Development of a Duplex TaqMan Real-Time Polymerase Chain Reaction for Accurate Identification and Quantification of Salmonella Enteritidis from Laboratory Samples and Contaminated Chicken Eggs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Bacterial Growth and Genomic DNA Extraction
2.3. PCR Primer Pairs and TaqMan Probes
2.4. Conventional PCR
2.5. SYBR-Based qRT-PCR
2.6. Duplex TaqMan Real-Time PCR System
2.7. Specificity of the Duplex Real-Time PCR
2.8. Standard Curve and Detection Limit of the Duplex Real-Time PCR
2.9. Evaluation of the Reproducibility of the Method in Detecting S. enteritidis
2.10. Real-Time PCR for Quantification of S. enteritidis in Organs in a Chicken Model
2.11. Real-Time PCR for the Detection of S. enteritidis in Clinical Chicken Eggs
2.12. Traditional Serotyping of Salmonella Isolates from Clinical Samples
2.13. Preparation of S. enteritidis Cell Suspension and Enumeration Methods
2.14. Traditional Plating and Traditional MPN Methods
2.15. TaqMan Real-Time PCR and MPN-qPCR-SIT Methods
2.16. Statistical Analysis
3. Results
3.1. Specificity Analysis of the Primers for the Amplification of lygD and invA
3.2. Specificity of the Duplex TaqMan Real-Time PCR Assay
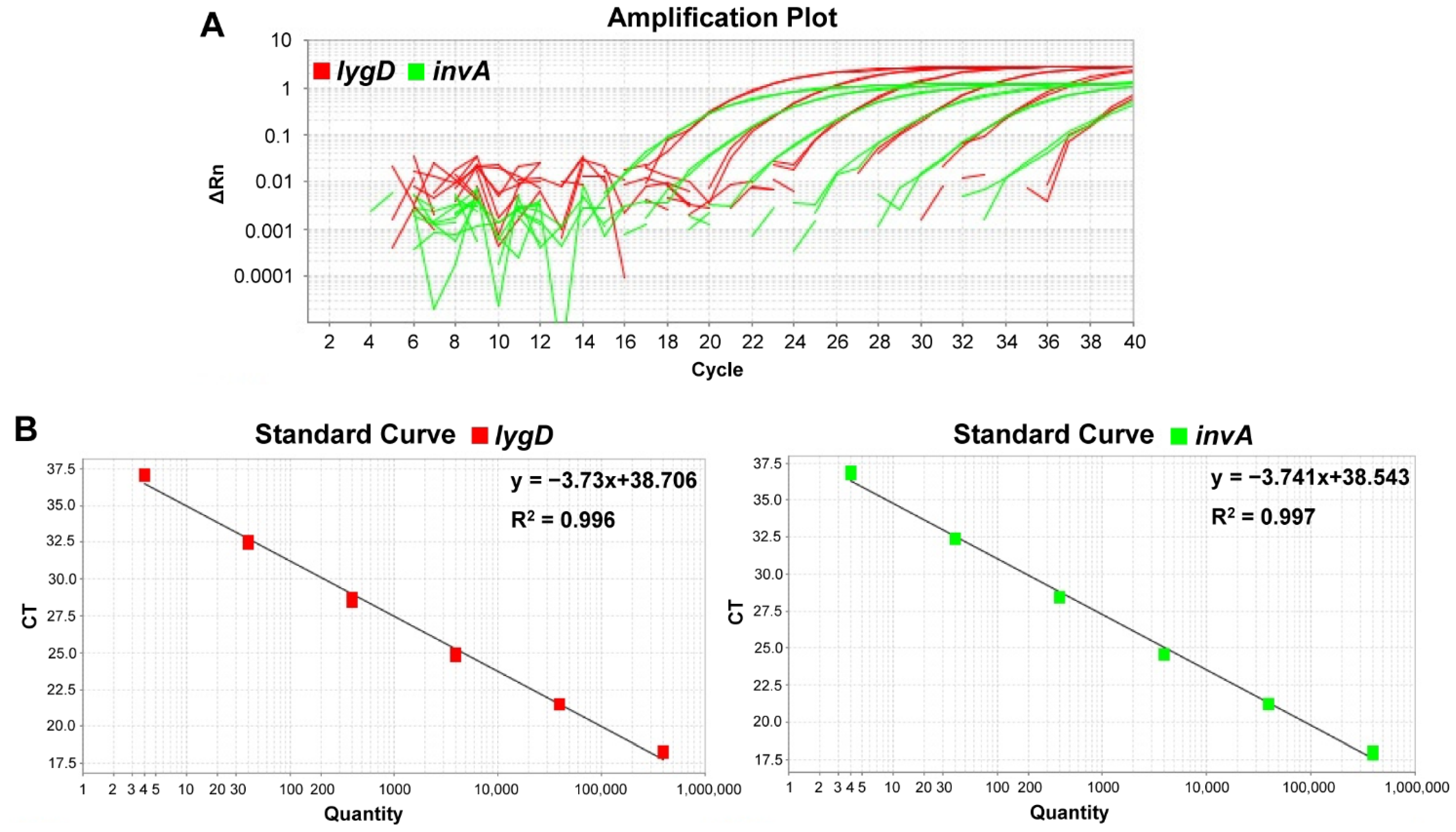
3.3. Standard Curves and Sensitivity of the Developed Real-Time PCR
3.4. Reproducibility of the TaqMan Real-Time PCR Assay
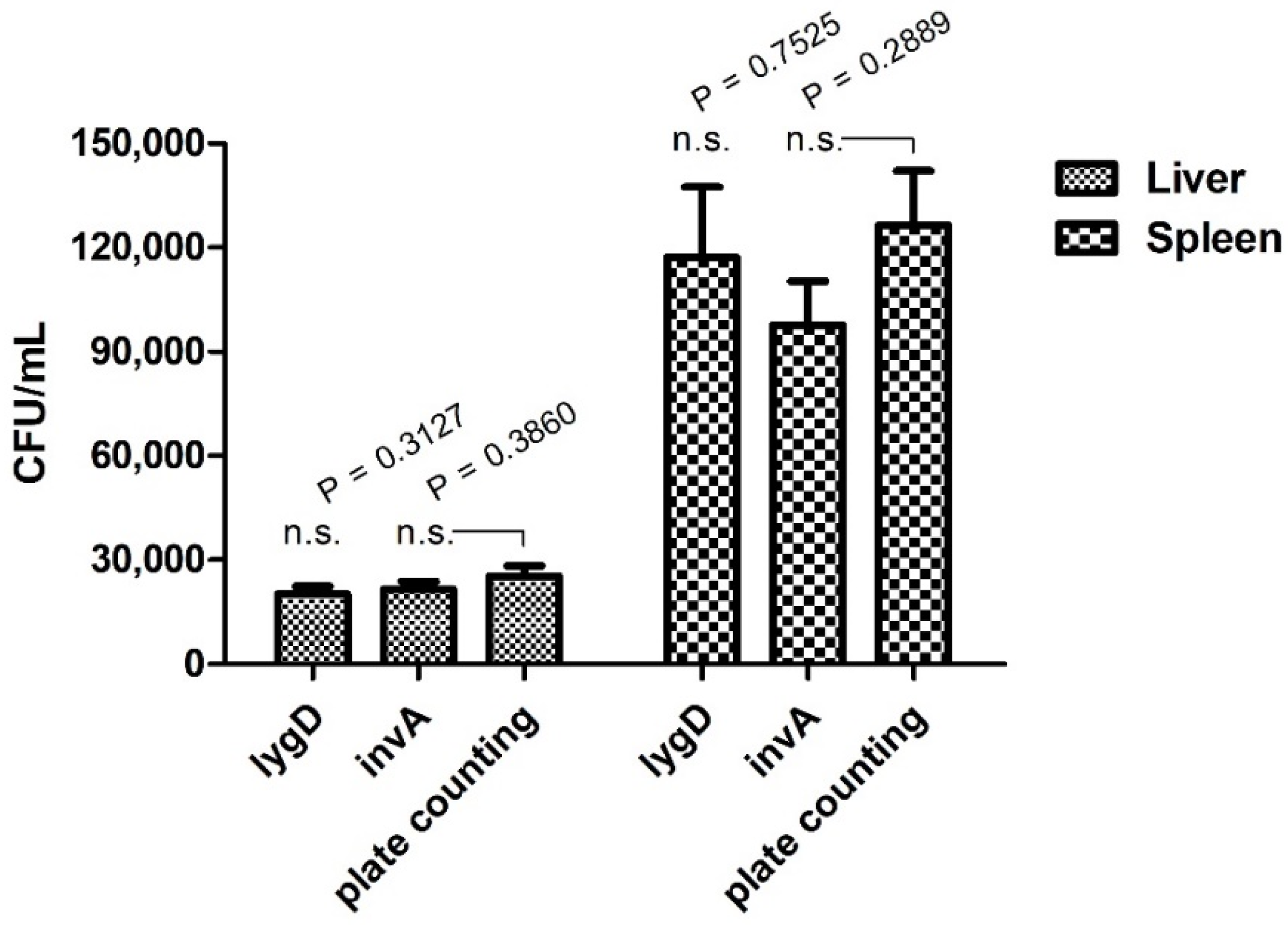
3.5. Quantification of S. enteritidis in a Chicken Model by Real-Time PCR Assay
3.6. Application of the Duplex Real-Time PCR for Clinical Chicken Eggs
3.7. Traditional Serotyping of Salmonella Isolates and Biochemical Identification
3.8. Comparison of MPN-qPCR-SIT with Other Enumeration Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Masi, L.; Yue, M.; Hu, C.; Rakov, A.V.; Rankin, S.C.; Schifferli, D.M. Cooperation of adhesin alleles in Salmonella-host tropism. mSphere 2017, 2, e00066-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, food safety and food handling practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella chronic carriage: Epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Raspoet, R.; Eeckhaut, V.; Vermeulen, K.; De Smet, L.; Wen, Y.; Nishino, K.; Haesebrouck, F.; Ducatelle, R.; Devreese, B.; Van Immerseel, F. The Salmonella Enteritidis TolC outer membrane channel is essential for egg white survival. Poult. Sci. 2019, 98, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, L.; Paoli, G.C.; Shi, C.; Tu, S.I.; Shi, X. A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int. J. Food Microbiol. 2010, 137, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Hugas, M.; Beloeil, P. Controlling Salmonella along the food chain in the European Union-progress over the last ten years. Eurosurveillance 2014, 19, 20804. [Google Scholar] [CrossRef] [Green Version]
