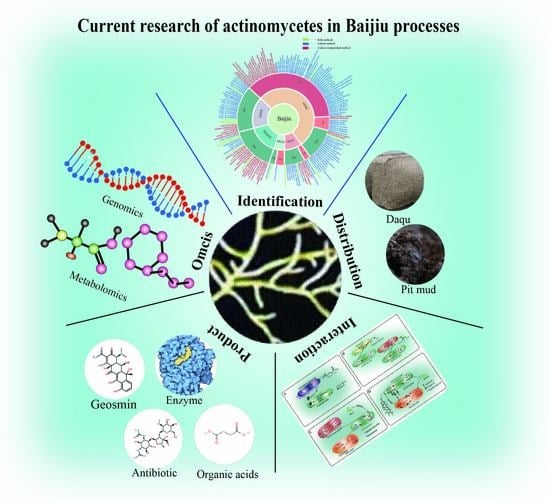
Systematic Review of Actinomycetes in the Baijiu Fermentation Microbiome
Abstract
1. Introduction
2. Isolation and Identification of Actinomycetes from Different Stages of Baijiu Production
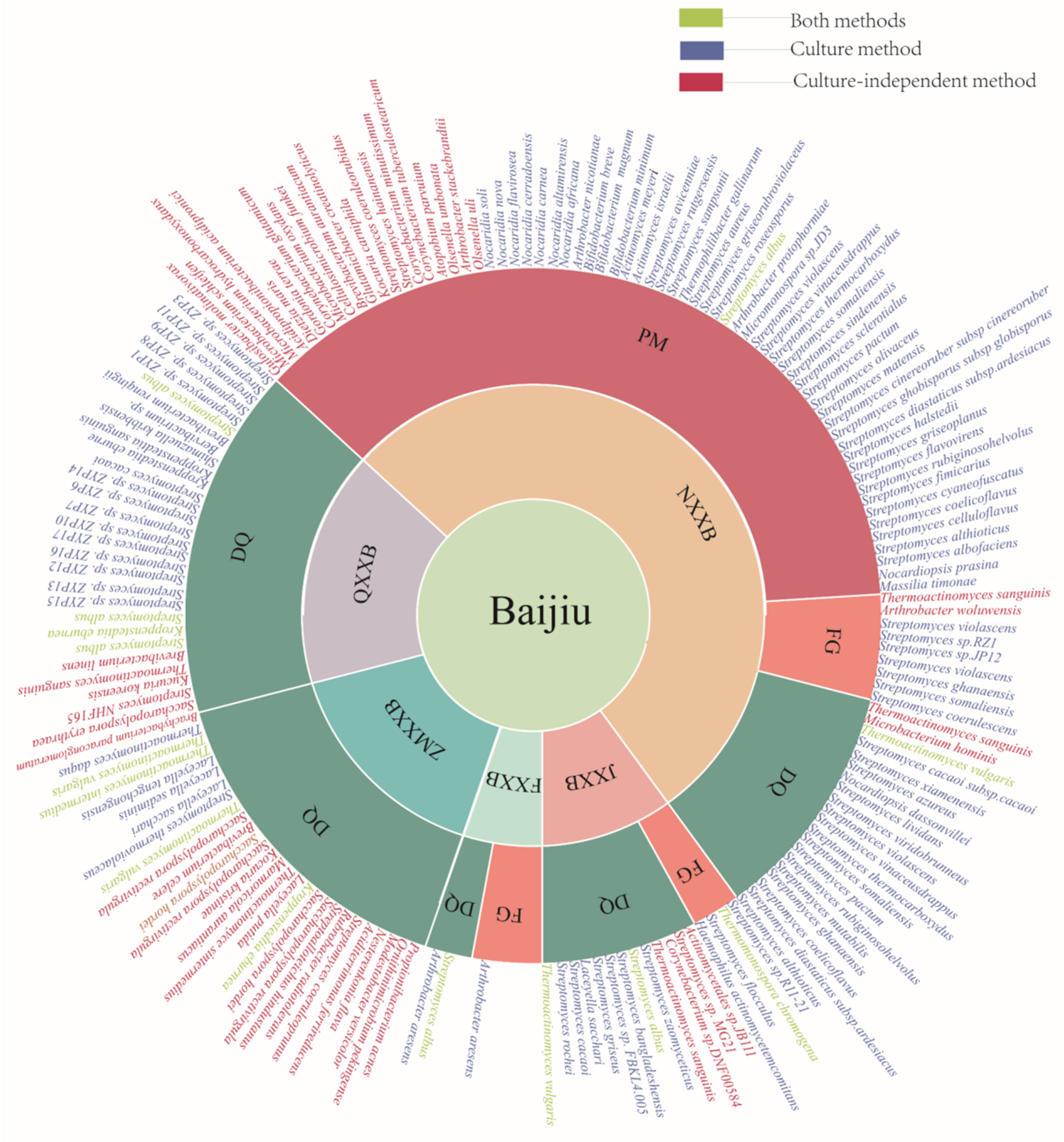
3. Methods for Identification of New Species of Microorganisms from Baijiu Production
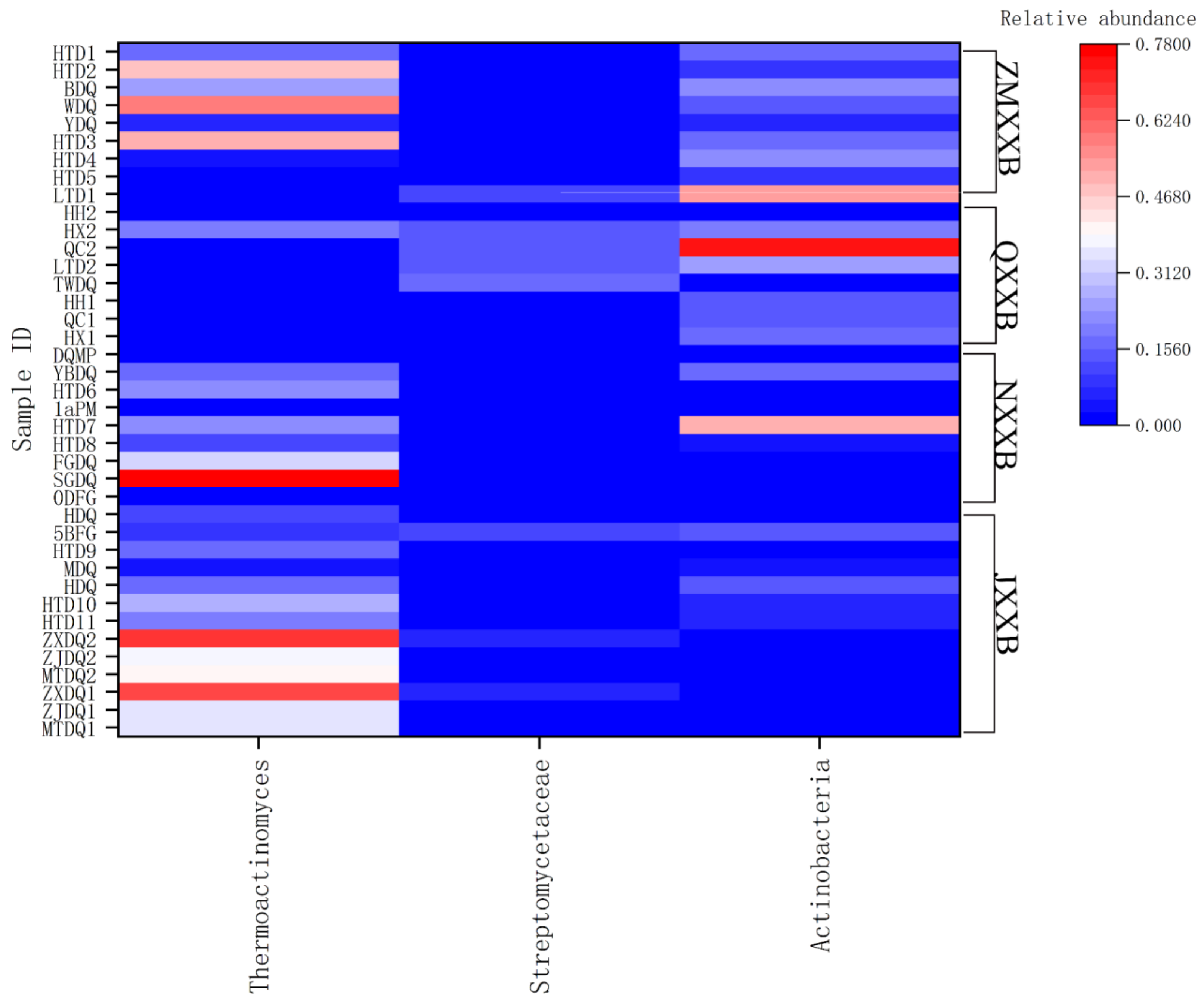
4. Distribution of Actinomycete Species from the Major Types of Baijiu
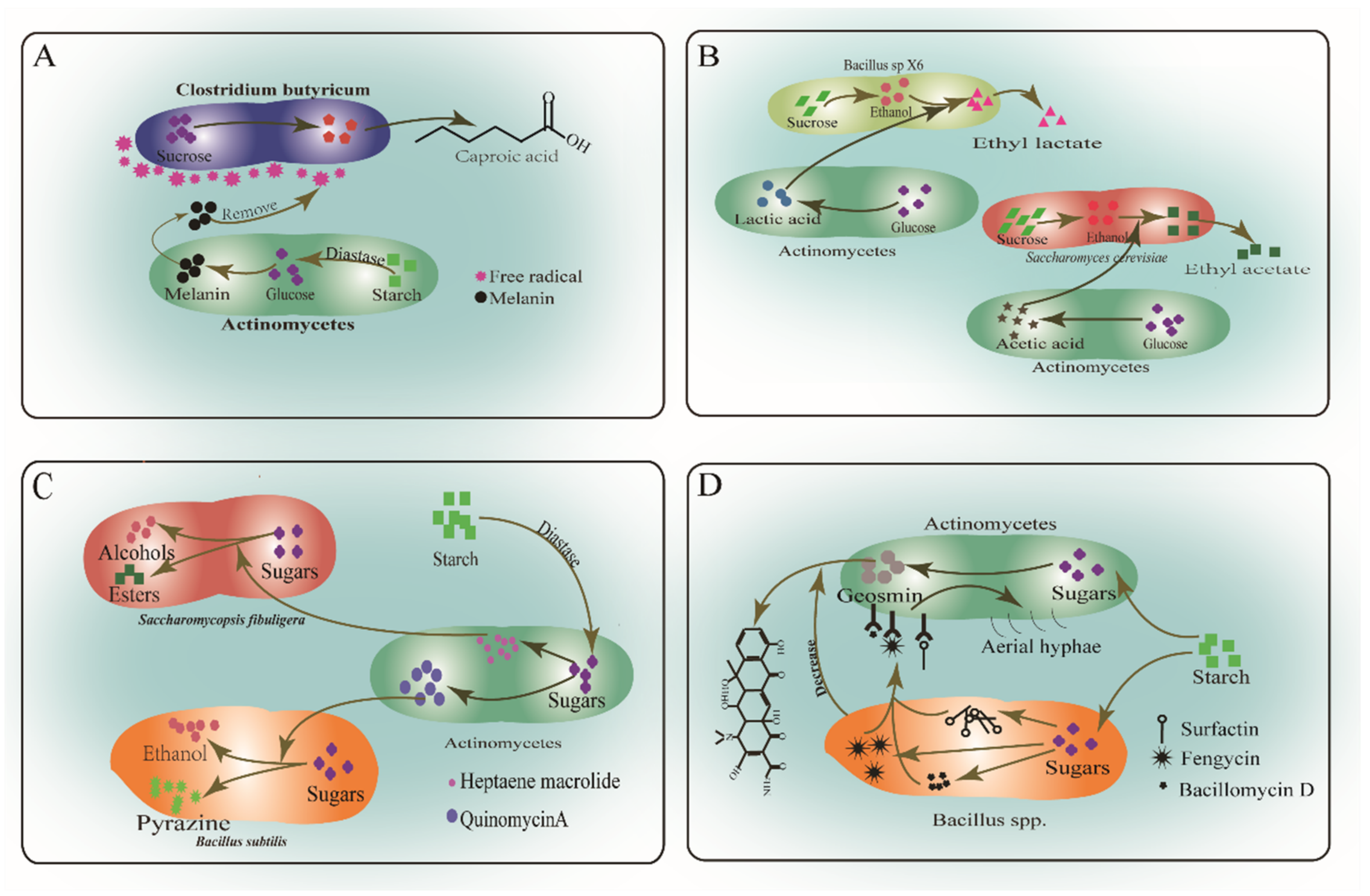
5. Interspecies Interactions of Actinomycetes and Other Microorganisms in Baijiu Fermentation
6. Actinomycete ‘Omics Research
7. Enzymes and Metabolites Produced by Baijiu Actinomycetes
8. Potential Applications of Actinomycetes in Baijiu Fermentation
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zheng, X.W.; Han, B.Z. Baijiu, Chinese liquor: History, classification and manufacture. J. Ethn. Foods 2016, 3, 19–25. [Google Scholar] [CrossRef]
- Xu, Y.R.; Wang, D.; Fan, W.L.; Mu, X.Q.; Chen, J. Traditional Chinese biotechnology. In Biotechnology in China II: Chemicals, Energy and Environment; Tsao, T.G., Ouyang, P., Chen, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 189–233. [Google Scholar]
- Ding, X.F.; Wu, C.; Zhang, L.Q.; Zheng, J.; Zhou, R.Q. Characterization of eubacterial and archaeal community diversity in the pit mud of Chinese Luzhou-flavor liquor by nested PCR DGGE. World J. Microbiol. Biotechnol. 2014, 30, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.S.; Zheng, J.; Zhou, R.Q.; Shi, B. Microbial community structure of pit mud in a Chinese strong aromatic liquor fermentation pit. J. Inst. Brew. 2012, 118, 356–360. [Google Scholar] [CrossRef]
- Zou, W.; Ye, G.B.; Zhang, K.Z. Diversity, Function, and Application of Clostridium in Chinese Strong Flavor Baijiu Ecosystem: A Review. J. Food Sci. 2018, 83, 1193–1199. [Google Scholar] [CrossRef]
- Zhao, L.; Mo, X.L.; Zhang, C.L.; Yang, L.; Wang, X.Y. Community diversity and succession in fermented grains during the stacking fermentation of Chinese moutai-flavored liquor making. Food Sci. Technol. 2021, 42, e61521. [Google Scholar] [CrossRef]
- Ma, S.Y.; Luo, H.B.; Zhao, D.; Qiao, Z.W.; Zheng, J.; An, M.Z.; Huang, D. Environmental factors and interactions among microorganisms drive microbial community succession during fermentation of Nongxiangxing daqu. Bioresour. Technol. 2021, 345, 126549. [Google Scholar] [CrossRef]
- Hong, J.X.; Wang, J.S.; Zhang, C.S.; Zhao, Z.G.; Tian, W.J.; Wu, Y.S.; Chen, H.; Zhao, D.R.; Sun, J.Y. Unraveling variation on the profile aroma compounds of strong aroma type of Baijiu in different regions by molecular matrix analysis and olfactory analysis. RSC Adv. 2021, 11, 33511–33521. [Google Scholar] [CrossRef]
- Tan, Y.; Du, H.; Zhang, H.; Fang, C.; Jin, G.; Chen, S.; Wu, Q.; Zhang, Y.; Zhang, M.; Xu, Y. Geographically associated fungus-bacterium interactions contribute to the formation of geography-dependent flavor during high-complexity spontaneous fermentation. Microbiol. Spectr. 2022, 22, e01844. [Google Scholar] [CrossRef]
- Wang, L. Research trends in Jiang-flavor baijiu fermentation: From fermentation microecology to environmental ecology. J. Food Sci. 2022, 87, 1362–1374. [Google Scholar] [CrossRef]
- Sun, W.N.; Xiao, H.Z.; Peng, Q.; Zhang, Q.G.; Li, X.X.; Han, Y. Analysis of bacterial diversity of Chinese Luzhou-flavor liquor brewed in different seasons by Illumina Miseq sequencing. Ann. Microbiol. 2016, 66, 1293–1301. [Google Scholar] [CrossRef]
- Wang, T.; Jiang, D.; Zejun, C.; Ruiping, Z.; Wen, T.; Xinghai, H. Isolation method of actinomycetes from pit mud. Liquor Mak. Sci. Technol. 2009, 26–28. [Google Scholar]
- Zhou, J.B.; Xu, Y.X.; Guo, W.; Zhang, M.C.; Xiong, X.M.; Fang, S.L. Research of Actinomycetes Enzyme Production Capacity in the Liquor Brewing. Liquor Mak. 2015, 42, 44–47. [Google Scholar]
- Pullen, C.B.; Schmitz, P.; Meurer, K.; Bamberg, D.D.v.; Lohmann, S.; de Castro França, S.; Groth, I.; Schlegel, B.; Möllmann, U.; Gollmick, F.; et al. New and bioactive compounds from Streptomyces strains residing in the wood of Celastraceae. Planta 2002, 216, 162–167. [Google Scholar] [CrossRef]
- Zhang, L.H.; An, Q.; Zhang, Y.P.; Zhang, X.H.; Lv, P.J.; Li, X.T.; Hu, Q.P. Evaluation of The Potential of Daqu- Derived Actinobacteria for Light-Flavour Chinese Liquor. Eur. J. Food Sci. Technol. 2019, 7, 1–9. [Google Scholar]
- Du, H.; Lu, H.M.; Xu, Y.; Du, X.W. Community of environmental streptomyces related to geosmin development in Chinese liquors. J. Agric. Food. Chem. 2013, 61, 1343–1348. [Google Scholar] [CrossRef]
- Du, H.; Fan, W.L.; Xu, Y. Characterization of geosmin as source of earthy odor in different aroma type Chinese liquors. J. Agric. Food. Chem. 2011, 59, 8331–8337. [Google Scholar] [CrossRef]
- Ren, Y.M.; Dai, S.; Fan, L.; Wei, M.; Shang, L.H.; Xie, W.; Zhuang, M.Y.; Hou, M.Z. Study on the isolation of actinomycetes and its application in the production of Lu-type liquor. Liquor Mak. Sci. Technol. 1997, 3, 13–15. [Google Scholar] [CrossRef]
- Guo, W.; Guan, J.; Chen, M.B.; Fang, S.L. Research on the Relationship between Actinomycetes Enzyme-producing Abilities and Its Ability to Promoting Caproic Acid Bacteria Producing Caproic Acid. Liquor Mak. 2016, 43, 62–65. [Google Scholar]
- Kou, M. Discussion on actinomycetes in solid-state fermentation of traditional liquor. Food Ind. 2020, 93–94. [Google Scholar]
- Zhang, J.M.; Huang Yong Guang Zhou, W.M.; Cheng, L.X.; Zhao, C.L. Research Advance in Actinobacterial in Traditional Liquor Solid Fermentation Process. Liquor Mak. Sci. Technol. 2013, 232, 73–79. [Google Scholar] [CrossRef]
- Cui, F.L.; Wang, Z.Y.; Wang, F.; Ping, Y.Z.Q.Z.S.; Hua, S.L. Report on the separation of fermented grains and Daqu microorganisms. J. Microbiol. 1981, 13–19. [Google Scholar]
- Sun, H.L. Isolation of actinomycetes from pit mud. China Brew. 1987, 33–35. [Google Scholar]
- Wang, F.R.; Zeng, H.; Xu, Z.X.; Xi, D.Z.; Wang, H.; Cao, W.T. Isolation&Identification of a Thermoactinomyces sp. Strain from High-temperature Jiangxiang Daqu. Liquor Mak. Sci. Technol. 2017, 4, 42–45. [Google Scholar] [CrossRef]
- Yu, H.; Huang, D.; Chen, Z.; Tang, J.; Mao, X.; Deng, L.; Liu, D.; Lu, K.B. Isolation of protease-producing actinomycetes from sauce-flavor Daqu and its protease-producing conditions. China Brew. 2017, 36, 64–68. [Google Scholar]
