Study on the Effect of Key Genes ME2 and adhE during Luzhou-Flavor Baijiu Brewing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Procedure
2.2. Screening Media Prediction
2.3. Enrichment and Screening of Functional Microorganisms in Pit Mud
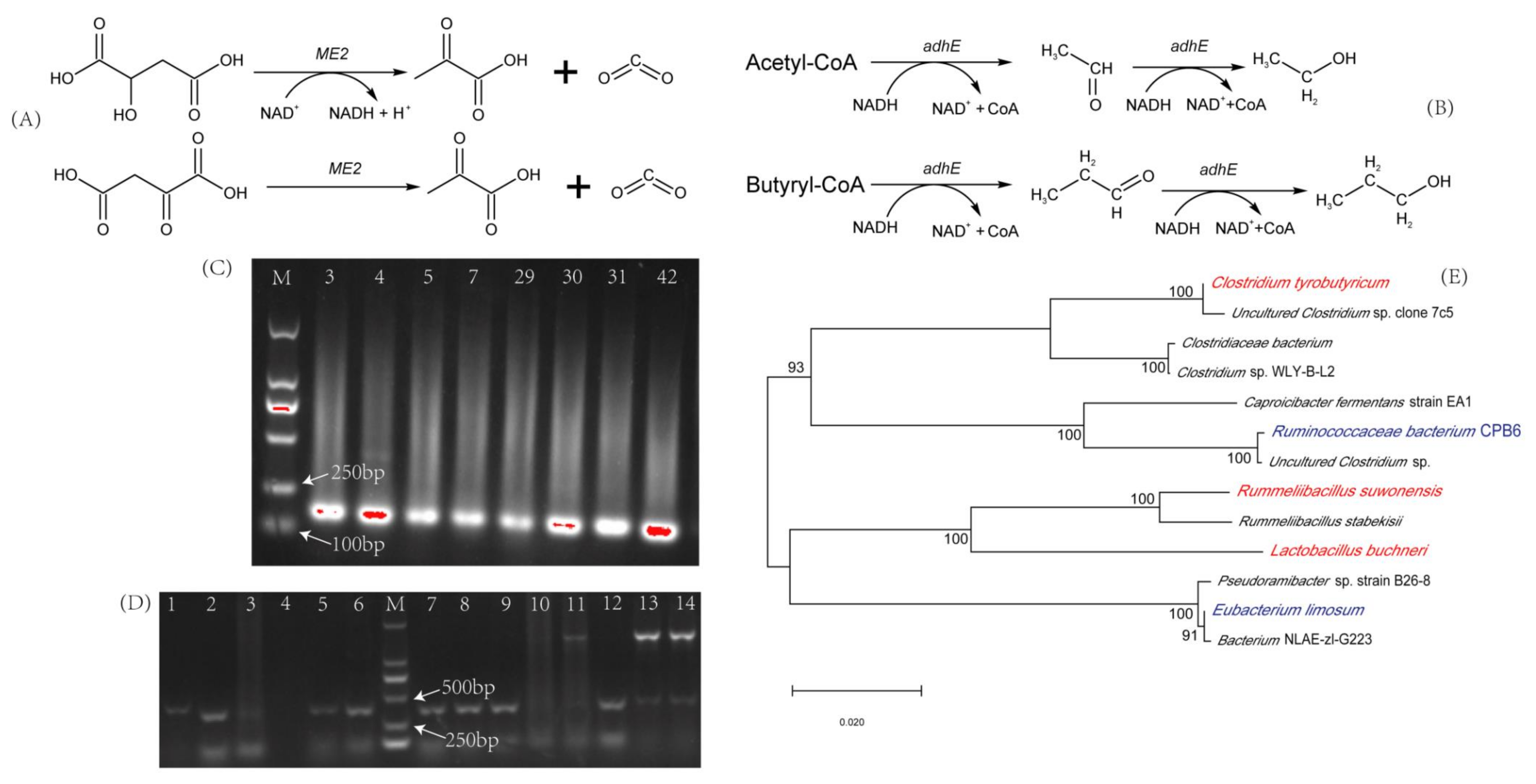
2.4. Colony PCR
2.5. Functional Strains Identification and Phylogenetic Tree Construction
2.6. Fermentation Test of Functional Microorganisms
2.7. Quantify the Expression Quantity of Gene ME2 and adhE
2.8. Detection of Volatile Flavor Substances
2.9. Detection of “Four Dominant Acids”
2.10. Statistical Analysis
3. Results
3.1. Screening Functional Microorganisms
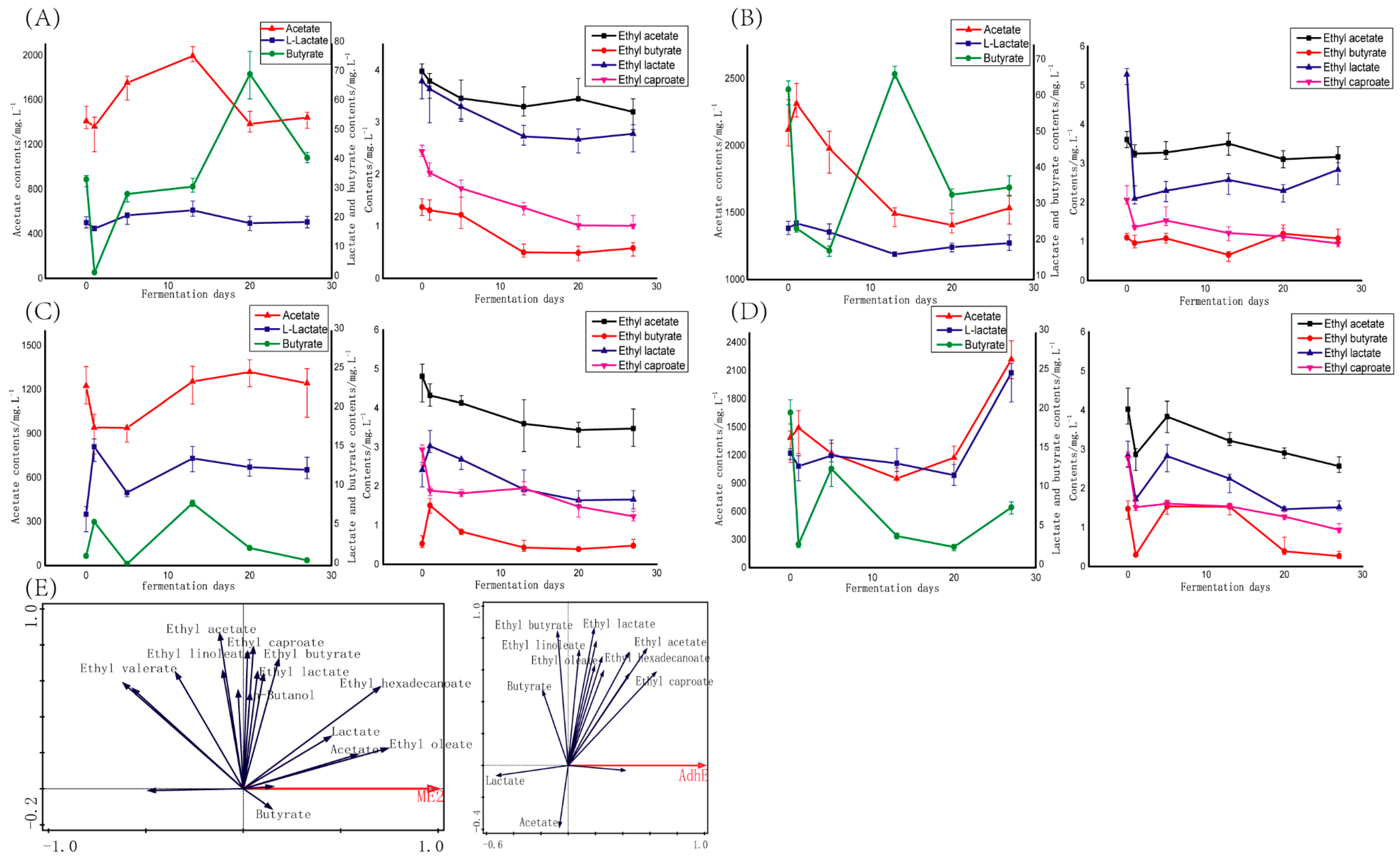
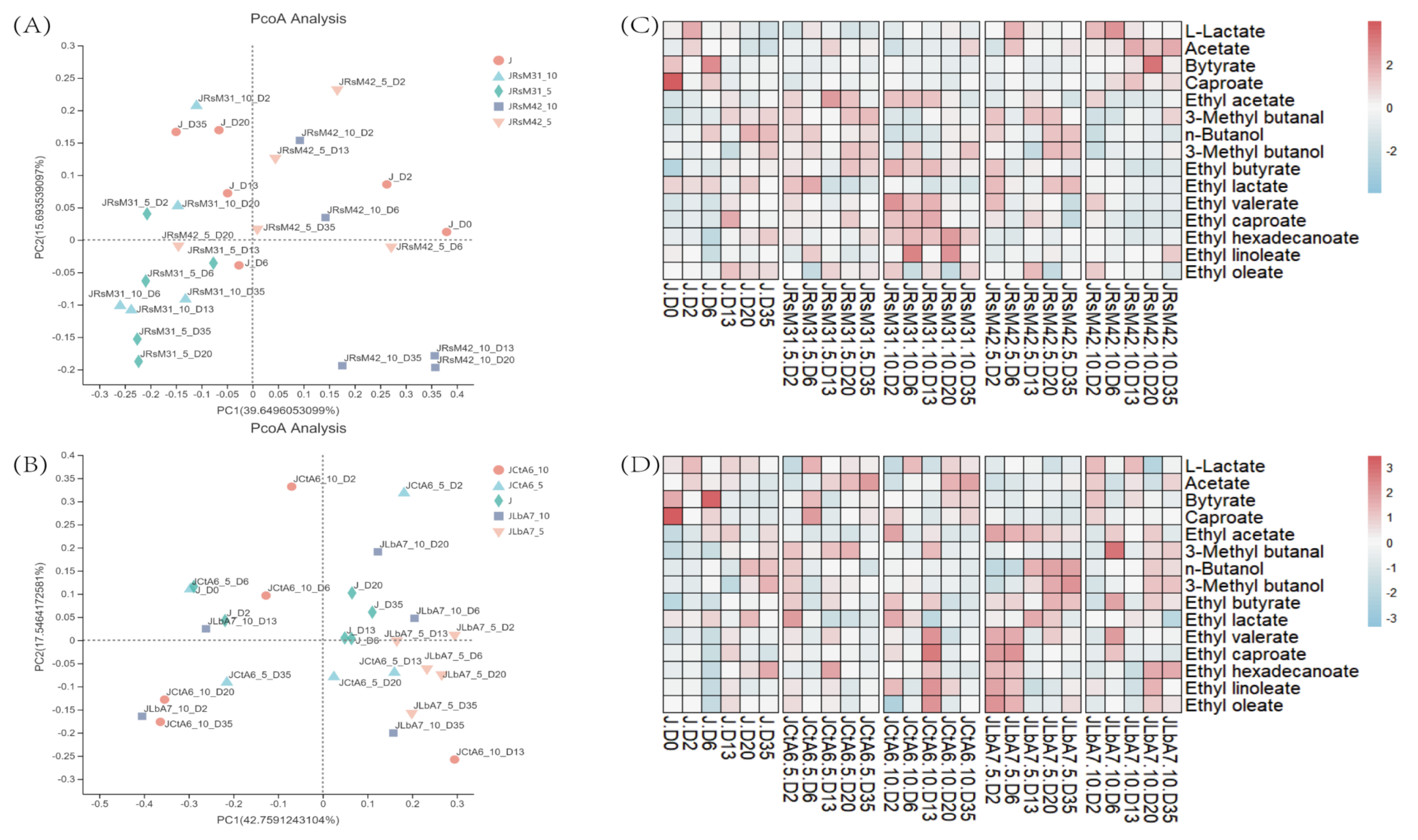
3.2. Liquid Fermentation of Functional Microorganisms
3.3. Solid Fermentation of Functional Microorganisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.X.; Wu, Z.Y.; Zhang, Q.S.; Wang, R.; Li, H. Combination of newly developed high quality Fuqu with traditional Daqu for Luzhou-flavor liquor brewing. World J. Microbiol. Biotechnol. 2009, 25, 1721–1726. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Yuan, Y.J.; Luo, W.; Zeng, L.Y.; Wu, Z.Y.; Zhang, W.X. Characterization of Prokaryotic Community Diversity in New and Aged Pit Muds from Chinese Luzhou-flavor Liquor Distillery. Food Sci. Technol. Res. 2017, 23, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, B.; Dong, H.; Huang, X.; Chen, R.; Chen, X.; Yang, L.; Peng, B.; Xie, G.; Cheng, W.; et al. Complete Genome Sequence of Clostridium kluyveri JZZ Applied in Chinese Strong-Flavor Liquor Production. Curr. Microbiol. 2018, 75, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wang, X.; Li, X.Z.; Wei, N.; Jin, H. The functional potential and active populations of the pit mud microbiome for the production of Chinese strong-flavor liquor. Microb. Biotechnol. 2017, 10, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.W.; Wu, Q.; Xu, Y.; Sun, B.G. Specific Volumetric Weight-Driven Shift in Microbiota Compositions with Saccharifying Activity Change in Starter for Chinese Baijiu Fermentation. Front. Microbiol. 2018, 9, 2349. [Google Scholar] [CrossRef]