- Kosa, K.M.; Cates, S.C.; Bradley, S.; Chambers, E., IV; Godwin, S. Consumer-reported handling of raw poultry products at home: Results from a national survey. J. Food Prot. 2015, 78, 180–186. [Google Scholar] [CrossRef]
- Nesbitt, A.; Ravel, A.; Murray, R.; McCormick, R.; Savelli, C.; Finley, R.; Parmley, J.; Agunos, A.; Majowicz, S.E.; Gilmour, M.; et al. Integrated surveillance and potential sources of Salmonella enteritidis in human cases in Canada from 2003 to 2009. Epidemiol. Infect. 2012, 140, 1757–1772. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Lin, Q.; Zhang, J.; Young, G.M.; Jiang, C.; Zhong, Y.; Zhang, J. Rapid identification of pathogens by using surface-enhanced Raman spectroscopy and multi-scale convolutional neural network. Anal. Bioanal. Chem. 2021, 413, 3801–3811. [Google Scholar] [CrossRef]
- Kagambèga, A.; Lienemann, T.; Aulu, L.; Traoré, A.S.; Barro, N.; Siitonen, A.; Haukka, K. Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol. 2013, 13, 253. [Google Scholar] [CrossRef] [Green Version]
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella serovars, 9th ed.; WHO Collaborating Center for Reference and Research on Salmonella; Institut Pasteur: Paris, France, 2007. [Google Scholar]
- Soria, M.C.; Soria, M.A.; Bueno, D.J. A comparative study of culture methods and PCR assay for Salmonella detection in poultry drinking water. Poult. Sci. 2013, 92, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Kasturi, K.N. A real-time PCR for rapid identification of Salmonella enterica Gaminara serovar. J. Microbiol. Methods 2020, 169, 105729. [Google Scholar] [CrossRef] [PubMed]
- Boukharouba, A.; González, A.; García-Ferrús, M.; Ferrús, M.A.; Botella, S. Simultaneous detection of four main foodborne pathogens in ready-to-eat food by using a simple and rapid multiplex PCR (mPCR) assay. Int. J. Environ. Res. Public Health 2022, 19, 1031. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.J.; Xiong, G.T.; Bai, M.Y.; Ge, Y.; Wu, Z.F. Detection of live Salmonella enterica in fresh-cut vegetables by a TaqMan-based one-step reverse transcription real-time PCR. Lett. Appl. Microbiol. 2018, 66, 447–454. [Google Scholar] [CrossRef]