- Luo, X.Y.; Wang, X.D.; Yi, Q.S. Isolation, screening and aroma component analysis of 3 strains of actinomycetes from Moutai-flavor Daqu. China Brew. 2018, 37, 62–67. [Google Scholar]
- Li, D.N.; Huang, W.; Wang, X.D.; Luo, X.Y.; Qiu, S.Y. Identification and Flavor Profile of a Thermoactinomycetaceae Strain Separated from Moutai-Flavor Daqu. Food Sci. 2018, 39, 171–176. [Google Scholar]
- Zhang, J.M.; Huang, Y.G.; Zhou, W.M.; You, X.L.; Zhao, C.L.; Zhou, J.T. Identification of 2 Streptococcus Strains and Their Tolerance of the Liquor-making Environment of Jiangxiang Baijiu(Liquor). Liquor Mak. Sci. Technol. 2014, 12, 8–12. [Google Scholar] [CrossRef]
- Li, D.N.; Wang, X.D.; Luo, X.Y.; Huang, W.; Qiu, S.Y. Whole Genome Sequencing and Sequence Analysis of Streptomyces sp.FBKL4.005 from Moutai-Flavor Daqu. Food Sci. 2018, 39, 206–212. [Google Scholar]
- Wang, X.D.; Luo, X.Y.; Qiu, S.Y. Isolation and screening of a thermophilic actinomycete from Moutai-flavor Daqu and its characterization. China Brew. 2018, 37, 51–56. [Google Scholar]
- Huang, Z.G.; Deng, J.; Wei, C.H.; Zhao, B.; Liu, Y.M. Separation and Identification of Bacteria Strains from Fermented Grains of Jiang-xiang Liquor. Liquor Mak. Sci. Technol. 2014, 6, 27–29. [Google Scholar] [CrossRef]
- Zhang, J.M.; Huang, Y.G.Z.; Wen, M.; Cheng, L.X.; Zhao, C.L.; You, X.L.; Zhou, J.T. Identification of a Streptomyces sp.Strain Separated from Stacking Fermented Grains of Jiangxiang Baijiu(Liquor). Liquor Mak. Sci. Technol. 2014, 6, 16–19. [Google Scholar] [CrossRef]
- Yang, F.; Chen, L.Q.; Lin, L.; Wang, H.Y.; Wang, L. Isolation & Identification of an Actinomycetes Strain and Analysis of Its Metabolites. Liquor Mak. Sci. Technol. 2014, 9, 42–44. [Google Scholar] [CrossRef]
- Bai, C.S.; Chen, L.; Lu, H.M.; Zhou, L.; Ren, X.K. Isolation and identification of thermoactinomycete from Moutai-flavor vinasse in Moutai region. Sci. Technol. Food Ind. 2017, 38, 199–202. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, D.; Wang, T.; You, L.; Feng, R.Z.; Wang, S.; He, J.R. Diversity of Actinomycetes in the Brewing Process of Luzhou-flavor Multiple-grains Liquor. Food Sci. 2011, 32, 192–196. [Google Scholar]
- Hou, X.G.; Sun, Z.K.; Li, X.S.; Chu, C.W.; Pan, Q.Q.; Chen, H.; Chen, Y.L.; Yuan, J.W. Isolation, identification and enzyme-production of culturable Actinomycetes from Daqu applied in making strong-flavor liquor. Food Sci. Technol. 2019, 44, 28–35. [Google Scholar] [CrossRef]
- Shi, S.; Zhang, X.; Yang, K.Z.; Liao, Q.J.; Qiao, Z.W.; Zheng, J.; Liu, D.T. Preliminary Study on the Regulation Effect of Actinomycetes on Brewing Microorganisms. Liquor Mak. Sci. Technol. 2021, 2, 17–20. [Google Scholar] [CrossRef]
- Luo, Q.C.; Zhao, D.; Qiao, Z.W.; Jia, Z. Isolation and Identification of an N-Propanol Degrading Actinomycete Strain. Liquor Mak. Sci. Technol. 2018, 12, 74–77. [Google Scholar] [CrossRef]
- Yao, Y.L.; Zheng, R.X.; Cheng, T.Y.; Deng, J.; Ren, Z.Q.; Wei, C.H.; Huang, Z.G. Study on the Fermentation Characteristics of Actinomycetes in Pit Mud of Luzhou-flavor Liquor. Food Res. Devel. 2020, 41, 191–197. [Google Scholar]
- Luan, X.S.; Hu, J.J.; Zhang, H.Y. Study on Facultatively Autotrophic Strepptimyces in Cellar Mud. Shandong Sci. 1991, 2, 9–15. [Google Scholar]
- Lu, L.F.; Yang, Y.; Zheng, L.; Zhang, R.; Liu, G.Q.; Tu, T.y.; Xu, T.; Luo, X.Z.; Ran, M.F.; Zhang, L.Q.; et al. Reclassification of Olsenella gallinarum as Thermophilibacter gallinarum comb. nov. and description of Thermophilibacter immobilis sp. nov., isolated from the mud in a fermentation cellar used for the production of Chinese Luzhou-flavour Baijiu. Int. J. Syst. Evol. Microbiol. 2021, 71, 5192. [Google Scholar] [CrossRef]
- Luo, B.X.; Zheng, R.X.; Cheng, T.Y.; Ren, Z.Q.; Wei, C.H.; Deng, J.; Huang, Z.G. In situ isolation and metabolic characteristics of actinomycetes from strong-flavor Baijiu pit mud. Food Ferment. Ind. 2021, 47, 75–83. [Google Scholar] [CrossRef]
- Guo, W.; Huang, Y.; Xie, Y.Q.; Fang, S.L.; Chen, M.B. Screening of Fine Actinomycetes that Promoting Caproic Acid Bacteria Producing Caproic Acid. Liquor Mak. 2016, 43, 47–51. [Google Scholar]
- Tang, Y.H.; Wu, W.R. Isolation of pit mud bacteria based on Biolog ECO technology. China Brew. 2014, 33, 121–125. [Google Scholar]
- Yao, S.; Xu, Y.Q.; Xin, C.H.; Xu, L.; Liu, Y.; Li, H.; Li, J.X.; Zhao, J.W.; Cheng, C.W. Genome Sequence of Thermoactinomyces daqus H-18, a Novel Thermophilic Species Isolated from High-Temperature Daqu. Genome Announc. 2015, 3, e01394-14. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, T.; Yao, S.; Ge, Y.Y.; Xin, C.H.; Xu, L.; Cheng, C. Identification on a Thermoactinomyces sp. Separated from High Temperature Daqu of Sesame Flavor Liquor. Biotechnol. Bull. 2012, 10, 210–216. [Google Scholar] [CrossRef]
- Zhang, M.; Yao, S.; Li, H.; Liu, Y.; Xin, C.H.; Xu, L.; Cheng, C. A specific PCR assay for rapid identifying Thermoactinomyces vulgaris. Food Ferment. Ind. 2014, 40, 64–66. [Google Scholar] [CrossRef]
- Ge, Y.Y.; Yao, S.; Liu, Y.; Cao, Y.H.; Zhang, F.G.; Xin, C.H.; Xu, L.; Cheng, C. Analysis on Thermophilic Bacterial Communities in High Temperature Daqu of Sesame Flavor Liquor. Food Ferment. Ind. 2012, 38, 16–19. [Google Scholar] [CrossRef]
- Yan, Y.; Xing, X.; Sun, Z.B.; Li, J.; Hao, S.Y.; Xu, J.L. Brevibacterium renqingii sp. nov., isolated from the Daqu of Baijiu. Arch. Microbiol. 2021, 203, 2291–2296. [Google Scholar] [CrossRef]
- Zhu, T.T. Analysis of Cultivable Microbe Diversity in Niulanshan Daqu. Liquor Mak. Sci. Technol. 2018, 5, 75–79. [Google Scholar] [CrossRef]
- Zheng, X.W.; Yan, Z.; Han, B.Z.; Zwietering, M.H.; Samson, R.A.; Boekhout, T.; Robert Nout, M.J. Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 2012, 31, 293–300. [Google Scholar] [CrossRef]
- Wei, J.W. Isolation & Identification of Cultivable Thermoactinomycetaceae in the Production of Qingxiang Baijiu. Liquor Mak. Sci. Technol. 2019, 1, 56–59. [Google Scholar] [CrossRef]
- Hu, J.X.; Yang, T.; Cai, G.L.; Wu, L.W.; Zhuang, M.Y.; Xu, W. Study on Microbial Communities in Feng & Composite-flavor Taibai Liquor (I). Microflora in Daqu. Liquor Mak. Sci. Technol. 2012, 5, 55–57. [Google Scholar]
