Effects of NaCl Concentration on the Behavior of Vibrio brasiliensis and Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Growth Conditions of V. brasiliensis in Salt Stress
2.3. Morphological Analysis of V. brasiliensis in Salt Stress
2.4. Swimming Motility of V. brasiliensis in Salt Stress
2.5. Extraction and Analysis of Outer Membrane Proteins (OMPs)
2.5.1. OMPs Extraction
2.5.2. Sodium Dodecyl Sulfate—Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.5.3. In Gel Tryptic Digest for Mass Spectrometry Analysis
2.6. Transcriptome Analysis
3. Results and Discussion
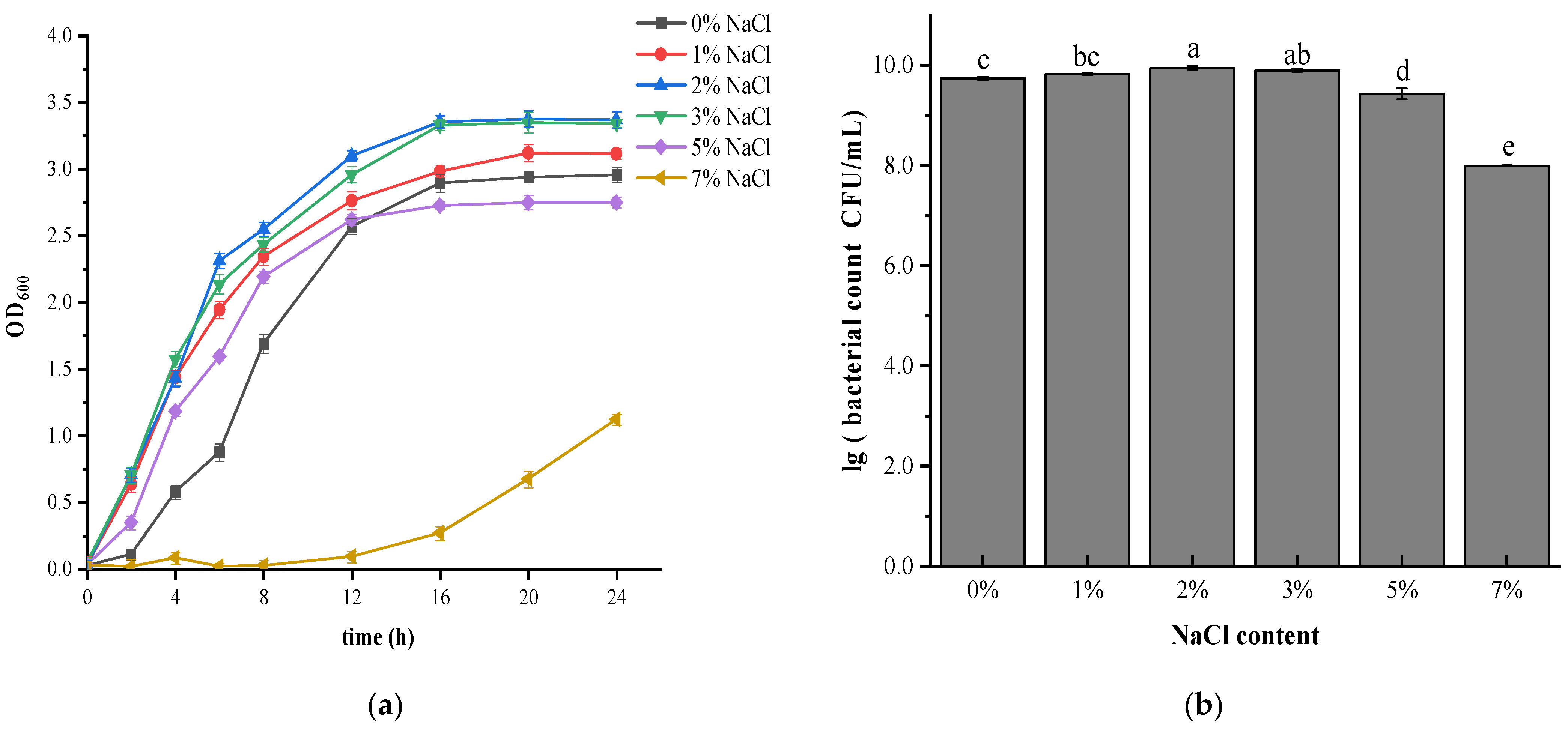
3.1. Effect of Salt Stress on Growth and Survival of V. brasiliensis
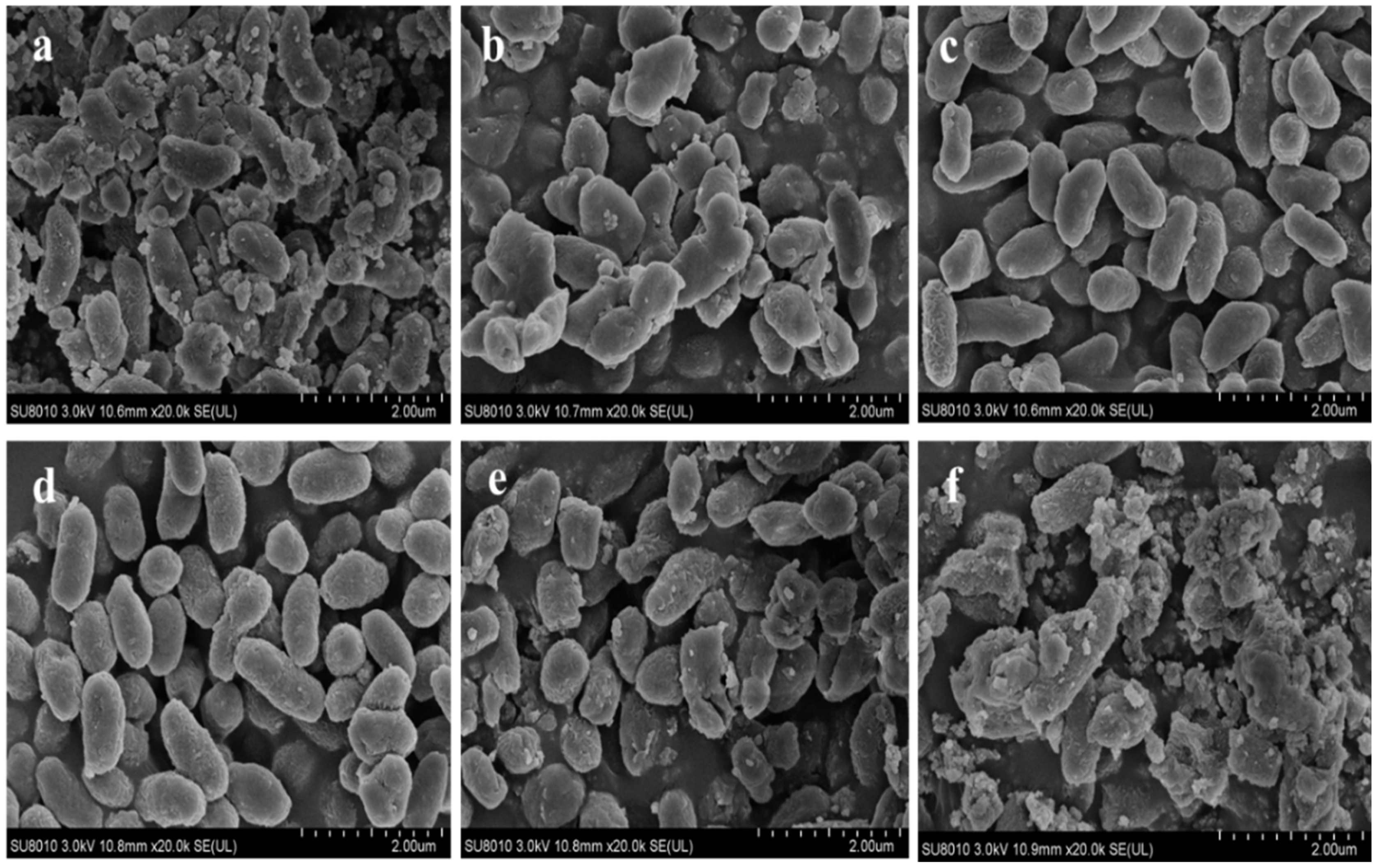
3.2. Morphological Analysis of V. brasiliensis in Response to Salt Stress
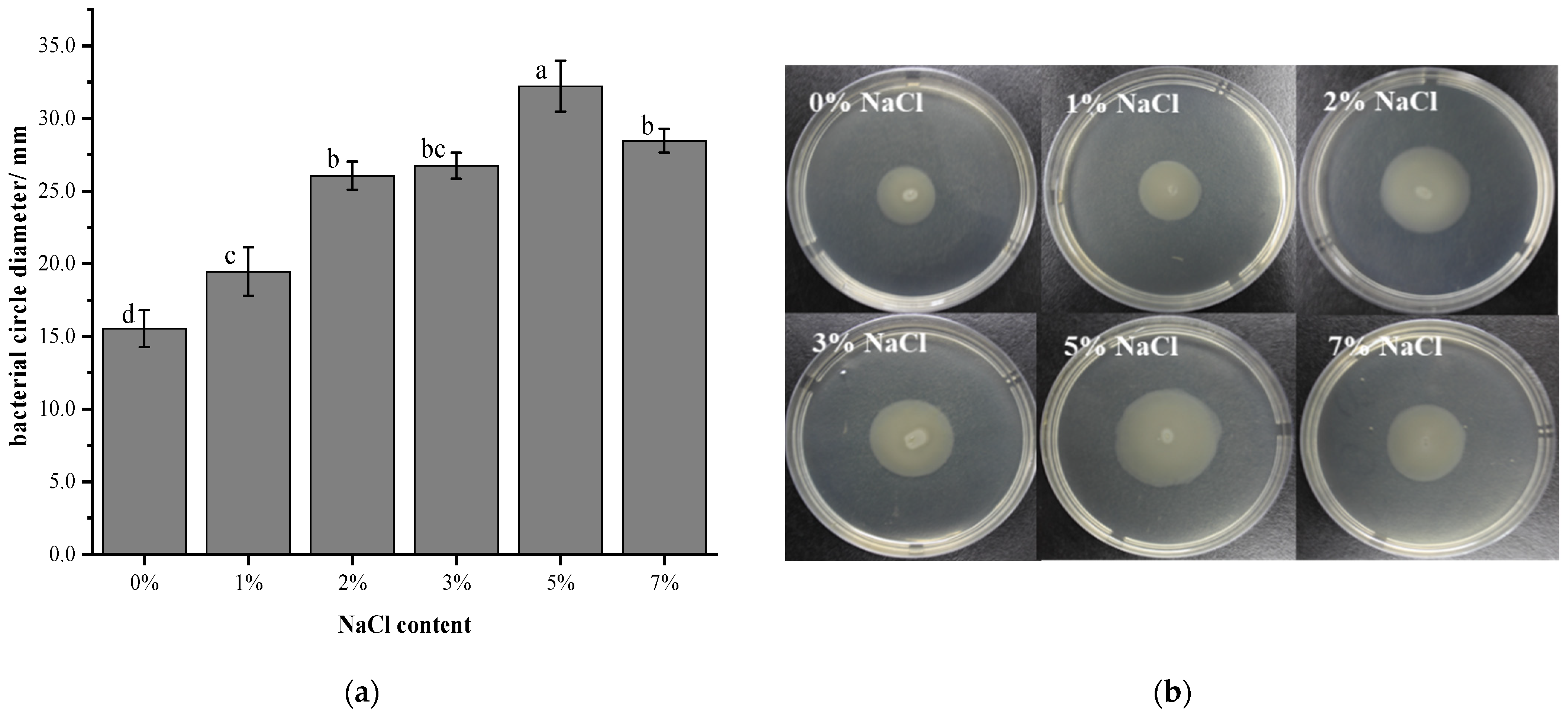
3.3. Effect of Salt Stress on the Swimming Ability of V. brasiliensis
3.4. Analysis of Differentially Expressed OMPs in Response to Salt Stress
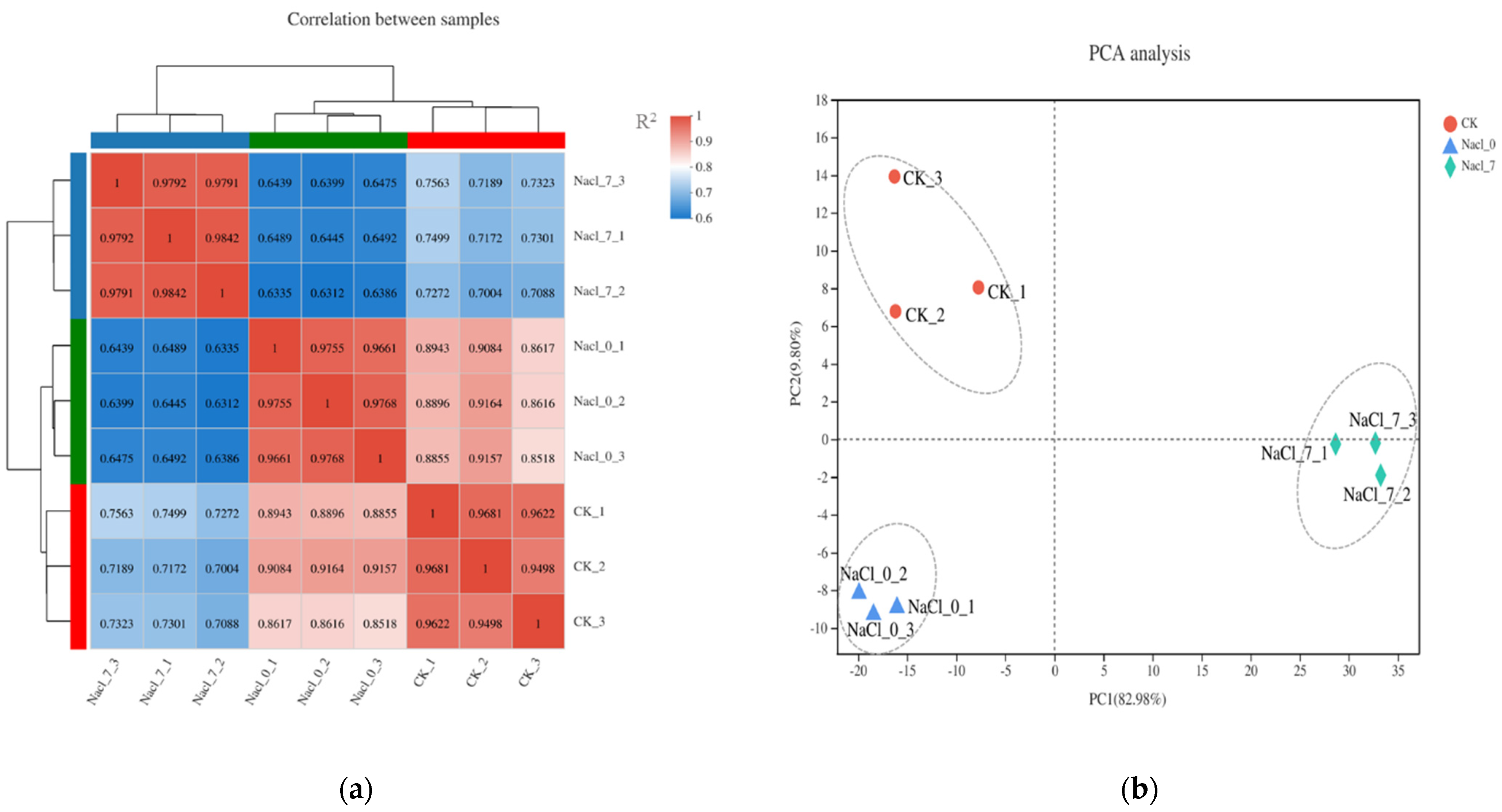
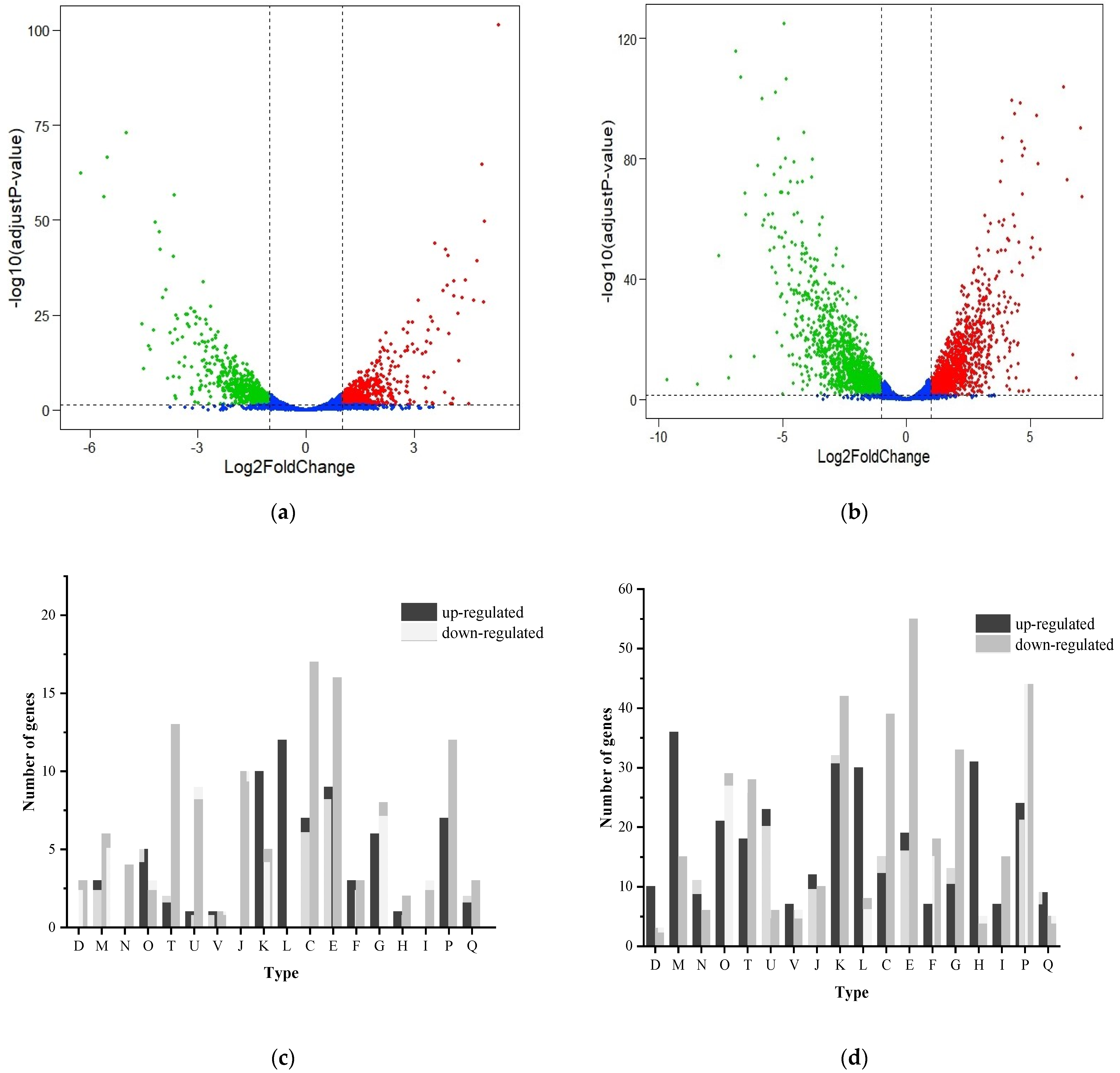
3.5. Transcriptome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hernandez-Perez, A.; Soderhall, K.; Sirikharin, R.; Jiravanichpaisal, P.; Soderhall, I. Vibrio areninigrae as a pathogenic bacterium in a crustacean. J. Invertebr. Pathol. 2021, 178, 107517. [Google Scholar] [CrossRef] [PubMed]
- Osunla, C.A.; Okoh, A.I. Vibrio Pathogens: A public health concern in rural water resources in Sub-Saharan Africa. Int. J. Environ. Res. Public Health 2017, 14, 1188. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.; Cheng, J.; Sharma, D.; Bitzikos, O.; Gustafson, R.; Fyfe, M.; Greve, R.; Murti, M.; Stone, J.; Honish, L.; et al. Outbreak of Vibrio parahaemolyticus associated with consumption of raw Oysters in Canada, 2015. Foodborne Pathog. Dis. 2018, 15, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.M.; Chalita, M.; Yang, S.J.; Yoon, S.H.; Cho, K.; Seong, W.K.; Hong, S.; Kim, J.; Kwak, H.S.; Chun, J. Comparative genomic analysis of the 2016 Vibrio cholerae outbreak in South Korea. Front. Public Health 2019, 7, 228. [Google Scholar] [CrossRef] [Green Version]
- Parola, P.; Anani, H.; Eldin, C.; Dubourg, G.; Lagier, J.C.; Levasseur, A.; Casalta, J.P.; Raoult, D.; Fournier, P.E. Case Report: Vibrio cholerae biliary tract infections in two North Africans in France. Am. J. Trop. Med. Hyg. 2020, 102, 1306–1308. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus. Trends Microbiol. 2020, 28, 81–82. [Google Scholar] [CrossRef]