- Ding, S.; Hu, H.; Yue, X.; Feng, K.; Gao, X.; Dong, Q.; Yang, M.; Tamer, U.; Huang, G.; Zhang, J. A fluorescent biosensor based on quantum dot-labeled streptavidin and poly-l-lysine for the rapid detection of Salmonella in milk. J. Dairy Sci. 2022, in press. [Google Scholar] [CrossRef]
- Feng, K.; Li, T.; Ye, C.; Gao, X.; Yue, X.; Ding, S.; Dong, Q.; Yang, M.; Huang, G.; Zhang, J. A novel electrochemical immunosensor based on Fe3O4@ graphene nanocomposite modified glassy carbon electrode for rapid detection of Salmonella in milk. J. Dairy Sci. 2022, 105, 2108–2118. [Google Scholar] [CrossRef]
- Bai, J.; Trinetta, V.; Shi, X.; Noll, L.W.; Magossi, G.; Zheng, W.; Porter, E.P.; Cernicchiaro, N.; Renter, D.G.; Nagaraja, T.G. A multiplex real-time PCR assay, based on invA and pagC genes, for the detection and quantification of Salmonella enterica from cattle lymph nodes. J. Microbiol. Methods 2018, 148, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Castillo, A.G.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Detection of Salmonella Typhimurium and Listeria monocytogenes biofilm cells exposed to different drying and pre-enrichment times using conventional and rapid methods. Int. J. Food Microbiol. 2020, 324, 108611. [Google Scholar] [CrossRef]
- Park, S.H.; Ricke, S.C. Development of multiplex PCR assay for simultaneous detection of Salmonella genus, Salmonella subspecies I, Salm. Enteritidis, Salm. Heidelberg and Salm. Typhimurium. J. Appl. Microbiol. 2015, 118, 152–160. [Google Scholar] [CrossRef]
- Worrall, L.J.; Vuckovic, M.; Strynadka, N.C. Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci. 2010, 19, 1091–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barletta, F.; Mercado, E.H.; Lluque, A.; Ruiz, J.; Cleary, T.G.; Ochoa, T.J. Multiplex real-time PCR for detection of Campylobacter, Salmonella, and Shigella. J. Clin. Microbiol. 2013, 51, 2822–2829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido, A.; Chapela, M.J.; Román, B.; Fajardo, P.; Vieites, J.M.; Cabado, A.G. In-house validation of a multiplex real-time PCR method for simultaneous detection of Salmonella spp., Escherichia coli O157 and Listeria monocytogenes. Int. J. Food Microbiol. 2013, 164, 92–98. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, T.; Yang, S.; Wang, X.; Guo, H. Response surface methodology to design a selective enrichment broth for rapid detection of Salmonella spp. by SYBR Green Ι real-time PCR. Appl. Microbiol. Biotechnol. 2013, 97, 4149–4158. [Google Scholar] [CrossRef] [PubMed]