- Fang, H.Z.; Yan, Z.K.; Fu, W.X.; Zhang, Y.L. The Rseach on Bad Flavor Substancest of Geosmin in Traditional Feng-flavor DaQu. Liquor Mak. 2016, 43, 56–60. [Google Scholar]
- Yang, T.; Hu, J.X.; Wu, L.W.; Cai, G.L.; Zhuang, M.Y.; Xu, W. Study on Microbial Communties during the Fermentation Process of Feng & Composite-flavor Taibai Liquor (II): Microbial Communities in Fermented grains. Liquor Mak. Sci. Technol. 2012, 6, 44–46. [Google Scholar]
- Yao, S.; Liu, Y.; Zhang, M.J.; Zhang, X.; Li, H.; Zhao, T.; Xin, C.H.; Xu, L.; Zhang, B.L.; Cheng, C. Thermoactinomyces daqus sp. nov., a thermophilic bacterium isolated from high-temperature Daqu. Int. J. Syst. Evol. Microbiol. 2014, 64, 206–210. [Google Scholar] [CrossRef]
- Ye, G.B.; Wang, C.H.; Qiao, X.Y.; Luo, H.B.; Zhuo, Y.C.; Bian, M.H. Study on the Structure of Bacterial Communities of Ultrahigh-temperature Daqu. Liquor Mak. Sci. Technol. 2014, 4, 5–8. [Google Scholar] [CrossRef]
- Shan, Q.; Liang, H.Z.; Zhang, C.X.; Zhang, L.Q.; Tan, X.; Liang, L.Q.; Li, C.W. Changes of Microbial Diversity in Stacked Fermentation for the Production of Moutai Flavor Liquor. J. Food Sci. Biotechnol 2016, 35, 330–335. [Google Scholar]
- Zhang, H.M.; Shu, Y.; Zhou, Q.W.; Li, A.J.; He, H.K.; Zhang, Z.Z. Analysis of the Microbial Community Structure of Gujinggong Liquor Starter through Culture-free Approach. Mod. Food Sci. Technol. 2014, 30, 44–49. [Google Scholar] [CrossRef]
- Ye, G.B.; Wang, C.H.; Wang, Y.; Zhen, P.; Wang, Y.; Luo, H. Comparative analysis of bacterial community structure of Chinese Fen-Daqus. Food Mach. 2015, 31, 11–15. [Google Scholar] [CrossRef]
- Liu, M.K.; Tang, Y.M.; Zhao, K.; Ren, D.Q.; Yao, W.C.; Tian, X.H. Analysis of actinobacteria community and diversity in the pit mud of chinese luzhou-flavour liquor. Acta Ecol. Sin. 2015, 35, 858–864. [Google Scholar]
- Xu, J.L.; Sun, L.P.; Xing, X.; Sun, Z.B.; Gu, H.Y.; Lu, X.; Li, Z.P.; Ren, Q. Culturing Bacteria From Fermentation Pit Muds of Baijiu With Culturomics and Amplicon-Based Metagenomic Approaches. Front. Microbiol. 2020, 11, 1223. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yang, L.; Hui, L.C.; Yuan Yuan, G.; Ming Juan, Z.; Chun Hui, X.; Ling, X.; Chi, C. Bacterial communities during the process of high-temperature Daqu production of roasted sesame-like flavour liquor. J. Inst. Brew. 2015, 121, 440–448. [Google Scholar] [CrossRef]
- Yu, X.J.; Feng, H.J.; Zhai, L.; Bai, X.B.; Xu, L.; Yu, P.P.; Cheng, C.; Yao, S. Dynamic changes of Thermoactinomyces and their functional genes in high temperature Daqu of Sesame-flavored Baijiu. Food Ferment. Ind. 2019, 45, 71–77. [Google Scholar] [CrossRef]
- Gao, Y.B.; Wang, H.Y.; Xu, Y. PCR-DGGE Analysis of the Bacterial Community of Chinese Liquor High and Medium Temperature Daqu. Microbiol. China 2010, 37, 999–1004. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Liu, X.F.; Yuan, Y.X.; Yan, Z.Y.; Liao, Y.Z.; Fu, S. Bacterial community structure in distiller’s yeast and accumulated fermented grains of Maotai-flavor liquor. Chin. J. Appl. Environ. Biol 2014, 20, 825–831. [Google Scholar]
- Huang, X.H.J.; Li, Z.; Han, B. Microbial diversity analysis in strong-flavor and sauce-flavor Daqu. China Brew. 2016, 35, 33–37. [Google Scholar]
- Fan, W.Y.; Zhao, X.R.; Du, G.C.; Chen, J.; Li, J.H.; Zheng, J.; Qiao, Z.W.; Zhao, D. Metaproteomic analysis of enzymatic composition in Baobaoqu fermentation starter for Wuliangye baijiu. Int. J. Food Sci. Technol. 2021, 56, 4170–4181. [Google Scholar] [CrossRef]
- Zhan, W.X.; Qiao, Z.W.; Hu, C.; Wang, Z.Y. Analysis of Bacterial Community in Fermented Grains During the Production of Chinese Strong Aromatic Spirits by PCR Technique. J. Sichuan. Univ (Eng. Sci. Ed) 2005, 5, 82–87. [Google Scholar]
- Li, D.; Suyi, Z.; Zhenyu, M.; Zonghua, A.; Songtao, W.; Liangyang, S.; Yan, Y.; Zhang, B. PCR-DGGE analysis of microbial community structure in fermented grains of Luzhou-flavor liquor. Liquor Mak. Sci. Technol. 2014, 25–27,31. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, J.; Cui, Y.; Zhao, W.; Zhang, J.; Bai, F. Microbial Diversity Analysis of Flight-flavor Daqu Using High-throughput Sequencing. J. Chin. Inst. Food Sci. Technol. 2019, 19, 244–250. [Google Scholar] [CrossRef]
- Hou, Q.C.; Wang, Y.R.; Cai, W.C.; Ni, H.J.; Zhao, H.J.; Zhang, Z.D.; Liu, Z.J.; Liu, J.M.; Zhong, J.A.; Guo, Z. Metagenomic and physicochemical analyses reveal microbial community and functional differences between three types of low-temperature Daqu. Food Res. Int. 2022, 156, 111167. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Ge, Y.; Li, H.; Liu, Y.; Zhao, J.W.; Zhang, F.G.; Xin, C.H.; Cheng, C. Analysis on Bacterial Communities in High Temperature Daqu of Sesame Flavor Liquor through Culture-free Approach. Food Ferment. Ind. 2012, 38, 1–6. [Google Scholar] [CrossRef]
- Zheng, X.; Yan, Z.; Robert Nout, M.J.; Boekhout, T.; Han, B.; Zwietering, M.H.; Smid, E.J. Characterization of the microbial community in different types of Daqu samples as revealed by 16S rRNA and 26S rRNA gene clone libraries. World J. Microbiol. Biotechnol. 2015, 31, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Jana, A.; Mondal, K.C.; Halder, S.K. Omics Approach to Understanding Microbial Diversity. Biotechnol. Adv. Microbiol. Mol. Biol. Nanotechnol. 2022, 26–36. [Google Scholar]
- Kumar, R.R.; Jadeja, V.J.; Shree, M.; Virani, N.A. Isolation of Actinomycetes: A Complete Approach. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 606–618. [Google Scholar] [CrossRef]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.E.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2020, 19, 225–240. [Google Scholar] [CrossRef]
- Gou, M.; Wang, H.Z.; Yuan, H.W.; Zhang, W.X.; Tang, Y.Q.; Kida, K. Characterization of the microbial community in three types of fermentation starters used for Chinese liquor production. J. Inst. Brew. 2015, 121, 620–627. [Google Scholar] [CrossRef]
- Wang, X.D.; Ban, S.D.; Zhou, H.X.; Hu, B.D.; Qiu, S.Y. Comparative Analysis of Bacterial Populations of Three Maotai-Flavored Daqus in Zunyi, Guizhou. Food Sci. 2016, 37, 110–116. [Google Scholar]
- Wang, X.D.; Ban, S.D.; Hu, B.L.; Qiu, S.Y.; Zhou, H.X. Bacterial diversity of Moutai-flavour Daqu based on high-throughput sequencing method. J. Inst. Brew. 2017, 123, 138–143. [Google Scholar] [CrossRef]