- Yoon, S.H.; Waters, C.M. Vibrio cholerae. Trends Microbiol. 2019, 27, 806–807. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-Granados, F.; Gallardo-Becerra, L.; Leonardo-Reza, M.; Ochoa-Romo, J.P.; Ochoa-Leyva, A. A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ 2018, 6, e5382. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Samanta, M.; Mohanty, S.; Maiti, N.K. Characterization of Vibrio species isolated from freshwater fishes by ribotyping. Indian J. Microbiol. 2010, 50, 101–103. [Google Scholar] [CrossRef] [Green Version]
- Duche, O.; Tremoulet, F.; Glaser, P.; Labadie, J. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 2002, 68, 1491–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Cao, W.; Hong, J.; Longwei, Z. Contamination status of Vibrio Parahaemolyticus in foods of wet markets in Chongming County of Shanghai. Occup. Health 2010, 26, 409–410. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, H.; Zhan, W.; Song, W. Isolation and characteristics of major outer membrane proteins of aquatic pathogens Vibrio anguillarum and Vibrio alginolyticus. J. Fish. Sci. China 2003, 10, 31–35. [Google Scholar]
- Marchesi, V.T.; Andrews, E.P. Glycoproteins-isolation from cell membranes with lithium diiodosalicylate. Science 1971, 174, 1247–1248. [Google Scholar] [CrossRef] [PubMed]
- Kao, D.Y.; Cheng, Y.C.; Kuo, T.Y.; Lin, S.B.; Lin, C.C.; Chow, L.P.; Chen, W.J. Salt-responsive outer membrane proteins of Vibrio anguillarum serotype 01 as revealed by comparative proteome analysis. J. Appl. Microbiol. 2009, 106, 2079–2085. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.G.; Wang, S.Y.; Ren, H.X.; Lin, X.M.; Wu, L.N.; Peng, X.X. Proteomic analysis on the expression of outer membrane proteins of Vibrio alginolyticus at different sodium concentrations. Proteomics 2005, 5, 3142–3152. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.X.; Ren, H.X.; Wang, S.Y.; Peng, X.X. Proteomic analysis of salt-sensitive outer membrane proteins of Vibrio parahaemolyticus. Res. Microbiol. 2004, 155, 835–842. [Google Scholar] [CrossRef]
- Jia, X.; Wang, J.B.; Rivera, S.; Duong, D.; Weinert, E.E. An O-2-sensing stressosome from a Gram-negative bacterium. Nat. Commun. 2016, 7, 12381. [Google Scholar] [CrossRef] [PubMed]