- Sun, W.N.; Xiao, H.Z.; Peng, Q.; Zhang, Q.; Li, X.; Han, Y. Analysis of bacterial diversity of Chinese Luzhou-flavor liquor brewed in different seasons by Illumina Miseq sequencing. Ann. Microbiol. 2016, 66, 1293–1301. [Google Scholar] [CrossRef]
- Zhao, C.Q.; Yan, X.L.; Yang, S.T.; Chen, F.F. Screening of Bacillus strains from Luzhou-flavor liquor making for high-yield ethyl hexanoate and low-yield propanol. LWT Food Sci. Technol. 2017, 77, 60–66. [Google Scholar] [CrossRef]
- Huang, X.N.; Fan, Y.; Meng, J.; Sun, S.; Han, B.Z. Laboratory-scale fermentation and multidimensional screening of lactic acid bacteria from Daqu. Food Biosci. 2021, 40, 100853. [Google Scholar] [CrossRef]
- Guo, J.H.; Jia, S.R. Effect of Cellulase-Producing Bacteria on Enzyme Activity and Ester Production in Fermented Grains of Chinese Liquor. J. Am. Soc. Brew Chem. 2015, 73, 130–136. [Google Scholar] [CrossRef]
- Xu, J.L.; Sun, L.P.; Xing, X.; Sun, Z.; Ren, Q. Culturing Bacteria from Fermentation Pit Muds of Baijiu with Culturomics and Amplicon-Based Metagenomic Approaches. Front. Microbiol. 2020, 11, 1223. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Huang, D.; Zhang, W.X. Combining culture-dependent and culture- independent molecular methods for the isolation and purification of a potentially novel anaerobic species from pit mud in a Chinese liquor distillery. J. Brew. 2016, 122, 754–762. [Google Scholar] [CrossRef]
- Chai, L.J.; Lu, Z.M.; Zhang, X.J.; Ma, J. Zooming in on Butyrate-Producing Clostridial Consortia in the Fermented Grains of Baijiu via Gene Sequence-Guided Microbial Isolation. Front. Microbiol. 2019, 10, 1397. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liao, Z.M.; Wu, Z.Y.; Taikei, S.; Zhang, W.X. Analysis of the difference between aged and degenerated pit mud microbiome in fermentation cellars for Chinese Luzhou-flavor baijiu by metatranscriptomics. J. Sci. Food Agric. 2021, 101, 4621–4631. [Google Scholar] [CrossRef]