- Wang, X.D.; Lei, A.L.; Ban, S.D.; Qiu, S.Y. Research on bacterial diversity of Maotai-flavor Daqu. Food Ferment. Ind. 2017, 43, 70–75. [Google Scholar] [CrossRef]
- Hou, Q.C.; Wang, Y.R.; Wang, W.P.; Tian, L.X.; Zhao, H.J.; Guo, Z. Difference of bacterial community structure and functional prediction in high-temperature Daqu of Maotai and Yaozhihe. Food Ferment. Ind. 2022, 48, 36–44. [Google Scholar] [CrossRef]
- Wang, Y.R.; Cai, W.C.; Wang, W.P.; Shu, N.; Zhang, Z.D.; Hou, Q.C.; Shan, C.H.; Guo, Z. Analysis of microbial diversity and functional differences in different types of high-temperature Daqu. Food Sci. Nutr. 2021, 9, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wu, X.; Xu, Y.; Zhang, Y.; Wang, Z.; Shen, L.; Yang, W.; Sun, J.; Liu, Y. Microbial composition and dynamic succession during the Daqu production process of Northern Jiang-flavored liquor in China. 3 Biotech. 2021, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.C.; Huang, Y.G.; Min, G. Evaluation of bacterial diversity during fermentation process: A comparison between handmade and machine-made high-temperature Daqu of Maotai-flavor liquor. Ann. Microbiol. 2020, 70, 57. [Google Scholar] [CrossRef]
- Wang, L.C.; Zhong, K.; Luo, A.M.; Chen, J.; Shen, Y.; Wang, X.; He, Q.; Gao, H. Dynamic changes of volatile compounds and bacterial diversity during fourth to seventh rounds of Chinese soy sauce aroma liquor. Food Sci. Nutr. 2021, 9, 3500–3511. [Google Scholar] [CrossRef]
- Mao, J.J.; Liu, X.L.; Gao, T.; Gu, S.B.; Wu, Y.; Zhao, L.N.; Ma, J.L.; Li, X.; Zhang, J. Unraveling the correlations between bacterial diversity, physicochemical properties and bacterial community succession during the fermentation of traditional Chinese strong-flavor Daqu. LWT 2021, 154, 112764. [Google Scholar] [CrossRef]
- Liu, Y.B.; Zhao, Z.J.; Chen, H.F.; Sun, X.Y.; Pan, C.M. Analysis of Bacterial Community Structure in Medium Temperature Daqu and High Temperature Daqu of Luzhou-flavor Liqu by High-throughput Sequencing. Mod. Food Sci. Technol. 2018, 34, 229–235. [Google Scholar] [CrossRef]
- He, M.W.; Jin, Y.; Zhou, R.Q.; Zhao, D.; Zheng, J.; Wu, C.D. Dynamic succession of microbial community in Nongxiangxing daqu and microbial roles involved in flavor formation. Food Res. Int. 2022, 159, 111559. [Google Scholar] [CrossRef]
- Wang, X.S.; Du, H.; Xu, Y. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int. J. Food Microbiol. 2017, 244, 27–35. [Google Scholar] [CrossRef]
- Wu, S.K.; Xie, J.; Wei, C.H.; Liu, Y.M.; Huang, Z.G.; Wan, S.L.; Deng, J. Comparison of Microbial Community Structure of Starter Cultures (Daqu) for Luzhou-Flavor Liquor in Different Regions of Sichuan. Food Sci. 2019, 40, 144–152. [Google Scholar]
- Su, G.; Wang, X.H.; Dong, D.W.; Ma, W.; Yang, J.M.; Ye, H. Application of High-Throughput Sequencing Technology in the Quality Grade Determination of Yanghe Daqu. Liquor Mak. Sci. Technol. 2019, 86–90. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Zhao, J.R.; Liu, X.; Zhang, C.S.; Zhao, Z.G.; Li, X.T.; Sun, B.G. Flavor mystery of Chinese traditional fermented baijiu: The great contribution of ester compounds. Food Chem. 2021, 369, 130920. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.L.; Zhao, T.; Wang, J.S.; Cai, K.Y.; Chen, P.; Deng, J.S.F.; Shang, L.; Cao, J.H.; Chen, M.B. Changes in the Microbial Community Structure during the Digitally Managed Fermentation of High-temperature Daqu. Food Sci. 2022, 43, 171–178. [Google Scholar]
- Tao, Y. Microbial community compositions and diversity in pit mud of Chinese Luzhou-flavor liquor. CIESC J. 2014, 65, 1800–1807. [Google Scholar]
- Zhang, Q.Y.; Yuan, Y.J.; Liao, Z.M.; Zhang, W.X. Use of microbial indicators combined with environmental factors coupled with metrology tools for discrimination and classification of Luzhou flavoured pit muds. J. Appl. Microbiol. 2017, 123, 933–943. [Google Scholar] [CrossRef]
- Tao, Y.; Li, J.B.; Rui, J.P.; Xu, Z.C.; Zhou, Y.; Hu, X.H.; Wang, X.; Liu, M.H.; Li, D.P.; Li, X.Z. Prokaryotic Communities in Pit Mud from Different-Aged Cellars Used for the Production of Chinese Strong-Flavored Liquor. Appl. Environ. Microbiol. 2014, 80, 2254–2260. [Google Scholar] [CrossRef]
- Hu, X.L.; Du, H.; Ren, C.; Xu, Y. Illuminating Anaerobic Microbial Community and Cooccurrence Patterns across a Quality Gradient in Chinese Liquor Fermentation Pit Muds. Appl. Environ. Microbiol. 2016, 82, 2506–2515. [Google Scholar] [CrossRef]
- Tao, Y.; Wang, X.; Li, X.Z.; Wei, N.; Jin, H.; Xu, Z.C.; Tang, Q.L.; Zhu, X.Y. The functional potential and active populations of the pit mud microbiome for the production of Chinese strong-flavour liquor. Microb. Biotechnol. 2017, 10, 1603–1615. [Google Scholar] [CrossRef]
- Liu, M.K.; Tang, Y.M.; Guo, X.J.; Zhao, K.; Tian, X.H.; Liu, Y.; Yao, W.C.; Deng, B.; Ren, D.Q.; Zhang, X.P. Deep sequencing reveals high bacterial diversity and phylogenetic novelty in pit mud from Luzhou Laojiao cellars for Chinese strong-flavor Baijiu. Food Res. Int. 2017, 102, 68–76. [Google Scholar] [CrossRef]
- Luo, Q.C.; Liu, C.L.; Li, W.F.; Wu, Z.Y.; Zhang, W.X. Comparison between Bacterial Diversity of Aged and Aging Pit Mud from Luzhou-flavor Liquor Distillery. Food Sci. Technol. Res. 2014, 20, 867–873. [Google Scholar] [CrossRef]
- Liang, H.P.; Luo, Q.C.; Zhang, A.Y.; Wu, Z.Y.; Zhang, W.X. Comparison of bacterial community in matured and degenerated pit mud from Chinese Luzho flavour liquor distillery in different regions. J. Inst. Brew. 2016, 122, 48–54. [Google Scholar] [CrossRef]
- Zhou, W.G.; Liao, Z.M.; Wu, Z.Y.; Suyama, T.K.; Zhang, W.X. Analysis of the difference between aged and degenerated pit mud microbiome in fermentation cellars for Chinese Luzhou-flavor baijiu by metatranscriptomics. J. Sci. Food Agric. 2021, 101, 4621–4631. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Y.Z.; Jin, L.; He, L.; Ao, X.L.; Liu, S.L.; Yang, Y.; Liu, A.P.; Chen, S.J.; Zou, L.K. Analyzing bacterial community in pit mud of Yibin Baijiu in China using high throughput sequencing. PeerJ 2020, 8, e9122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Meng, Y.J.; Wang, Y.L.; Zhou, Q.W.; Li, A.J.; Liu, G.Y.; Li, J.X.i.; Xing, X.H. Prokaryotic communities in multidimensional bottom-pit-mud from old and young pits used for the production of Chinese Strong-Flavor Baijiu. Food Chem. 2019, 312, 126084. [Google Scholar] [CrossRef]