- Kalburge, S.S.; Whitaker, W.B.; Boyd, E.F. High-salt preadaptation of Vibrio parahaemolyticus enhances survival in response to lethal environmental stresses. J. Food Prot. 2014, 77, 246–253. [Google Scholar] [CrossRef]
- Xiong, X.P.; Zhang, B.W.; Yang, M.J.; Ye, M.Z.; Peng, X.X.; Li, H. Identification of vaccine candidates from differentially expressed outer membrane proteins of Vibrio alginolyticus in response to NaCl and iron limitation. Fish Shellfish Immunol. 2010, 29, 810–816. [Google Scholar] [CrossRef]
- Xu, C.X.; Wang, S.Y.; Wu, L.N.; Ren, H.X.; Peng, X.X. Proteomic analysis on the expression of the outer membrane proteins of Vibrio alginolyticus at different Na+ concentrations. Mol. Cell. Proteom. 2004, 3, S161. [Google Scholar]
- Yang, L.; Zhan, L.; Han, H.; Gao, H.; Guo, Z.; Qin, C.; Yang, R.; Liu, X.; Zhou, D. The low-salt stimulon in Vibrio parahaemolyticus. Int. J. Food Microbiol. 2010, 137, 49–54. [Google Scholar] [CrossRef]
- Arias, C.R.; Lafrentz, S.; Cai, W.; Olivares-Fuster, O. Adaptive response to starvation in the fish pathogen Flavobacterium columnare: Cell viability and ultrastructural changes. BMC Microbiol. 2012, 12, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringgaard, S.; Zepeda-Rivera, M.; Wu, X.; Schirner, K.; Davis, B.M.; Waldor, M.K. ParP prevents dissociation of CheA from chemotactic signaling arrays and tethers them to a polar anchor. Proc. Natl. Acad. Sci. USA 2014, 111, E255–E264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.B.; Liu, H.H.; Gu, D.; Lu, X.X.; Zhou, X.H.; Xia, X.D. Transcriptomic analysis of PhoR reveals its role in regulation of swarming motility and T3SS expression in Vibrio parahaemolyticus. Microbiol. Res. 2020, 235, 126448. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Ouyang, M.; Zhang, H.; Rao, S.; Yin, Y. Biological characteristics of Vibrio parahaemolyticus during growth in adverse environment. Food Sci. 2018, 39, 177–182. [Google Scholar] [CrossRef]
- Lutwyche, P.; Exner, M.M.; Hancock, R.E.W.; Trust, T.J. A conserved Aeromonas-salmonicida porin provides protective immunity to rainbow-trout. Infect. Immun. 1995, 63, 3137–3142. [Google Scholar] [CrossRef] [Green Version]