- Ramtahal, M.A.; Somboro, A.M.; Amoako, D.G.; Abia, A.L.K.; Perrett, K.; Bester, L.A.; Essack, S.Y. Molecular epidemiology of Salmonella enterica in poultry in South Africa using the farm-to-fork approach. Int. J. Microbiol. 2022, 2022, 5121273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yue, M.; Rankin, S.; Weill, F.X.; Frey, J.; Schifferli, D.M. One-step identification of five prominent chicken Salmonella serovars and biotypes. J. Clin. Microbiol. 2015, 53, 3881–3883. [Google Scholar] [CrossRef] [Green Version]
- Xiong, D.; Song, L.; Tao, J.; Zheng, H.; Zhou, Z.; Geng, S.; Pan, Z.; Jiao, X. An efficient multiplex PCR-based assay as a novel tool for accurate inter-serovar discrimination of Salmonella Enteritidis, S. Pullorum/Gallinarum and S. Dublin. Front. Microbiol. 2017, 8, 420. [Google Scholar] [CrossRef] [Green Version]
- Sutton, S. The most probable number method and its uses in enumeration, qualification, and validation. J. Valid. Technol. 2010, 16, 35–38. [Google Scholar]
- Rodgers, C.; Parveen, S.; Chigbu, P.; Jacobs, J.; Rhodes, M.; Harter-Dennis, J. Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland Coastal Bays. J. Appl. Microbiol. 2014, 117, 1198–1209. [Google Scholar] [CrossRef]
- Pauly, N.; Wichmann-Schauer, H.; Ballhausen, B.; Torres Reyes, N.; Fetsch, A.; Tenhagen, B.A. Detection and quantification of methicillin-resistant Staphylococcus aureus in fresh broiler meat at retail in Germany. Int. J. Food Microbiol. 2019, 292, 8–12. [Google Scholar] [CrossRef]
- Rao, M.; Tamber, S. Microbiological analysis of frozen profiteroles and mini chocolate eclairs implicated in a national salmonellosis outbreak. Food Microbiol. 2021, 100, 103871. [Google Scholar] [CrossRef]
- Jones, J.L.; Lüdeke, C.H.; Bowers, J.C.; DeRosia-Banick, K.; Carey, D.H.; Hastback, W. Abundance of Vibrio cholerae, V. vulnificus, and V. parahaemolyticus in oysters (Crassostrea virginica) and clams (Mercenaria mercenaria) from Long Island Sound. Appl. Environ. Microbiol. 2014, 80, 7667–7672. [Google Scholar] [CrossRef] [Green Version]
- Russo, P.; Botticella, G.; Capozzi, V.; Massa, S.; Spano, G.; Beneduce, L. A fast, reliable, and sensitive method for detection and quantification of Listeria monocytogenes and Escherichia coli O157:H7 in ready-to-eat fresh-cut products by MPN-qPCR. Biomed Res. Int. 2014, 2014, 608296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.A.; Park, S.H.; Lee, S.I.; Ricke, S.C. Development of a rapid method to quantify Salmonella Typhimurium using a combination of MPN with qPCR and a shortened time incubation. Food Microbiol. 2017, 65, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Wigley, P.; Hulme, S.; Powers, C.; Beal, R.; Smith, A.; Barrow, P. Oral infection with the Salmonella enterica serovar Gallinarum 9R attenuated live vaccine as a model to characterize immunity to fowl typhoid in the chicken. BMC Vet. Res. 2005, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Tao, J.; Jiao, Y.; Fei, X.; Zhou, L.; Wang, Y.; Zheng, H.; Pan, Z.; Jiao, X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 2016, 222, 56–64. [Google Scholar] [CrossRef]