- Thompsom, H.; Tersteegen, A.; Thauer, R.K.; Hedderich, R. Two malate dehydrogenases in Methanobacterium thermoautotrophicum. Arch Microbiol. 1998, 170, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Bartholomae, M.; Meyer, F.M.; Commichau, F.M.; Burkovski, A.; Hillen, W. Complex formation between malate dehydrogenase and isocitrate dehydrogenase from Bacillussubtilis is regulated by tricarboxylic acid cycle metabolites. FEBS J. 2014, 281, 132–1143. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.W.; Lipscomb, G.L.; Nguyen, D.M.; Crowley, A.T.; Schut, G.T.; Scott, I.; Kelly, R.M.; Adams, M.W. Ethanol production by the hyperthermophilic archaeon Pyrococcus furiosus by expression of bacterial bifunctional alcohol dehydrogenases. Microb. Biotechnol. 2017, 6, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kasahara, K.; Hirose, Y.; Morimoto, Y.; Izawa, M.; Ochi, K. Enhancement of butanol production by sequential introduction of mutations conferring butanol tolerance and streptomycin resistance. J. Biosci. Bioeng. 2017, 4, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zong, W.M.; Hong, W.; Zhang, Z.T.; Wang, Y. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab. Eng. 2018, 47, 49–59. [Google Scholar] [CrossRef]
- Zhu, J.B.; Long, M.N.; Xu, F.C.; Wu, X.B.; Xu, H.J. Enhanced hydrogen production by insertional inactivation of adhE gene in Klebsiella oxytoca HP1. Chin. Sci. Bull. 2007, 52, 492–496. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Li, J.W.; He, H.K.; Cao, R.J.; Li, A.J.; Zhang, Z.Z. Significant difference of the culturable anaerobic microbial species in GujingTribute pitmud with different ages. In Proceedings of the 5th International Conference on Advanced Composite Materials and Manufacturing Engineering, Xishuangbanna, China, 16–17 June 2018; Volume 394, p. 022025. [Google Scholar]