- Levin, J.Z.; Yassour, M.; Adiconis, X.; Nusbaum, C.; Thompson, D.A.; Friedman, N.; Gnirke, A.; Regev, A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 2010, 7, 709–715. [Google Scholar] [CrossRef] [Green Version]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.J.; Jiang, Z.Y.; Zhang, S.M.; Chen, Q.Y.; Tong, S.Q.; Liu, X.D.; Jiang, Q.; Yang, H.; Wei, W.H.; Zhang, X.J. Transcriptome analysis and immune-related genes expression reveals the immune responses of Macrobrachium rosenbergii infected by Enterobacter cloacae. Fish Shellfish Immun. 2020, 101, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.Y.; Ni, Y.; Gao, L. Biological characteristics of Vibrio parahaemolyticus growing in the medium of different salinity. Mod. Food 2018, 6, 91–94. [Google Scholar] [CrossRef]
- Fu, X.P.; Liang, W.L.; Du, P.C.; Yan, M.Y.; Kan, B. Transcript changes in Vibrio cholerae in response to salt stress. Gut Pathog. 2014, 6, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsen, C.; Paulsen, S.M.; Tunsjo, H.S.; Krinner, S.; Sorum, H.; Haugen, P.; Willassen, N.P. Motility and flagellin gene expression in the fish pathogen Vibrio salmonicida: Effects of salinity and temperature. Microb. Pathog. 2008, 45, 258–264. [Google Scholar] [CrossRef]
- Boehm, A.; Kaiser, M.; Li, H.; Spangler, C.; Kasper, C.A.; Ackermann, M.; Kaever, V.; Sourjik, V.; Roth, V.; Jenal, U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 2010, 141, 107–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atsumi, T.; McCartert, L.; Imae, Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 1992, 355, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Homma, M. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta (BBA)-Bioenerg. 2001, 1505, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Kojima, S.; Homma, M. Structure, gene regulation and environmental response of flagella in Vibrio. Front. Microbiol. 2013, 4, 410. [Google Scholar] [CrossRef] [Green Version]
- Konovalova, A.; Kahne, D.E.; Silhavy, T.J. Outer membrane biogenesis. Annu. Rev. Microbiol. 2017, 71, 539–556. [Google Scholar] [CrossRef] [PubMed]
- Jeeves, M.; Knowles, T.J. A novel pathway for outer membrane protein biogenesis in Gram-negative bacteria. Mol. Microbiol. 2015, 97, 607–611. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, S.; Chaudhuri, K.; Sen, K.; Das, J. Porins of Vibrio cholerae: Purification and characterization of OmpU. J. Bacteriol. 1996, 178, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, J.; Davis, B.M.; Waldor, M.K. Antimicrobial peptides activate the Vibrio cholerae sigma(E) regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 2007, 63, 848–858. [Google Scholar] [CrossRef]