- Hu, X.L.; Tian, R.J.; Wang, K.L.; Cao, Z.H.; Yan, P.X.; Li, F.Q.; Li, X.S.; Li, S.L.; He, P.X. The prokaryotic community, physicochemical properties and flavors dynamics and their correlations in fermented grains for Chinese strong-flavor Baijiu production. Food Res. Int. 2021, 148, 110626. [Google Scholar] [CrossRef]
- Xiao, C.; Yang, Y.; Lu, Z.M.; Chai, L.J.; Zhang, X.J.; Wang, S.T.; Shen, C.H.; Shi, J.S.; Xu, Z.H. Daqu microbiota exhibits species-specific and periodic succession features in Chinese baijiu fermentation process. Food Microbiol. 2021, 98, 103766. [Google Scholar] [CrossRef]
- Feng, J.T.; Lu, Z.M.; Shi, W.; Xiao, C.; Zhang, X.J.; Chai, L.J.; Wang, S.T.; Shen, C.H.; Shi, J.S.; Xu, Z.H. Effects of different culture temperatures on microbial community structure, enzyme activity, and volatile compounds in Daqu. Chin. J. Appl. Environ. Biol. 2021, 27, 760–767. [Google Scholar] [CrossRef]
- Li, Z.J.; Fan, Y.H.; Huang, X.N.; Han, B.Z. Microbial Diversity and Metabolites Dynamic of Light-Flavor Baijiu with Stacking Process. Ferment 2022, 8, 67. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Zhao, J.; Du, X.Q. Barcoded pyrosequencing analysis of the bacterial community of Daqu for light flavour Chinese liquor. Lett. Appl. Microbiol. 2014, 58, 549–555. [Google Scholar] [CrossRef]
- Li, C.; Mu, L.; Wang, J.Y.; Lei, Z.H.; Chen, J.Y.; Han, B.Z. Physiochemical and microbiological analysis of Fen-type Daqu. China Brew. 2009, 140–142. [Google Scholar]
- Cai, W.C.; Wang, Y.R.; Ni, H.; Liu, Z.J.; Liu, J.M.; Zhong, J.A.; Hou, Q.C.; Shan, C.H.; Yang, X.Q.; Guo, Z. Diversity of microbiota, microbial functions, and flavor in different types of low-temperature Daqu. Food Res. Int. 2021, 150, 110734. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Xiao, Y.W.; Li, X.R.; Ma, E.B.; Du, X.S.; Quan, Z.X. Analyses of microbial consortia in the starter of Fen Liquor. Lett. Appl. Microbiol. 2009, 48, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.W.; Lv, F.X.; Ma, G.D.; Farooq, A.; Li, H.H.; Du, Y.; Liu, Y. High throughput sequencing of the bacterial composition and dynamic succession in Daqu for Chinese sesame flavour liquor. J. Inst. Brew. 2019, 126, 98–104. [Google Scholar] [CrossRef]
- Fan, G.S.; Du, Y.H.; Fu, Z.L.; Chen, M.; Wang, Z.; Liu, P.X.; Li, X.T. Characterisation of physicochemical properties, flavour components and microbial community in Chinese Guojing roasted sesame-like flavour Daqu. J. Inst. Brew. 2019, 126, 105–115. [Google Scholar] [CrossRef]
- Wu, X.Y.; Jing, R.X.; Chen, W.H.; Geng, X.J.; Li, M.; Yang, F.Z.; Yan, Y.Z.; Liu, Y. High-throughput sequencing of the microbial diversity of roasted-sesame-like flavored Daqu with different characteristics. 3 Biotech. 2020, 10, 502. [Google Scholar] [CrossRef]
- Wang, H.Y.; Zhang, X.J.; Zhao, L.P.; Xu, Y. Analysis and comparison of the bacterial community in fermented grains during the fermentation for two different styles of Chinese liquor. J. Ind. Microbiol. Biotechnol. 2008, 35, 603–609. [Google Scholar] [CrossRef]
- Yun, J.L.; Yan, X.; Zhu, B.; Ju, W.T.; Wei, G.H.; Hu, J.X.; Li, H.W.; Chen, X. Analysis of Microflora in Fermented Grains during the Fermentation of Taibai Liquor. Liquor Mak. Sci. Technol. 2006, 12, 40–42. [Google Scholar]
- Liu, C.J.; Gong, X.W.; Zhao, G.; Soe Htet, M.N.; Jia, Z.Y.; Yan, Z.K.; Liu, L.; Zhai, Q.H.; Huang, T.; Deng, X.P.; et al. Liquor Flavour Is Associated With the Physicochemical Property and Microbial Diversity of Fermented Grains in Waxy and Non-waxy Sorghum (Sorghum bicolor) During Fermentation. Front. Microbiol. 2021, 12, 618458. [Google Scholar] [CrossRef]
- Huang, Y.H.; Yi, Z.L.; Jin, Y.L.; Zhao, Y.G.; He, K.Z.; Liu, D.Y.; Zhao, D.; He, H.; Luo, H.B.; Zhang, W.X.; et al. New microbial resource: Microbial diversity, function and dynamics in Chinese liquor starter. Sci. Rep. 2017, 7, 14577. [Google Scholar] [CrossRef]
- Tian, N.; Guo, X.; Wang, M.Z.; Chen, C.; Cui, H.H.; Zhang, L.P.; Tang, H. Bacterial community diversity of shilixiang baijiu Daqu based on metagenomics. J. Food Biochem. 2020, 44, e13410. [Google Scholar] [CrossRef]
- Hu, Y.L.; Dun, Y.H.; Li, S.N.; Fu, B.; Xiong, X.M.; Peng, N.; Liang, Y.X.; Zhao, S.M. Changes in microbial community during fermentation of high temperature Daqu used in the production of Chinese BaiyunbiaŽ liquor. J. Inst. Brew. 2017, 123, 594–599. [Google Scholar] [CrossRef]
- Guo, X.W.; Fan, E.D.; Ma, B.T.; Li, Z.X.; Zhang, Y.X.; Zhang, Z.M.; Chen, Y.F.; Xiao, D.G. Research progress in functional bacteria in solid-state fermented Baijiu in China. Food Ferment. Ind. 2020, 46, 280–286. [Google Scholar] [CrossRef]
- Zou, W.; Zhao, C.; Luo, H.-b. Diversity and Function of Microbial Community in Chinese Strong-Flavor Baijiu Ecosystem: A Review. Front. Microbiol. 2018, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.Q.; Jin, Z.Y.; Ali, A.; Wang, C.J.; Liu, J. Caproic Acid-Producing Bacteria in Chinese Baijiu Brewing. Front. Microbiol. 2022, 13, 883142. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Guan, J.; Chen, M.B.; Xie, Y.Q.; Zhang, Y.; Fang, S.L. Mechanism of Actinomycetes Promoting Caproic Acid Bacteria to Produce Caproic Acid. Liquor Mak. Sci. Technol. 2016, 10, 48–52. [Google Scholar] [CrossRef]
- Cao, X.Z.; Huang, C.P.; Xiong, L.; Zhou, Z.B. Effect of co-cultivation on growth and metabolism of caproic acid bacteria. China Brew. 2010, 11, 35–38. [Google Scholar]
- Du, H.; Lu, H.M.; Xu, Y. Influence of geosmin-producing Streptomyces on the growth and volatile metabolites of yeasts during chinese liquor fermentation. J. Agric. Food. Chem. 2015, 63, 290–296. [Google Scholar] [CrossRef]
- You, X.L.; Huang, Y.L.; Huang, Y.G.; Zhou, W.M. Study on the biological characteristics of actinomycetes on functional bacteria producing maotai-flavor during fermentation. Liquor Mak. Sci. Technol. 2015, 8, 1–5. [Google Scholar] [CrossRef]
- Huang, B.; Liu, N.; Huang, Y.; Chen, J.C. Coculture of actinomycetes with Bacillus subtilis and its effect on the bioactive secondary metabolites. Chin. J. Biotechnol. 2009, 25, 932–940. [Google Scholar]
- Zhi, Y.; Wu, Q.; Du, H.; Xu, Y. Biocontrol of geosmin-producing Streptomyces spp. by two Bacillus strains from Chinese liquor. Int. J. Food Microbiol. 2016, 231, 1–9. [Google Scholar] [CrossRef]
