNNNNGAACCCGCTGAACTTAAGC 3′ | |
| LROR f2 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNTNNNNGAACCCGCTGAACTTAAGC 3′ | |
| LROR f3 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNCTNNNNGAACCCGCTGAACTTAAGC 3′ | |
| LROR f4 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNACTNNNNGAACCCGCTGAACTTAAGC 3′ | |
| LROR f5 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNGACTNNNNGAACCCGCTGAACTTAAGC 3′ | |
| LROR f6 | 5′GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNTGACTNNNNGAACCCGCTGAACTTAAGC 3′ | |
| LR3 f1 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNCACCGTGTTTCAAGACGGG 3′ | |
| LR3 f2 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNTNNNCACCGTGTTTCAAGACGGG 3′ | |
| LR3 f3 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNCTNNNCACCGTGTTTCAAGACGGG 3′ | |
| LR3 f4 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNACTNNNCACCGTGTTTCAAGACGGG 3′ | |
| LR3 f5 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNGACTNNNCACCGTGTTTCAAGACGGG 3′ | |
| LR3 f6 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNTGACTNNNCACCGTGTTTCAAGACGGG 3′ | |
| 806R f1 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNACGGACTACHVGGGTWTCTAAT 3′ | |
| 806R f2 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNTNNNACGGACTACHVGGGTWTCTAAT 3′ | |
| 806R f3 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNCTNNNACGGACTACHVGGGTWTCTAAT 3′ | |
| 806R f4 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNACTNNNACGGACTACHVGGGTWTCTAAT 3′ | |
| 806R f5 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNGACTNNNACGGACTACHVGGGTWTCTAAT 3′ | |
| 806R f6 | 5′GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNTGACTNNNACGGACTACHVGGGTWTCTAAT 3′ | |
| 341F_f1 | 5′GCC TCC CTC GCG CCA TCA GAG ATG TGT ATA AGA GAC AGN NNN NNN NAG CCT ACG GGN GGC WGC AG 3′ | |
| 341F_f2 | 5′GCC TCC CTC GCG CCA TCA GAG ATG TGT ATA AGA GAC AGN NNN TNN NNA GCC TAC GGG NGG CWG CAG 3′ | |
| 341F_f3 | 5′ GCC TCC CTC GCG CCA TCA GAG ATG TGT ATA AGA GAC AGN NNN CTN NNN AGC CTA CGG GNG GCW GCA G 3′ | |
| 341F_f4 | 5′GCC TCC CTC GCG CCA TCA GAG ATG TGT ATA AGA GAC AGN NNN ACT NNN NAG CCT ACG GGN GGC WGC AG 3′ | |
| 341F_f5 | 5′GCC TCC CTC GCG CCA TCA GAG ATG TGT ATA AGA GAC AGN NNN GAC TNN NNA GCC TAC GGG NGG CWG CAG 3′ | |
| 341F_f6 | 5′GCC TCC CTC GCG CCA TCA GAG ATG TGT ATA AGA GAC AGN NNN TGA CTN NNN AGC CTA CGG GNG GCW GCA G 3′ | |
| PCR_F | 5′AATGATACGGCGACCACCGAGATCTACACGCCTCCCTCGCGCCATCAGAGATGTG 3′ | 3 |
| PCR_R_bc | 5′CAAGCAGAAGACGGCATACGAGAT XXXXXXXXXGTGACTGGAGTTCAGACGTGTGCTC 3′ 1 |
| Microorganism | Fermentation Time (h) | Growth Condition (Aerobic or Anaerobic) |
|---|---|---|
| Saccharomyces cerevisiae | 0, 24 | Aerobic and Anaerobic |
| Liquorilactobacillus nagelii | Aerobic and Anaerobic | |
| Acetobacter pasteurianus | Aerobic and Anaerobic | |
| Saccharomyces cervicae | 48 | Aerobic |
| Acetobacter ghanensis | Aerobic and Anaerobic | |
| Limosilactobacillus fermentum | Anaerobic | |
| Acetobacter syzygii | Aerobic and Anaerobic | |
| Candida metapsilosis | Aerobic and Anaerobic | |
| Bacillus amyloliquefaciens | Aerobic and Anaerobic | |
| Saccharomyces cerevisiae | 72 | Aerobic and Anaerobic |