- Wang, W.; Guo, S.; Gao, Y.; Liang, X.; Liu, L.; Pan, S. Comparative immunogenicity of outer membrane protein K and whole-cell antigens of Vibrio parahaemolyticus for diagnosis. Lett. Appl. Microbiol. 2021, 73, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.M.; Engle, M.; Oliver, J.D. Effect of osmotic variation on the outer-membrane proteins of Vibrio vulnificus identification of a major heat-modifiable protein. Microbios 1994, 80, 209–214. [Google Scholar] [PubMed]
- Dorr, T.; Lam, H.; Alvarez, L.; Cava, F.; Davis, B.M.; Waldor, M.K. A novel peptidoglycan binding protein crucial for PBP1A-mediated cell wall biogenesis in Vibrio cholerae. PLoS Genet. 2014, 10, e1004433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanabe, T.; Naka, A.; Aso, H.; Nakao, H.; Narimatsu, S.; Inoue, Y.; Ono, T.; Yamamoto, S. A novel aerobactin utilization cluster in Vibrio vulnificus with a gene involved in the transcription regulation of the iutA homologue. Microbiol. Immunol. 2005, 49, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.C.; Drake, E.J.; Grant, T.D.; Gulick, A.M. Structural and functional characterization of aerobactin synthetase IucA from a hypervirulent pathotype of Klebsiella pneumoniae. Biochemistry 2016, 55, 3559–3570. [Google Scholar] [CrossRef]
- Ben Abdallah, F.; Lagha, R.; Ellafi, A.; Namane, A.; Rousselle, J.C.; Lenormand, P.; Kallel, H. Identification of outer membrane proteins altered in response to UVC-radiation in Vibrio parahaemolyticus and Vibrio alginolyticus. Indian J. Microbiol. 2012, 52, 660–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abromaitis, S.; Koehler, J.E. The bartonella quintana extracytoplasmic function sigma factor RpoE has a role in bacterial adaptation to the arthropod vector environment. J. Bacteriol. 2013, 195, 2662–2674. [Google Scholar] [CrossRef] [Green Version]
- Tettmann, B.; Dotsch, A.; Armant, O.; Fjell, C.D.; Overhage, J. Knockout of extracytoplasmic function Sigma factor ECF-10 affects stress resistance and biofilm formation in Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2014, 80, 4911–4919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebens, M.; Lundquist, P.; Soderlund, L.; Todorovic, M.; Carlin, N.I.A. The nptA gene of Vibrio cholerae encodes a functional sodium-dependent phosphate cotransporter homologous to the type II cotransporters of eukaryotes. J. Bacteriol. 2002, 184, 4466–4474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staley, C.; Harwood, V.J. Differential expression of a sodium-phosphate cotransporter among Vibrio vulnificus strains. Microb. Ecol. 2014, 67, 24–33. [Google Scholar] [CrossRef]