- Lu, S.W.; Jin, H.; Wang, Y.; Tao, Y. Genome-wide transcriptomic analysis of n-caproic acid production in Ruminococcaceae bacterium CPB6 with lactate supplementation. J. Microbiol. Biotechnol. 2021, 31, 1533–1544. [Google Scholar] [CrossRef] [PubMed]
- Her, J.; Kim, J. Rummeliibacillus suwonensis sp nov. Isolated from Soil Collected in a Mountain Area of South Korea. J. Microbiol. 2013, 51, 268–272. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.; Fan, X.J.; Qin, Y.H.; Lv, Y.K. Draft Genome Sequence of Rummeliibacillus sp. Strain TYF005, a Physiologically Recalcitrant Bacterium with High Ethanol and Salt Tolerance Isolated from Spoilage Vinegar. Microbiol. Resour. Announc. 2019, 8, e00244-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Yasin, M.; Jeong, J.; Cha, M.; Kang, H.; Jang, N.; Choi, I.G.; Chang, I.S. Acetate-assisted increase of butyrate production by Eubacterium limosum KIST612 during carbon monoxide fermentation. Bioresour. Technol. 2017, 245, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Komori, K.; Ohkubo, Y.; Katano, N.; Motoshima, H. One year investigation of the prevalence and diversity of clostridial spores in raw milk from the Tokachi area of Hokkaido. Anim. Sci. J. 2019, 90, 135–139. [Google Scholar] [CrossRef]
- Li, R.R.; Jiang, D.; Zheng, M.L.; Xu, C. Microbial community dynamics during alfalfa silage with or without clostridial fermentation. Sci. Rep. 2020, 10, 17782. [Google Scholar] [CrossRef] [PubMed]
- Podrzaj, L.; Burtscher, J.; Domig, K.J. Draft Genome Sequences of 12 Clostridium tyrobutyricum Strains Isolated from Raw Milk and Cheese. Microbiol. Resour. Announc. 2021, 10, e0073521. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Lin, M.; Jiang, W.Y.; Zhao, J.B.; Li, W.M.; Yang, S.T. Development of an in vivo fluorescence based gene expression reporter system for Clostridium tyrobutyricum. J. Biotechnol. 2019, 305, 18–22. [Google Scholar] [CrossRef] [PubMed]