- Huang, Y.L.; Zhang, J.M.; Huang Yong Guang Hu, J.F.; Hu, F.; Zhong, F.D. Flavor regulation of Moutai-flavor Baijiu brewing by Streptomyces sp.A22. China Brew. 2016, 35, 27–31. [Google Scholar]
- Schneider, J.; Yepes, A.; García-Betancur, J.-C.; Westedt, I.; Mielich, B.; López, D. Streptomycin-Induced Expression in Bacillus subtilis of YtnP, a Lactonase-Homologous Protein That Inhibits Development and Streptomycin Production in Streptomyces griseus. Appl. Environ. Microbiol. 2011, 78, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.H.; Liu, S.P.; Zhang, S.Y.; Qin, H.; Han, X.; Mao, J. Multi-Omics Reveals Microbial Roles and Metabolic Functions at the Spatiotemporal Niche in Pit Mud. Res. Sq. 2022, 1–33. [Google Scholar]
- Ye, C.; Wei, X.Y.; Shi, T.Q.; Sun, X.; Xu, N.; Gao, C.; Zou, W. Genome-scale metabolic network models: From frst-generation to next-generation. Appl. Microbiol. Biotechnol. 2022, 106, 4907–4920. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.J.; Zhai, L.; Yu, X.J.; Cheng, K.; Liu, Y.; Yao, S. Fatty acids metabolism changes of Thermoactinomyces daqus CICC 10681 at different temperatures. Food Ferment. Ind. 2018, 44, 49–54. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef]
- Kim, J.N.; Kim, Y.J.; Jeong, Y.J.; Roe, J.H.; Kim, B.G.; Cho, B.K. Comparative Genomics Reveals the Core and Accessory Genomes of Streptomyces Species. J. Microbiol. Biotechnol. 2015, 25, 1599–1605. [Google Scholar] [CrossRef]
- Zhao, C.; Su, W.; Mu, Y.; Mu, Y.C.; Jiang, L. Integrative Metagenomic—Metabolomics for Analyzing the Relationship Between Microorganisms and Non-volatile Profiles of Traditional Xiaoqu. Front. Microbiol. 2020, 11, 617030. [Google Scholar] [CrossRef]
- Liu, M.K.; Tang, Y.M.; Guo, X.J.; Zhao, K.; Penttinen, P.; Tian, X.H.; Zhang, X.Y.; Ren, D.Q.; Zhang, X.P. Structural and Functional Changes in Prokaryotic Communities in Artificial Pit Mud during Chinese Baijiu Production. mSystems 2020, 5, e00829-19. [Google Scholar] [CrossRef]
- Song, Z.W.; Du, H.; Zhang, Y.; Xu, Y. Unraveling Core Functional Microbiota in Traditional Solid-State Fermentation by High-Throughput Amplicons and Metatranscriptomics Sequencing. Front. Microbiol. 2017, 8, 1294. [Google Scholar] [CrossRef]
- Gan, S.; Yang, F.; Sahu, S.K.; Luo, R.; Liao, S.L.; Wang, H.Y.; Jin, T.; Wang, L.; Zhang, P.F.; Liu, X.; et al. Deciphering the Composition and Functional Profile of the Microbial Communities in Chinese Moutai Liquor Starters. Front. Microbiol. 2019, 10, 1540. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, S.T.; Lu, Z.M.; Zhang, X.J.; Chai, L.J.; Shen, C.H.; Shi, J.S.; Xu, Z.H. Metagenomics unveils microbial roles involved in metabolic network of flavor development in medium-temperature daqu starter. Food Res. Int. 2021, 140, 110037. [Google Scholar] [CrossRef] [PubMed]
- Schöller, C.; Gürtler, H.; Pedersen, R.; Molin, S.; Wilkins, K. Volatile metabolites from actinomycetes. J. Agric. Food. Chem. 2002, 50, 2615–2621. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, D.Y.; Ai, M.; Tang, Q.X.; Huang, J.; Ding, X.F.; Wu, C.D.; Zhou, R.Q. Correlation between volatile profiles and microbial communities: A metabonomic approach to study Jiang-flavor liquor Daqu. Food Res. Int. 2019, 121, 422–432. [Google Scholar] [CrossRef]
- Wang, T.; You, L.; Zhao, D.; Feng, R.Z.; Wang, S.; Feng, X.Y.; Lin, Q. Preliminary Analysis of Volatiles in Fermentation Broths of Actinomycetes Isolated from Luzhou-Flavor Liquor Brewing Environments. Food Sci. 2012, 33, 184–187. [Google Scholar]
- Li, H.; Lian, B.; Ding, Y.; Nie, C.; Zhang, Q. Bacterial diversity in the central black component of Maotai Daqu and its flavor analysis. Ann. Microbiol. 2014, 64, 1659–1669. [Google Scholar] [CrossRef]
- You, X.L.; Huang, Y.G.H.; Yun, L.; Hu, F.; Hu, J.F.; Zhong, F.D. Activity of the Metabolites of an Actinomycetes Strain from Jiangxiang Baijiu Production Process and Its Effects on Other Functional Bacteria in Liquor-Making Environment. Liquor Mak. Sci. Technol. 2018, 17–23. [Google Scholar] [CrossRef]
- Shi, S.; Hu, C.; Zhang, W.X. Preliminary study on an actinomycete producing brown-pigment and its 16S rDNA sequence analysis. Sci. Technol. Food Ind. 2010, 31, 239–240. [Google Scholar] [CrossRef]
- Wang, X.D.; Yang, Y.H.; Li, Y.L.; Chen, H.M. Biological Characteristics and Screening and Identification of PL Producing Strain. Food Ferment. Ind. 2007, 1, 40–42. [Google Scholar]
- Chaudhary, H.S.; Soni, B.; Shrivastava, A.; Shrivastava, S.R. Diversity and versatility of actinomycetes and its role in antibiotic production. J. Appl. Pharm. Sci. 2013, 3, 83–94. [Google Scholar]
- You, L.; Wang, T. Volatile Products of 4 Streptomyces Strains Isolated from Strong-flavor Liquor Factories. Food Ind. 2012, 33, 118–120. [Google Scholar]
- Bezuidt, O.; Gomri, M.A.; Pierneef, R.; Van Goethem, M.W.; Kharroub, K.; Cowan, D.A.; Makhalanyane, T.P. Draft genome sequence of Thermoactinomyces sp. strain AS95 isolated from a Sebkha in Thamelaht, Algeria. Stand. Genom. Sci. 2016, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.B.; Zheng, P. Spirit-based distillers’ grain as a promising raw material for succinic acid production. Biotechnol. Lett. 2013, 35, 679–684. [Google Scholar] [CrossRef]
- Luo, Q.C.; Qiao, Z.W.; Zheng, J.; Zhang, X.; Lei, X.J.; Shi, S.; Liu, D.T. Screening and metabolic kinetic analysis of Arthrobacter protophormiae for lactic acid degradation. China Brew. 2021, 40, 82–86. [Google Scholar]
- Xiao, C.; Lu, Z.M.; Zhang, X.J.; Wang, S.T.; Ao, L.; Shen, C.H.; Shi, J.S.; Xu, Z.H. Bio-Heat Is a Key Environmental Driver Shaping the Microbial Community of Medium-Temperature Daqu. Appl. Environ. Microbiol. 2017, 83, e01550-17. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.S. Isolation and characteristics of facultative autotrophic Streptomyces in the fermentation tank of Qu liquor. Food. Ferment. Ind. 2001, 11, 17–20. [Google Scholar]
| Samples | Places | Types | Species | Media | References |
|---|---|---|---|---|---|
| High-temperature DQ 1 | Maotai, Guizhou | JXXB | Thermoactinomyces vulgaris | GTY medium | [24] |
| High-temperature DQ | Sichuan | JXXB | Streptomyces rochei | Casein medium | [25] |
| High-temperature DQ | Guizhou | JXXB | Streptomyces cacaoi, Streptomyces zaomyceticus | Gause No. 1 medium, ISP2 medium | [26] |