- Nachin, L.; Nannmark, U.; Nystrom, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.N.; Lin, X.M.; Wang, F.P.; Ye, D.Z.; Xiao, X.; Wang, S.Y.; Peng, X.X. OmpW and OmpV are required for NaCl regulation in Photobacterium damsela. J. Proteome Res. 2006, 5, 2250–2257. [Google Scholar] [CrossRef] [PubMed]
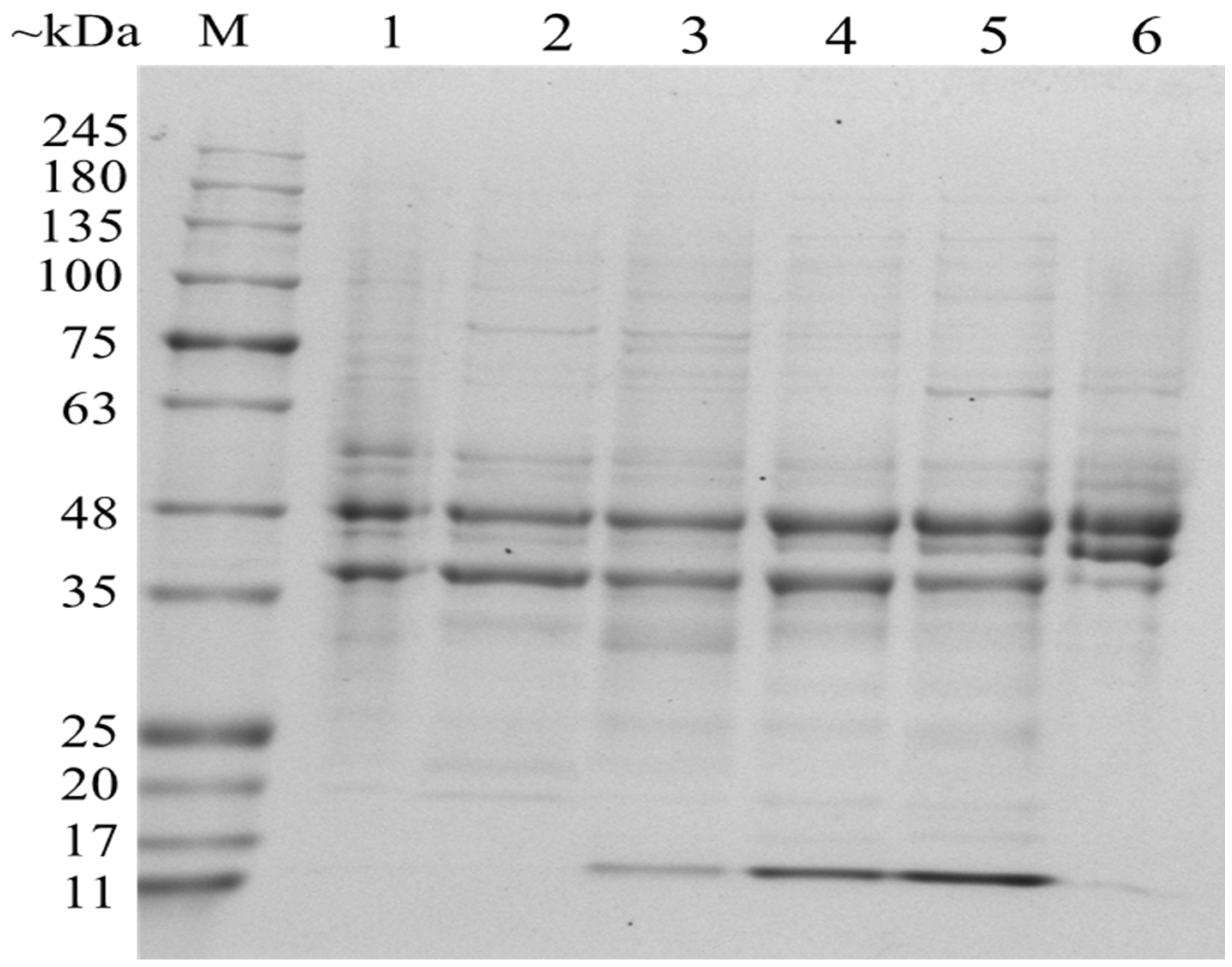
| NaCl (%) | Accession | Gene | Proteins | Mw [KDa] | Molecular Function |
|---|---|---|---|---|---|
| 2 | E8LNG0 | lpoA | Penicillin-binding protein activator LpoA | 67.3 | enzyme regulator activity |
| 2 | E8LPR8 | VIBR0546_18016 | Flp pilus assembly protein CpaB | 27.2 | NA |
| 2 | E8LQ55 | VIBR0546_05478 | MSHA biogenesis protein MshL | 59.2 | NA |
| 3 | E8LNG0 | lpoA | Penicillin-binding protein activator LpoA | 67.3 | enzyme regulator activity |
| 3 | E8LS16 | VIBR0546_10974 | Sodium-type flagellar protein MotY | 33.5 | NA |
| 5 | E8LS16 | VIBR0546_10974 | Sodium-type flagellar protein MotY | 33.5 | NA |
| 5 | E8M0I9 | VIBR0546_09489 | Outer membrane protein OmpA | 35.1 | porin activity |
| 7 | E8LS16 | VIBR0546_10974 | Sodium-type flagellar protein MotY | 33.5 | NA |
| 7 | E8M0I9 | VIBR0546_09489 | Outer membrane protein OmpA | 35.1 | porin activity |
| 7 | E8LX27 | VIBR0546_17898 | Ferric aerobactin receptor | 77.7 | siderophore uptake transmembrane transporter activity; signaling receptor activity |
| Gene Name | Gene Description | FC | Log2FC | p Value | Regulate |
|---|---|---|---|---|---|
| VIBR0546_12042 | proton/glutamate symporter | 5.165 | 2.369 | 1.15 × 10−13 | up |
| nhaA | pH-dependent sodium/proton antiporter | 3.672 | 1.877 | 1.61 × 10−12 | up |
| VIBR0546_00894 | NptA protein | 11.824 | 3.564 | 5.86 × 10−12 | up |
| VIBR0546_21825 | putative outer membrane protein | 7.040 | 2.816 | 6.8 × 10−11 | up |
| VIBR0546_00270 | universal stress protein UspE | 2.950 | 1.561 | 6.62 × 10−8 | up |
| VIBR0546_01129 | putative Na+/H+ antiporter | 3.182 | 1.670 | 1.85 × 10−7 | up |
| VIBR0546_09929 | universal stress protein family 8 | 2.511 | 1.328 | 4.65 × 10−6 | up |
| VIBR0546_10589 | universal stress protein A | 2.881 | 1.527 | 2.87 × 10−5 | up |
| ompR | osmolarity response regulator | 2.471 | 1.305 | 9.71 × 10−5 | up |
| VIBR0546_03992 | putative salt-induced outer membrane protein | 2.567 | 1.360 | 4.52 × 10−4 | up |
| VIBR0546_17778 | OmpR family two-component response regulator | 2.039 | 1.028 | 9.87 × 10−3 | up |