| Acetobacter ghanensis | Aerobic and Anaerobic | |
| Liquorilactobacillus nagelii | Aerobic | |
| Limosilactobacillus fermentum | Aerobic and Anaerobic | |
| Acetobacter pasteurianus | 96 | Aerobic and Anaerobic |
| Candida metapsilosis | Aerobic and Anaerobic | |
| Bacillus amyloliquefaciens | Aerobic and Anaerobic | |
| Bacillus subtilis | Aerobic and Anaerobic |
| Fermentation Timepoint (h) | Total | Genera | Species | % Genus | % Species |
|---|---|---|---|---|---|
| 0 | 357,753 | 270,603 | 266,123 | 75.64% | 74.39% |
| 24 | 296,897 | 87,324 | 82,059 | 29.41% | 27.64% |
| 48 | 248,527 | 124,598 | 111,888 | 50.13% | 45.02% |
| 72 | 86,069 | 15,834 | 12,879 | 18.40% | 14.96% |
| 96 | 275,299 | 215,588 | 105,137 | 78.31% | 38.19% |
| 120 | 420,132 | 246,800 | 224,339 | 58.74% | 53.40% |
| Fermentation Timepoint (h) | Shannon Index (H) |
|---|---|
| 0 | 1.967 |
| 24 | 3 |
| 48 | 1.898 |
| 72 | 3.065 |
| 96 | 1.45 |
| 120 | 2.03 |
| Fermentation Timepoint | Inoculated Microbial Species | Growth Condition (Aerobic or Anaerobic) | Compounds Produced after Inoculation | Log2 FC 1 | Aroma Descriptor |
|---|---|---|---|---|---|
| 0 h | Liquorilactobacillus nagelii | Aerobic | Ethanol | NDC | Alcoholic |
| 1-[2-(2-Methylbutyl)phenyl] ethanone | −1.14 | NF 2 | |||
| 2,3-Butanediol | 1.76 | Fruity, creamy, buttery | |||
| Benzene ethanol | 2.43 | Floral | |||
| Benzaldehyde | NDC 3 | Almond | |||
| Anaerobic | Ethanol | 2.69 | Alcoholic | ||
| 9-propylphenanthrene | 3.77 | NF | |||
| (3R)-3-Phenyl-2,3-dihydro-1H-isoindol | 1.83 | NF | |||
| Benzene ethanol | 1.64 | Floral | |||
| Acetophenone | 0.63 | Floral | |||
| Benzaldehyde | NDC | Almond | |||
| S. cerevisiae | Aerobic | Benzene ethanol | 2.24 | Floral | |
| benzyl 2,2′-di-O-methyldivaricatate | 1.39 | NF | |||
| Anaerobic | Benzaldehyde | NDC | Almond | ||
| A. pasteurianus | Aerobic | Butanal, 3-methyl- | 1.86 | Chocolate | |
| Benzaldehyde | NDC | Almond | |||
| Acetaldehyde | 1.02 | Fruity | |||
| Acetophenone | 2.84 | Floral | |||
| Benzene ethanol | 3.58 | Floral | |||
| Acetic acid | 4.42 | Sour | |||
| 24 h | Liquorilactobacillus nagelii | Anaerobic | Benzene ethanol | 1.64 | Floral |
| S. cerevisiae | Aerobic | 2,3-Butanediol | 0.13 | Fruity, creamy, buttery | |
| Anaerobic | 2-Propyldecan-1-ol | 1.00 | Floral | ||
| A. pasteurianus | Aerobic | Acetophenone | −1.48 | Floral | |
| Acetaldehyde | −0.40 | Fruity | |||
| Anaerobic | Benzaldehyde | NDC | Almond | ||
| 48 h | C. metapsilosis | Aerobic | Benzaldehyde | −2.68 | Almond |
| Butanoic acid | −3.22 | Cheesy | |||
| Anaerobic | 2 -Butanone, 3-hydroxy- | −0.98 | NF | ||
| Butanal, 3-methyl- | −2.94 | Chocolate | |||
| B. amyloliquefaciens | Aerobic | Benzaldehyde | −1.50 | Almond | |
| S. cerevisiae | Aerobic | Benzaldehyde | NDC | Almond | |
| A. syzygii | Aerobic | Benzaldehyde | −1.09 | Almond | |
| Anaerobic | Benzaldehyde | −1.85 | Almond | ||
| Limosilactobacillus fermentum | Anaerobic | Butanal, 3-methyl | 0.00 | Chocolate | |
| Acetaldehyde | 0.00 | Fruity | |||
| 72 h | A. ghanensis | Aerobic | Benzene ethanol | −1.77 | Floral |