- Li, Y.; Cai, Y.; Tao, J.; Kang, X.; Jiao, Y.; Guo, R.; Wang, G.; Pan, Z.; Jiao, X. Salmonella isolated from the slaughterhouses and correlation with pork contamination in free market. Food Control 2016, 59, 591–600. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Li, J.; Zheng, H.; Jin, X.; Shen, Y.; Lei, T.; Sun, X.; Pan, Z.; Jiao, X. Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian, China. Food Control 2017, 78, 238–246. [Google Scholar] [CrossRef]
- Costard, S.; Pouzou, J.G.; Belk, K.E.; Morley, P.S.; Schmidt, J.W.; Wheeler, T.L.; Arthur, T.M.; Zagmutt, F.J. No change in risk for antibiotic-resistant salmonellosis from beef, United States, 2002–2010. Emerg. Infect. Dis. 2020, 26, 2108–2117. [Google Scholar] [CrossRef]
- Parichehr, M.; Mohammad, K.; Abbas, D.; Mehdi, K. Developing a multiplex real-time PCR with a new pre-enrichment to simultaneously detect four foodborne bacteria in milk. Future Microbiol. 2019, 14, 885–898. [Google Scholar] [CrossRef]
- Siala, M.; Barbana, A.; Smaoui, S.; Hachicha, S.; Marouane, C.; Kammoun, S.; Gdoura, R.; Messadi-Akrout, F. Screening and detecting Salmonella in different food matrices in Southern Tunisia using a combined enrichment/real-time PCR method: Correlation with conventional culture method. Front. Microbiol. 2017, 8, 2416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, F.; Gao, J.; Zhang, W.; Xu, X. Development of multiplex TaqMan qPCR for simultaneous detection and differentiation of eight common swine viral and bacterial pathogens. Braz. J. Microbiol. 2021, 53, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Y.; Kang, X.L.; Meng, C.; Xiong, D.; Xu, Y.; Geng, S.Z.; Pan, Z.M.; Jiao, X.A. Multiple PCR assay based on the cigR gene for detection of Salmonella spp. and Salmonella Pullorum/Gallinarum identification. Poult. Sci. 2020, 99, 5991–5998. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain a | Serovar/Species | Source | Duplex PCR Results | ||
|---|---|---|---|---|---|
| lygD | invA | ||||
| Salmonella | C50041 | Enteritidis | Laboratory stock | + | + |
| C50336 | Enteritidis | Laboratory stock | + | + | |
| Z11 | Enteritidis | Laboratory stock | + | + | |