- He, F.F.; Qin, S.W.; Yang, Z.; Bai, X.H.; Sun, Y.K.; Wang, J.F. Butyric acid production from spent coffee grounds by engineered Clostridium tyrobutyricum overexpressing galactose catabolism genes. Bioresour. Technol. 2020, 34, 122977. [Google Scholar] [CrossRef]
- Rabelo, C.H.S.; Harter, C.J.; Avila, C.L.D.; Reis, R.A. Meta-analysis of the effects of Lactobacillus plantarum and Lactobacillus buchneri on fermentation, chemical composition and aerobic stability of sugarcane silage. Grassl. Sci. 2019, 65, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Cheon, M.J.; Lim, S.M.; Lee, N.K.; Paik, H.D. Probiotic Properties and Neuroprotective Effects of Lactobacillus buchneri KU200793 Isolated from Korean Fermented Foods. Int. J. Mol. Sci. 2020, 21, 1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.Q.; Skory, C.; Qureshi, N. Ethanol tolerance assessment in recombinant E. coli of ethanol responsive genes from Lactobacillus buchneri NRRL B-30929. World J. Microb. Biot. 2020, 36, 179. [Google Scholar] [CrossRef] [PubMed]
| Gene | Reagent | Volume of Addition | Primer | PCR Procedure |
|---|---|---|---|---|
| ME2 | 2 × T5 Super PCR Mix (Colony) (Tsingke Biotechnology Co., Ltd.) | 12.5 µL | Forward Primer:(ME2-F): 5′-CTATTGCGAAGCACCTG-3′ Reverse Primer (ME2-R): 5′-AAACTCCCCTGTTTATGTT-3′ Product length: 148 bp | Initial denaturation at 98 °C for 180 s; followed by 32 cycles of denaturation at 98 °C for 10 s, annealing at 49 °C for 30 s and elongation at 72 °C for 30 s; then final elongation at 72 °C for 120 s. |
| Forward Primer (ME2-F, 10 µM) | 1 µL | |||
| Reverse Primer (ME2-R, 10 µM) | 1 µL | |||
| Template | 2 µL | |||
| ddH2O up to | 25 µL | |||
| adhE | 2 × T5 Super PCR Mix (Colony) (Tsingke Biotechnology Co., Ltd.) | 12.5 µL | Forward Primer (adhE-F): 5-GATGCTTTGATTGCCCTTGG-3′ Reverse Primer (adhE-R): 5-AAACGGGTTGTTGTTGGTG-3′ Product length: 354 bp | Initial denaturation at 98 °C for 180 s; followed by 32 cycles of denaturation at 98 °C for 10 s, annealing at 58 °C for 30 s and elongation at 72 °C for 30 s; then final elongation at 72 °C for 120 s. |