| Anaerobic | 1-Butanol, 3-methyl | −2.67 | Malty, bitter, chocolate | ||
| Benzene ethanol | −4.26 | Floral | |||
| Limosilactobacillus fermentum | Aerobic | 1 butanol-3 methyl | −2.31 | Malty, bitter, chocolate | |
| Anaerobic | Ethanol, 2-nitro | −2.06 | NF | ||
| Liquorilactobacillus nagelii | Aerobic | Acetic acid, ethyl ester | −3.31 | Fruity, sweet | |
| S. cerevisiae | Aerobic | Acetic acid, ethyl ester | −2.77 | Fruity, sweet | |
| Anaerobic | Benzene ethanol | −2.45 | Floral | ||
| 1 butanol–3 methyl | −2.52 | Malty, bitter, chocolate | |||
| 96 h | B. subtilis | Aerobic | Phenylvinylacetylene | −1.59 | NF |
| Benzeneacetic acid, ethyl ester | −1.83 | Floral | |||
| Acetic acid | 0.78 | Sour | |||
| Benzaldehyde | NDC | Almond | |||
| Anaerobic | 3-Penten-2-ol | 0.84 | Green vinyl | ||
| Caffeine | 2.33 | NF | |||
| C. metapsilosis | Aerobic | B-Phenylethyl formate | 5.11 | Floral | |
| Hexadecane | NDC | NF | |||
| Anaerobic | Ethylphenyl acetate | 5.28 | Floral | ||
| B-Phenylethyl formate | NDC | Floral | |||
| A. ghanensis | Aerobic | Benzene acetaldehyde | −2.45 | Green | |
| Pentadecane | NDC | Waxy | |||
| Acetic acid | −1.93 | Sour | |||
| Anaerobic | Benzene ethanol | 1.38 | Floral | ||
| Acetic acid, 2-phenylethyl ester | −2.57 | NF | |||
| A. pasteurianus | Aerobic | 2-Naphthalene-sulfonic acid | 2.59 | NF | |
| 1-phenyl-(2,2,2-2H3)ethanone | 5.12 | NF | |||
| Benzene acetaldehyde | −4.31 | Green | |||
| Anaerobic | (Z)-But-2-enyl benzoate | NDC | NF | ||
| B. amyloliquefaciens | Aerobic | Ethylphenyl acetate | 2.13 | Floral | |
| Ethanol | NDC | Alcoholic | |||
| Benzophenone | NDC | Balsamic, rose, herbal | |||
| Anaerobic | Ethylphenyl acetate | 5.69 | Floral | ||
| Ethanol | 7.96 | Alcoholic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tigrero-Vaca, J.; Maridueña-Zavala, M.G.; Liao, H.-L.; Prado-Lince, M.; Zambrano-Vera, C.S.; Monserrate-Maggi, B.; Cevallos-Cevallos, J.M. Microbial Diversity and Contribution to the Formation of Volatile Compounds during Fine-Flavor Cacao Bean Fermentation. Foods 2022, 11, 915. https://doi.org/10.3390/foods11070915
Tigrero-Vaca J, Maridueña-Zavala MG, Liao H-L, Prado-Lince M, Zambrano-Vera CS, Monserrate-Maggi B, Cevallos-Cevallos JM. Microbial Diversity and Contribution to the Formation of Volatile Compounds during Fine-Flavor Cacao Bean Fermentation. Foods. 2022; 11(7):915. https://doi.org/10.3390/foods11070915
Chicago/Turabian StyleTigrero-Vaca, Joel, María Gabriela Maridueña-Zavala, Hui-Ling Liao, Mónica Prado-Lince, Cynthia Sulay Zambrano-Vera, Bertha Monserrate-Maggi, and Juan M. Cevallos-Cevallos. 2022. "Microbial Diversity and Contribution to the Formation of Volatile Compounds during Fine-Flavor Cacao Bean Fermentation" Foods 11, no. 7: 915. https://doi.org/10.3390/foods11070915
APA StyleTigrero-Vaca, J., Maridueña-Zavala, M. G., Liao, H.-L., Prado-Lince, M., Zambrano-Vera, C. S., Monserrate-Maggi, B., & Cevallos-Cevallos, J. M. (2022). Microbial Diversity and Contribution to the Formation of Volatile Compounds during Fine-Flavor Cacao Bean Fermentation. Foods, 11(7), 915. https://doi.org/10.3390/foods11070915