| Pi9 | Enteritidis | Isolate from pig | + | + | |
| Ch17 | Enteritidis | Isolate from chicken | + | + | |
| S06004 | Pullorum | Laboratory stock | – | + | |
| 6508 | Pullorum | Isolate from chicken | – | + | |
| SG9 | Gallinarum | [35] | – | + | |
| SL5928 | Dublin | Laboratory stock | – | + | |
| T3 | Uganda | [36] | – | + | |
| T9 | Meleagridis | [37] | – | + | |
| T8 | Anatis | [37] | – | + | |
| G2 | London | [36] | – | + | |
| Pi16 | London | Laboratory stock | – | + | |
| ZX | Rissen | [36] | – | + | |
| Y7 | Derby | [36] | – | + | |
| Pi12 | Derby | Isolate from pig | – | + | |
| ZHJ5 | Derby | Laboratory stock | – | + | |
| Y8 | Typhimurium | [37] | – | + | |
| Pi14 | Typhimurium | Laboratory stock | – | + | |
| Pi24 | Typhimurium | Laboratory stock | – | + | |
| C500 | Choleraesuis | Laboratory stock | – | + | |
| ZH65 | Indiana | [36] | – | + | |
| ZH5 | Sinstorf | Laboratory stock | – | + | |
| ZH10 | Newlands | Isolate from cattle | – | + | |
| ZZH24 | Muenster | Laboratory stock | – | + | |
| ZH82 | Yoruba | Isolate from pig | – | + | |
| G449 | Dumfries | Laboratory stock | – | + | |
| G241 | Kentucky | Laboratory stock | – | + | |
| G382 | Agona | Laboratory stock | – | + | |
| ZMH35 | Newport | Laboratory stock | – | + | |
| TJ42 | Thompson | [36] | – | + | |
| Ch15 | Thompson | Laboratory stock | – | + | |
| P192 | Senftenberg | Laboratory stock | – | + | |
| G439 | Blockley | Laboratory stock | – | + | |
| G86 | Inchpark | Laboratory stock | – | + | |
| P122 | Virchow | Laboratory stock | – | + | |
| P74 | Farsta | Laboratory stock | – | + | |
| G85 | Dabou | Laboratory stock | – | + | |
| GS3 | Potsdam | Laboratory stock | – | + | |
| Non-Salmonella | H37Rv | Mycobacterium tuberculosis | ATCC 27294 | – | – |
| 11168 | Campylobacter jejuni | ATCC 700819 | – | – | |
| TH5 | Campylobacter jejuni | Isolate from chicken | – | – | |
| cj18 | Campylobacter jejuni | Laboratory stock | – | – | |
| S19 | Brucella abortus | Laboratory stock | – | – | |
| 51592 | Shigella flexneri | Laboratory stock | – | – | |
| EGDe | Listeria monocytogenes | ATCC BAA-679 | – | – | |
| LM23 | Listeria monocytogenes | Laboratory stock | – | – | |
| 1314 | Escherichia coli | Isolate from chicken | – | – | |
| E10 | Escherichia coli | Laboratory stock | – | – | |
| 8-1-6 | Escherichia coli | Isolate from chicken | – | – | |
| 27217 | Staphylococcus aureus | ATCC 27217 | – | – | |
| Gene | Primer Name | Sequence (5′-3′) a | Amplicon Size (bp) | Location |
|---|---|---|---|---|
| lygD | lygD-F | CTTTCTCAGATTCAGGGAGTATATCA | 111 | CP013097.1 1469293–1469403 |