| VIBR0546_02830 | choline/carnitine/betaine transporter | 0.079 | −3.660 | 3.18 × 10−60 | down |
| VIBR0546_00510 | outer membrane protein | 0.081 | −3.631 | 7.45 × 10−24 | down |
| VIBR0546_06992 | sodium/proline symporter | 0.097 | −3.366 | 8.10 × 10−14 | down |
| VIBR0546_03902 | putative sodium/sulfate symporter | 0.204 | −2.295 | 2.04 × 10−13 | down |
| VIBR0546_13277 | Outer membrane protein | 0.263 | −1.926 | 5.11 × 10−12 | down |
| VIBR0546_09484 | outer membrane protein OmpA | 0.250 | −2.000 | 4.01 × 10−11 | down |
| VIBR0546_07217 | outer membrane protein W | 0.253 | −1.984 | 4.35 × 10−11 | down |
| VIBR0546_05877 | OmpA family protein | 0.293 | −1.772 | 1.04 × 10−10 | down |
| VIBR0546_18581 | sodium/alanine symporter | 0.383 | −1.385 | 8.86 × 10−9 | down |
| VIBR0546_13282 | outer membrane protein | 0.396 | −1.338 | 1.58 × 10−6 | down |
| VIBR0546_03420 | putative outer membrane protein | 0.370 | −1.434 | 2.10 × 10−5 | down |
| VIBR0546_02985 | Outer membrane protein | 0.262 | −1.933 | 9.81 × 10−5 | down |
| VIBR0546_16166 | putative arginine/ornithine antiporter | 0.478 | −1.066 | 2.62 × 10−3 | down |
| Gene Name | Gene Description | FC | Log2FC | p Value | Regulate |
|---|---|---|---|---|---|
| VIBR0546_17048 | putative threonine efflux protein | 26.152 | 4.709 | 1.9 × 10−43 | up |
| VIBR0546_21150 | Na+/H+ antiporter | 9.496 | 3.247 | 4.47 × 10−36 | up |
| VIBR0546_01064 | Ca2+/Na+ antiporter | 10.100 | 3.336 | 5.15 × 10−32 | up |
| VIBR0546_01411 | outer membrane protein | 4.147 | 2.052 | 1.00 × 10−30 | up |
| VIBR0546_01241 | proton/glutamate symport protein | 4.243 | 2.085 | 2.23 × 10−17 | up |
| VIBR0546_05214 | LPS assembly outer membrane complex protein LptD | 2.683 | 1.424 | 6.98 × 10−9 | up |
| VIBR0546_21825 | putative outer membrane protein | 3.554 | 1.830 | 1.01 × 10−7 | up |
| VIBR0546_10974 | sodium-type flagellar protein MotY precursor | 2.906 | 1.539 | 1.09 × 10−7 | up |
| VIBR0546_21600 | putative sodium-type flagellar protein MotY precursor | 6.971 | 2.801 | 5.54 × 10−6 | up |
| VIBR0546_03992 | putative salt-induced outer membrane protein | 2.468 | 1.304 | 3.57 × 10−5 | up |
| VIBR0546_06697 | sodium/glutamate symporter | 2.705 | 1.435 | 1.36 × 10−4 | up |
| VIBR0546_12742 | sodium/alanine symporter | 2.326 | 1.218 | 2.10 × 10−4 | up |
| VIBR0546_12482 | putative outer membrane protein | 2.272 | 1.184 | 7.17 × 10−4 | up |