| High-temperature DQ | Maotai, Guizhou | JXXB | Laceyella sacchari | Modified Gause No. 2 Medium, ISP2 medium | [27] |
| High-temperature DQ | Guizhou | JXXB | Streptomyces griseus, Streptomyces albus | Gause No. 1 medium | [28] |
| High-temperature DQ | Maotai, Guizhou | JXXB | Streptomyces sp. FBKL4.005 | ISP2 medium | [29] |
| High-temperature DQ | Maotai, Guizhou | JXXB | Streptomyces bangladeshensis | GTY medium | [30] |
| FG 2 | Gulin, Sichuan | JXXB | Aggregatibacter actinomycetemcomitans | Beef extract peptone medium | [31] |
| Alcoholic fermentative material | Huaihua, Guizhou | JXXB | Streptomyces flocculus | Gause No. 1 medium | [32] |
| Soilof baijiu production environment | Guizhou | JXXB | Streptomyces sp. R11-21 | Gause No. 1 medium | [33] |
| FG | Huaihua, Guizhou | JXXB | Thermostaphylospora chromogena | Gause No. 1 medium | [34] |
| DQ | Yibin, Sichuan | NXXB | Streptomyces althioticus, Streptomycescoelicoflavus, Streptomyces diastaticus subsp. ardesiacus, Streptomyces ghanaensis, Streptomyces mutabilis, Streptomyces pactum, Streptomyces rubiginosohelvolus, Streptomyces somaliensis, Streptomyces thermocarboxydus, Streptomyces vinaceusdrappus, Streptomyces violascens, Streptomyces viridobrunneus | Modified Gause No. 1 Medium | [35] |
| FG | Yibin, Sichuan | NXXB | Streptomyces coerulescens, Streptomyces somaliensis, Streptomyces ghanaensis, Streptomyces violascens, | ||
| PM 3 | Yibin, Sichuan | NXXB | Massilia timonae, Nocardiopsis prasina, Streptomyces albofaciens, Streptomyces althioticus, Streptomyces celluloflavus, Streptomyces cinereoruber subsp. cinereoruber, Streptomyces coelicoflavus, Streptomyces cyaneofuscatus, Streptomyces diastaticus subsp. ardesiacus, Streptomyces fimicarius, Streptomyces flavovirens, Streptomyces ghobisporus subsp. globisporus, Streptomyces griseoplanus, Streptomyces halstedii, Streptomyces matensis, Streptomyces olivaceus, Streptomyces pactum, Streptomyces rubiginosohelvolus, Streptomyces sclerotialus, Streptomyces sindenensis, Streptomyces somaliensis, Streptomyces thermocarboxydus, Streptomyces vinaceusdrappus, Streptomyces violascens | ||
| Mature medial-temperature DQ | Henan | NXXB | Streptomyces lividans, Nocardiopsis dassonvillei, Streptomyces azureus, Streptomyces xiamenensis, Streptomyces cacaoi subsp. Cacaoi | Gause No. 1 medium | [36] |
| Sealing mud | Yibin, Sichuan | NXXB | Streptomyces sp. JP12 | Gause No. 1 medium | [37] |
| FG | Yibin, Sichuan | NXXB | Streptomyces sp. RZ1 | ||
| PM | Yibin, Sichuan | NXXB | Micromonospora sp. JD3 | ||
| The air of fermentation pit | Yibin, Sichuan | NXXB | Streptomyces vinaceusdrappus, | ||
| PM (20 years) | Yibin, Sichuan | NXXB | Arthrobacter protophormiae | enriched medium and Inorganic salt medium | [38] |
| PM | Yibin, Sichuan | NXXB | Streptomyces albus | Gause No. 1 medium | [39] |
| PM | Luzhou, Sichuan | NXXB | Streptomyces roseosporus, Streptomyces griseorubroviolaceus, Streptomyces aureus | Gause No. 1 medium and complete Inorganic Basal Medium | [40] |
| PM | Sichuan | NXXB | Thermophilibacter gallinarum | R2A medium | [41] |
| PM (50 years) | Southern of Sichuan | NXXB | Streptomyces sampsonii, Streptomyces rutgersensis | Situ-medium | [42] |
| PM | Hubei | NXXB | Streptomyces avicenniae | Gause No. 1 medium | [43] |
| PM (50 years) | Anhui | NXXB | Actinomyces israelii, Actinomyces meyeri, Bifidobacterium minimum, Bifidobacterium magnum, Bifidobacterium breve, Arthrobacter nicotianae, Nocardia africana, Nocardia altamirensis, Nocardia carnea, Nocardia cerradoensis, Nocardia flavirosea, Nocardia nova, Nocardia xishanensis | Isolation medium | [44] |
| DQ | Zibo, Shandong | ZMXXB | Thermoactinomyces daqus | R2A medium | [45] |
| High-temperature DQ | Shandong | ZMXXB | Thermoactinomyces vulgaris | R2A medium | [46] |
| High-temperature DQ | Shandong | ZMXXB | Thermoactinomyces intermedius, Laceyella tengchongensis, Laceyella sediminis, Laceyella sacchari, Laceyella putida | ISP2 medium | [47] |
| High-temperature DQ | Shandong | ZMXXB | Thermoactinomyces vulgaris, Streptomyces thermoviolaceus | R2A medium | [48] |
| DQ | Xinghuacun, Shanxi | QXXB | Streptomyces sp. ZYP3, Streptomyces sp. ZYP6, Streptomyces sp. ZYP7, Streptomyces sp. ZYP10, Streptomyces sp. ZYP12 Streptomyces sp. ZYP13, Streptomyces sp. ZYP15, Streptomyces sp. ZYP16, Streptomyces sp. ZYP17, Streptomyces sp. ZYP18 | GS medium | [15] |
| DQ | Xinghuacun, Shanxi | QXXB | Streptomyces sp. ZYP11 | GW1 medium | |
| DQ | Xinghuacun, Shanxi | QXXB | Streptomyces sp. ZYP9 | R2A medium | |
| DQ | Xinghuacun, Shanxi | QXXB | Streptomyces sp. ZYP8 | GMKA medium | |
| DQ | Xinghuacun, Shanxi | QXXB | Streptomyces sp.ZYP1, Streptomyces sp. ZYP14 | HV medium | |
| DQ | Shanxi | QXXB | Brevibacterium renqingii | LSA medium | [49] |
| DQ | Beijing | QXXB | Streptomyces albus, Streptomyces cacaoi | ISP2 medium | [50] |
| Out part of DQ | Shanxi Xinghuacun | QXXB | Bervibactrium sp. Micrococcus lutens | MRSA medium | [51] |
| DQ, FG | Beijing | QXXB | Shimazuella kribbensis, Kroppenstedtia sanguinis, Kroppenstedtia eburnea | Modified Gause No. 2 Medium | [52] |
| DQ | Shaanxi | FXXB | Arthrobacter aresens | Gause No. 1 medium | [53] |
| DQ | Shaanxi | FXXB | Streptomyces albus | PDA medium | [54] |
| FG | Shaanxi | FXXB | Arthrobacter aresens | Gause No. 1 medium | [55] |
| Samples | Types | Places | Methods | Species | References |
|---|---|---|---|---|---|
| High-temperature DQ 1 | JXXB | Sichuan | PCR-DGGE | Thermoactinomyces sanguinis | [57] |
| DQ | JXXB | Jiangsu | PCR-DGGE | Thermoactinomyces sanguinis | [65] |
| FG 2 | JXXB | Guizhou | PCR-DGGE | Thermoactinomyces sanguinis | [58] |
| FG | JXXB | Renhuai, Guizhou | PCR-DGGE | Uncultured Actinomycete colone 4-306, Corynebacterium sp. DNF00584, Streptomyces sp. MG21, Actinomycetales sp. JB111, Thermoactinomyces sanguinis | [66] |
| DQ | NXXB | South | PCR-DGGE | Thermoactinomyces vulgaris | [67] |
| Baobaoqu | NXXB | Sichuan | LC-MS/MS | Microbacterium hominis, Thermoactinomyces vulgaris | [68] |
| High-temperature DQ | NXXB | Anhui | 16S rDNA | Thermoactinomyces sanguinis | [59] |
| PM 3 | NXXB | Sichuan | PCR-DGGE | Olsenella uli, Olsenella profusa, Lancefieldellaparvuium, Corynebacterium tuberculostearicum, Corynebacterium minutissimum, Streptomyces coeruleorubidus, Streptomyces hainanensis | [61] |