| lygD-R | GTTCTTCTGGTACTTACGATGACAAC | |||
| lygD-P | Cy5-CCTGTTGTCTGCTCACCATTCGCC-BHQ2 | |||
| invA | invA-F | GCGTTCTGAACCTTTGGTAATAA | 104 | CP013097.1 2915046–2915149 |
| invA-R | CGTTCGGGCAATTCGTTA | |||
| invA-P | FAM-TGGCGGTGGGTTTTGTTGTCTTCT-TAMRA |
| Concentration of Template (Copies/μL) | Intra-Assay Reproducibility | Inter-Assay Reproducibility | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean Ct ± SD | CV (%) | Mean Ct ± SD | CV (%) | |||||
| lygD | invA | lygD | invA | lygD | invA | lygD | invA | |
| 4 × 106 | 14.97 ± 0.2177 | 14.59 ± 0.2059 | 1.45 | 1.41 | 14.75 ± 0.3277 | 14.43 ± 0.3670 | 2.22 | 2.54 |
| 4 × 105 | 18.24 ± 0.2303 | 17.97 ± 0.2878 | 1.26 | 1.60 | 18.13 ± 0.3571 | 17.74 ± 0.3541 | 1.97 | 2.00 |
| 4 × 104 | 21.68 ± 0.4199 | 21.21 ± 0.0367 | 1.94 | 0.17 | 21.63 ± 0.1721 | 21.12 ± 0.2388 | 0.80 | 1.13 |
| 4 × 103 | 25.55 ± 0.1740 | 24.79 ± 0.0759 | 0.68 | 0.31 | 25.32 ± 0.3970 | 24.62 ± 0.1324 | 1.57 | 0.54 |
| 4 × 102 | 29.59 ± 0.0991 | 29.16 ± 0.1973 | 0.33 | 0.68 | 29.04 ± 0.4371 | 28.91 ± 0.4013 | 1.51 | 1.39 |
| 4 × 101 | 33.19 ± 0.3454 | 32.74 ± 0.1991 | 1.04 | 0.61 | 32.90 ± 0.2988 | 32.65 ± 0.2808 | 0.91 | 0.86 |
| Sample | Real-Time PCR | Traditional Serotyping | Sample | Real-Time PCR | Traditional Serotyping | ||
|---|---|---|---|---|---|---|---|
| lygD | invA | lygD | invA | ||||
| A1 | + | + | SE | C12 | – | + | SL |
| A2 | – | – | – | C13 | – | – | – |
| A3 | + | + | SE | C14 | – | – | – |
| A4 | + | + | SE | E1 | – | – | – |
| A5 | + | + | SE | E2 | + | + | SE |
| A6 | + | + | SE | E3 | – | – | – |
| A7 | + | + | SE | E4 | – | – | – |
| A8 | + | + | – | E5 | – | – | – |
| A9 | + | + | SE | E6 | – | – | – |
| A10 | + | + | SE | E7 | – | – | – |
| A11 | + | + | SE | E8 | – | – | – |
| A12 | + | + | SE | E9 | + | + | SE |
| A13 | + | + | SE | E10 | + | + | SE |
| B1 | – | – | – | E11 | – | – | – |
| B2 | – | – | – | E12 | – | + | SL |
| B3 | – | – | – | E13 | – | – | – |
| B4 | + | + | SE | E14 | + | + | SE |
| B5 | – | – | – | E15 | + | + | SE |
| B6 | – | – | – | E16 | – | – | – |
| B7 | + | + | SE | F1 | – | – | – |
| B8 | – | – | – | F2 | – | – | – |
| B9 | + | + | SE | F3 | – | – | – |
| B10 | – | – | – | F4 | – | – | – |
| B11 | – | – | – | F5 | + | + | SE |
| B12 | – | – | – | F6 | – | – | – |
| C1 | – | – | – | F7 | – | – | – |
| C2 | – | – | – | F8 | – | – | – |
| C3 | + | + | – | F9 | – | – | – |
| C4 | – | – | – | F10 | – | – | – |
| C5 | – | + | SW | F11 | + | + | – |
| C6 | – | – | – | F12 | – | – | – |
| C7 | – | – | – | F13 | – | – | – |
| C8 | + | + | SE | F14 | – | + | SL |