| VIBR0546_07217 | outer membrane protein W | 0.008 | −6.889 | 9.60 × 10−120 | down |
| VIBR0546_05309 | RNA polymerase sigma factor RpoS | 0.028 | −5.163 | 7.74 × 10−90 | down |
| VIBR0546_00510 | outer membrane protein | 0.038 | −4.726 | 7.75 × 10−47 | down |
| VIBR0546_20323 | outer membrane protein | 0.062 | −4.007 | 3.32 × 10−37 | down |
| VIBR0546_00505 | putative anti-sigma regulatory factor | 0.030 | −5.066 | 5.08 × 10−36 | down |
| VIBR0546_18581 | sodium/alanine symporter | 0.171 | −2.552 | 1.12 × 10−19 | down |
| VIBR0546_09929 | universal stress protein family 8 | 0.160 | −2.647 | 3.93 × 10−19 | down |
| VIBR0546_00270 | universal stress protein UspE | 0.221 | −2.179 | 3.5 × 10−15 | down |
| VIBR0546_12042 | proton/glutamate symporter | 0.230 | −2.123 | 8.13 × 10−15 | down |
| VIBR0546_09484 | outer membrane protein OmpA | 0.232 | −2.110 | 1.90 × 10−13 | down |
| VIBR0546_10579 | universal stress protein UspB | 0.249 | −2.005 | 1.88 × 10−11 | down |
| VIBR0546_06992 | sodium/proline symporter | 0.111 | −3.171 | 6.54 × 10−10 | down |
| VIBR0546_10589 | universal stress protein A | 0.245 | −2.026 | 3.43 × 10−7 | down |
| ompR | osmolarity response regulator | 0.365 | −1.452 | 5.85 × 10−7 | down |
| VIBR0546_00894 | NptA protein | 0.231 | −2.116 | 1.03 × 10−4 | down |
| VIBR0546_05877 | OmpA family protein | 0.427 | −1.229 | 1.20 × 10−4 | down |
| VIBR0546_05598 | outer membrane protein OmpK | 0.420 | −1.252 | 1.06 × 10−3 | down |
| VIBR0546_01821 | Outer membrane protein | 0.217 | −2.202 | 2.26 × 10−3 | down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Li, Y.; Wang, B.; Yin, L.; Jia, X. Effects of NaCl Concentration on the Behavior of Vibrio brasiliensis and Transcriptome Analysis. Foods 2022, 11, 840. https://doi.org/10.3390/foods11060840
Hu S, Li Y, Wang B, Yin L, Jia X. Effects of NaCl Concentration on the Behavior of Vibrio brasiliensis and Transcriptome Analysis. Foods. 2022; 11(6):840. https://doi.org/10.3390/foods11060840
Chicago/Turabian StyleHu, Shuyang, Yuwei Li, Boran Wang, Lijun Yin, and Xin Jia. 2022. "Effects of NaCl Concentration on the Behavior of Vibrio brasiliensis and Transcriptome Analysis" Foods 11, no. 6: 840. https://doi.org/10.3390/foods11060840
APA StyleHu, S., Li, Y., Wang, B., Yin, L., & Jia, X. (2022). Effects of NaCl Concentration on the Behavior of Vibrio brasiliensis and Transcriptome Analysis. Foods, 11(6), 840. https://doi.org/10.3390/foods11060840