| PM | NXXB | Sichuan | 16S rRNA | Arthrobacter stackebrandtii, Kocuria carniphila, Glutamicibacter creatinolyticus, Brevibacterium aurantiacum, Cellulosimicrobium funkei, Microbacterium oxydans, Corynebacterium glutamicum, Gordonia terrae, Dietzia maris, Acidipropionibacterium acidipropionici, Microbacterium hydrocarbonoxydans, Microbacterium schleiferi, Gulosibacter molinativorax | [62] |
| FG | NXXB | Sichuan | PCR-DGGE | Arthrobacter woluwensis | [69] |
| Alcoholic fermentative materials | NXXB | Sichuan | PCR-DGGE | Thermoactinomyces sanguinis | [70] |
| Low-temperature DQ | QXXB | Beijing, Shanxi, Taiwan, Heilongjiang, Hebei | 16S rDNA | Streptomyces albus, Kroppenstedtia eburnea | [71] |
| Mature DQ | QXXB | Shanxi | 16SrRNA | Thermoactinomyces sanguinis, Streptomyces albus, Brevibacterium linens, Brachybacterium paraconglomeratum, Rothia koreensis | [60] |
| DQ | QXXB | Xiangyang, Hubei | WGS | Streptomyces albus, Streptomyces NHF165, Saccharopolyspora erythraea | [72] |
| High-temperature DQ | ZMXXB | Shandong | 16S rDNA | Thermoactinomyces vulgaris | [73] |
| DQ (8 days of fermentation) | ZMXXB | Shandong | 16S rDNA | Saccharopolyspora rectivirgula, Brevibacterium celere | [63] |
| DQ (24 days of fermentation) | ZMXXB | Shandong | 16S rDNA | Saccharopolyspora hordei, Saccharopolyspora rectivirgula, Rothiakristinae, Marmoricola aurantiacus | |
| DQ (49 days of fermentation) | ZMXXB | Shandong | 16S rDNA | Thermoactinomycesintermedius, Thermoactinomyces vulgaris, Laceyella putida, Kroppenstedtia eburnea, Saccharopolyspora hordei, Saccharopolyspora rectivirgula, Streptoalloteichus hindustanus, Rubrobacter radiotolerans, Streptomyces coeruleoprunus, Aciditerrimonasferrireducens, Nesterenkonia flava, Modestobacter versicolor, Ornithinimicrobium pekingense, Cutibacterium acnes | |
| DQ | Northern and southwestern | 16S rRNA and 26S rRNA | Saccharypolyspora rosea, Saccharopolyspora rectivirgula, Saccharopolyspora hordei, Saccharopolyspora spinosa, Streptomyces albus, Streptomyces cacaoi, Thermoactinomyces sanguinis, Thermoactinomyces vulgaris, Thermobisporabispora, Thermostaphylospora chromogena, Actinopolyspora erythraea | [74] |
| Species | Strain | NCBI Access No. | Size (Mb) | GC% | Proteins | rRNA Operons | tRNA Genes | References |
|---|---|---|---|---|---|---|---|---|
| Thermoactinomyces daqus | H-18 | NZ_JPST01000000 | 3.44 | 48.8 | 3440 | 5 | 58 | [45] |
| Streptomyces mutabilis | Z9A-32 | HQ238326 | 7.83 | 71.6 | 6711 | 19 | 82 | [35] |
| Streptomyces vinaceusdrappus | W8A-43 | HQ238406 | 8.46 | 72.5 | 7332 | 5 | 67 | [35] |
| Streptomyces violascens | S11A-6 | HQ238298 | 8.92 | 70.5 | 7,754 | 24 | 71 | [35] |
| Streptomyces griseus | A2 | JX007982 | 8.55 | 72.2 | 6968 | 18 | 67 | [28] |
| Streptomyces albus | NRRL B-2365 | DQ026669.1 | 7.59 | 72.7 | 6166 | 4 | 59 | [28] |
| Thermoactinomyces vulgaris | ATCC15734 | AF089892 | 2.62 | 48 | 2590 | 21 | 72 | [24] |
| Thermophilibacter gallinarum | LZLJ-2T | NZ_JADCJZ000000000.1 | 1.85 | 65.2 | 1599 | 6 | 49 | [41] |
| Streptomyces cacaoi | NBRC12748 | NR041061 | 8.57 | 73.4 | 6679 | 8 | 61 | [50] |
| Arthrobacter stackebrandtii | NG3 | MT269547.1 | 4.43 | 65.6 | 3683 | 15 | 54 | [62] |
| Microorganisms | Substrate | Product | References |
|---|---|---|---|
| Streptomyces sp. R11-21 | Flour | 2-Methyl isobornyl alcohol and terpenes | [33] |
| Wheat or sorghum | 3-Hydroxy-2-butanone and terpenes | ||
| Streptomyces bangladeshensis | Glucose | Terpenes and pyrazine | [30] |
| Streptomyces albus Streptomyces sampsonii Streptomyces rutgersensis | Wheat bran | 3-Hydroxy-2-butanone, 2,3-butanediol, ethanol and ethyl acetate | [39] |
| Streptomyces mutabilis Streptomyces vinaceusdrappus Streptomyces coelicoflavus Streptomyces violascens | Glucose | Butyric acid, hexanoic acid, ethyl butyrate, ethyl hexanoate, ethyl lactate and furfural | [152] |
| Thermoactinomycetes FBKL4.010 | Wheat | Furfuryl alcohol, phenethyl alcohol, 2,6-dimethylpyrazine, and 2,3,5,6-tetramethylpyrazine | |
| Streptomyces sp. A22 | Wheat bran | Ethyl hexanoate and phenethyl alcohol | [132] |
| Streptomyces aureus | Tyrosine | Brown-pigment | [149] |
| Streptomyces albus S5 | Glucose | Salinomycin | [148] |
| Streptomyces cacaoi Streptomyces zaomyceticus | Sorghum | Lipids | [26] |
| Laceyellasacchari | Sorghum | Tetramethylpyrazine | [27] |
| Streptomyces sampsonii Streptomyces rutgersensis | Oat | 3-Hydroxy-2-butanone and 2,3-butanediol | [42] |
| Glucose | Ethyl caproate, and geosmin | ||
| Streptomyces fradiae, Streptomyces radiopugnans, Streptomyces sampsonii, Streptomyces albus | Starch | Geosmin | [16] |
| Streptomyces spp. | Starch | Ethyl lactate and caproic acid | [146] |
| Streptomyces avicenniae | Starch | Melanin | [126] |
| Thermoactinomyces sp. | Starch | Butanoate, Aceticacid, hexyl ester | [143] |
| Streptomyces sampsonii | Starch | Heptaene macrolide antibiotics | [128] |
| Streptomyces albus Streptomyces griseus | Cellulose | Bultanol, acetone, 3-methyl-3-buten-1-ol, dimethyl disulfide | [144] |
| Thermoactinomyces sp. | Starch | Pyrazines, aromatics and and alcohol | [145] |
| Actinomycetales sp. | |||
| Thermophilibacter gallinarum | Glucose | Lactic acid and acetic acid | [41] |
| Streptomyces albus | Glucose | Poly-lysine | [150] |
| Actinomycetes | DQ | Enzymes, pyrazine and aromatic substances | [123] |
| FG | Salinomycin and terpenes | ||
| PM | Acids, esters and terpenes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Yang, H.; Liu, J.; Luo, H.; Zou, W. Systematic Review of Actinomycetes in the Baijiu Fermentation Microbiome. Foods 2022, 11, 3551. https://doi.org/10.3390/foods11223551
Chen C, Yang H, Liu J, Luo H, Zou W. Systematic Review of Actinomycetes in the Baijiu Fermentation Microbiome. Foods. 2022; 11(22):3551. https://doi.org/10.3390/foods11223551
Chicago/Turabian StyleChen, Cong, Haiquan Yang, Jie Liu, Huibo Luo, and Wei Zou. 2022. "Systematic Review of Actinomycetes in the Baijiu Fermentation Microbiome" Foods 11, no. 22: 3551. https://doi.org/10.3390/foods11223551
APA StyleChen, C., Yang, H., Liu, J., Luo, H., & Zou, W. (2022). Systematic Review of Actinomycetes in the Baijiu Fermentation Microbiome. Foods, 11(22), 3551. https://doi.org/10.3390/foods11223551