| Forward Primer (adhE-F, 10 µM) | 1 µL | |||
| Reverse Primer (adhE-R, 10 µM) | 1 µL | |||
| Template | 2 µL | |||
| ddH2O up to | 25 µL |
| Gene | Primer | Sequence 5’-3’ | Annealing Temperature | Products Length |
|---|---|---|---|---|
| adhE | adhE-2F | GTATTCCAAATGTCAGCG | 48.6 °C | 73 bp |
| adhE-2R | TTGATTTCTTTACAGAGGGT | 49.0 °C | ||
| ME2 | ME2-F | CTATTGCGAAGCACCTG | 49.6 °C | 148 bp |
| ME2-R | AAACTCCCCTGTTTATGTT | 48.6 °C |
| Genes | Strains | Key Genes Expression Quantities (log10 Copies/mL) | |||||
|---|---|---|---|---|---|---|---|
| 0 d | 1 d | 5 d | 13 d | 20 d | 27 d | ||
| ME2 | RsM31 | 5.96 ± 0.02 f | 5.50 ± 0.02 g | 4.58 ± 0.02 h | 4.13 ± 0.02 i | 3.84 ± 0.02 j | 3.64 ± 0.01 k |
| RsM42 | 6.90 ± 0.01 a | 6.86 ± 0.01 ab | 6.81 ± 0.01 b | 6.72 ± 0.01 c | 6.64 ± 0.01 d | 6.50 ± 0.01 e | |
| adhE | CtA6 | 12.30 ± 0.03 a | 11.44 ± 0.02 b | 11.33 ± 0.01 c | 11.22 ± 0.02 d | 11.21 ± 0.01 d | 11.20 ± 0.01 d |
| LbA7 | 11.48 ± 0.01 b | 11.09 ± 0.01 e | 10.99 ± 0.01 f | 10.93 ± 0.01 g | 10.91 ± 0.01 g | 10.91 ± 0.01 g | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, W.; Xia, Y.; Zhao, Y.; Wang, Y.; Wu, Z.; Suyama, T.; Zhang, W. Study on the Effect of Key Genes ME2 and adhE during Luzhou-Flavor Baijiu Brewing. Foods 2022, 11, 700. https://doi.org/10.3390/foods11050700
Zhou W, Xia Y, Zhao Y, Wang Y, Wu Z, Suyama T, Zhang W. Study on the Effect of Key Genes ME2 and adhE during Luzhou-Flavor Baijiu Brewing. Foods. 2022; 11(5):700. https://doi.org/10.3390/foods11050700
Chicago/Turabian StyleZhou, Wen, Yu Xia, Yajiao Zhao, Yan Wang, Zhengyun Wu, Taikei Suyama, and Wenxue Zhang. 2022. "Study on the Effect of Key Genes ME2 and adhE during Luzhou-Flavor Baijiu Brewing" Foods 11, no. 5: 700. https://doi.org/10.3390/foods11050700
APA StyleZhou, W., Xia, Y., Zhao, Y., Wang, Y., Wu, Z., Suyama, T., & Zhang, W. (2022). Study on the Effect of Key Genes ME2 and adhE during Luzhou-Flavor Baijiu Brewing. Foods, 11(5), 700. https://doi.org/10.3390/foods11050700