| C9 | + | + | SE | F15 | – | – | – |
| C10 | – | – | – | Total | 26/70 | 30/70 | 23/70 (SE) 1/70 (SW) 3/70 (SL) |
| C11 | + | + | SE | ||||
| Trial | MPN-qPCR-SIT (log MPN/mL) | Traditional MPN (log MPN/mL) | Traditional Plating (log CFU/mL) | Real-Time PCR (log Copies/mL) | ||||
|---|---|---|---|---|---|---|---|---|
| MPN | Lower Limit of the 95% CI | Upper Limit of the 95% CI | MPN | Lower Limit of the 95% CI | Upper Limit of the 95% CI | |||
| 1 | 0.64 | −1.33 | 2.60 | 0.67 | −1.74 | 3.08 | ND | ND |
| 2 | 0.68 | −0.79 | 2.14 | 0.72 | −1.25 | 2.68 | ND | ND |
| 3 | 0.80 | −2.25 | 3.85 | 0.76 | −1.78 | 3.30 | 1.00 | ND |
| 4 | 0.96 | −0.13 | 2.04 | 0.92 | 0.34 | 1.49 | 1.48 | 1.22 |
| 5 | 1.09 | −1.65 | 3.82 | 0.87 | −3.07 | 4.81 | 1.65 | 1.29 |
| 6 | 1.47 | 0.07 | 2.87 | 1.50 | −0.22 | 3.21 | 1.95 | 1.65 |
| 7 | 2.18 | −0.43 | 4.78 | 1.97 | 1.97 | 1.97 | 2.26 | 1.80 |
| 8 | 2.85 | 0.44 | 5.26 | 2.71 | −1.48 | 6.90 | 2.72 | 2.52 |
| 9 | 2.86 | 0.50 | 5.21 | 2.85 | 0.44 | 5.26 | 3.03 | 2.59 |
| 10 | 2.68 | −1.89 | 7.25 | 2.85 | 0.44 | 5.26 | 3.26 | 2.76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, D.; Zhou, Y.; Song, L.; Liu, B.; Matchawe, C.; Chen, X.; Pelle, R.; Jiao, X.; Pan, Z. Development of a Duplex TaqMan Real-Time Polymerase Chain Reaction for Accurate Identification and Quantification of Salmonella Enteritidis from Laboratory Samples and Contaminated Chicken Eggs. Foods 2022, 11, 742. https://doi.org/10.3390/foods11050742
Xiong D, Zhou Y, Song L, Liu B, Matchawe C, Chen X, Pelle R, Jiao X, Pan Z. Development of a Duplex TaqMan Real-Time Polymerase Chain Reaction for Accurate Identification and Quantification of Salmonella Enteritidis from Laboratory Samples and Contaminated Chicken Eggs. Foods. 2022; 11(5):742. https://doi.org/10.3390/foods11050742
Chicago/Turabian StyleXiong, Dan, Yi Zhou, Li Song, Bowen Liu, Chelea Matchawe, Xiang Chen, Roger Pelle, Xinan Jiao, and Zhiming Pan. 2022. "Development of a Duplex TaqMan Real-Time Polymerase Chain Reaction for Accurate Identification and Quantification of Salmonella Enteritidis from Laboratory Samples and Contaminated Chicken Eggs" Foods 11, no. 5: 742. https://doi.org/10.3390/foods11050742
APA StyleXiong, D., Zhou, Y., Song, L., Liu, B., Matchawe, C., Chen, X., Pelle, R., Jiao, X., & Pan, Z. (2022). Development of a Duplex TaqMan Real-Time Polymerase Chain Reaction for Accurate Identification and Quantification of Salmonella Enteritidis from Laboratory Samples and Contaminated Chicken Eggs. Foods, 11(5), 742. https://doi.org/10.3390/foods11050742






