Heterologous Expression of the Lactobacillus sakei Multiple Copper Oxidase to Degrade Histamine and Tyramine at Different Environmental Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Reagents, and Materials
2.2. Cloning and Heterologous Expression of the MCO Gene
2.3. Purification of rMCO
2.4. rMCO Activity Assay
2.5. Amine-Oxidizing Activity of rMCO
3. Results and Discussion
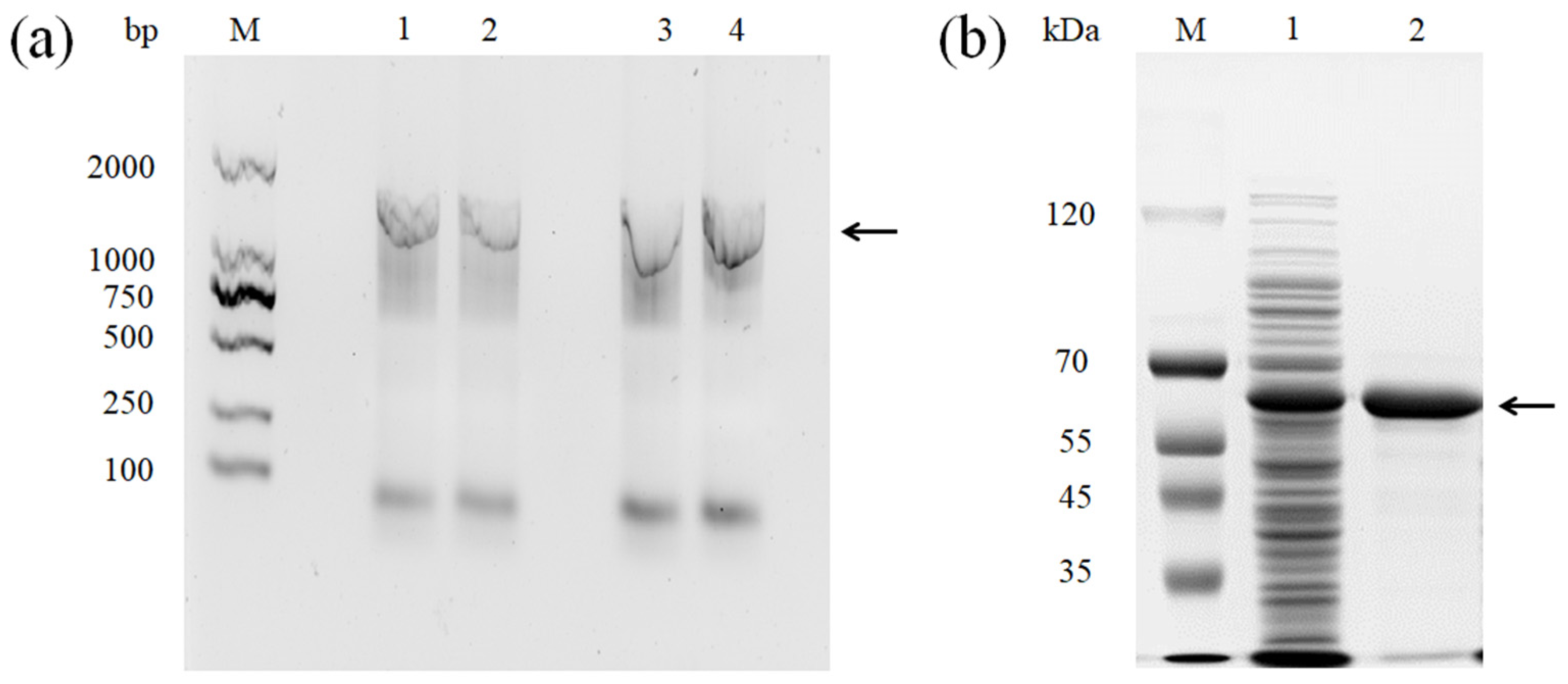
3.1. Sequence Analysis and Heterologous Expression of the rMCO Gene from L. sakei Ls
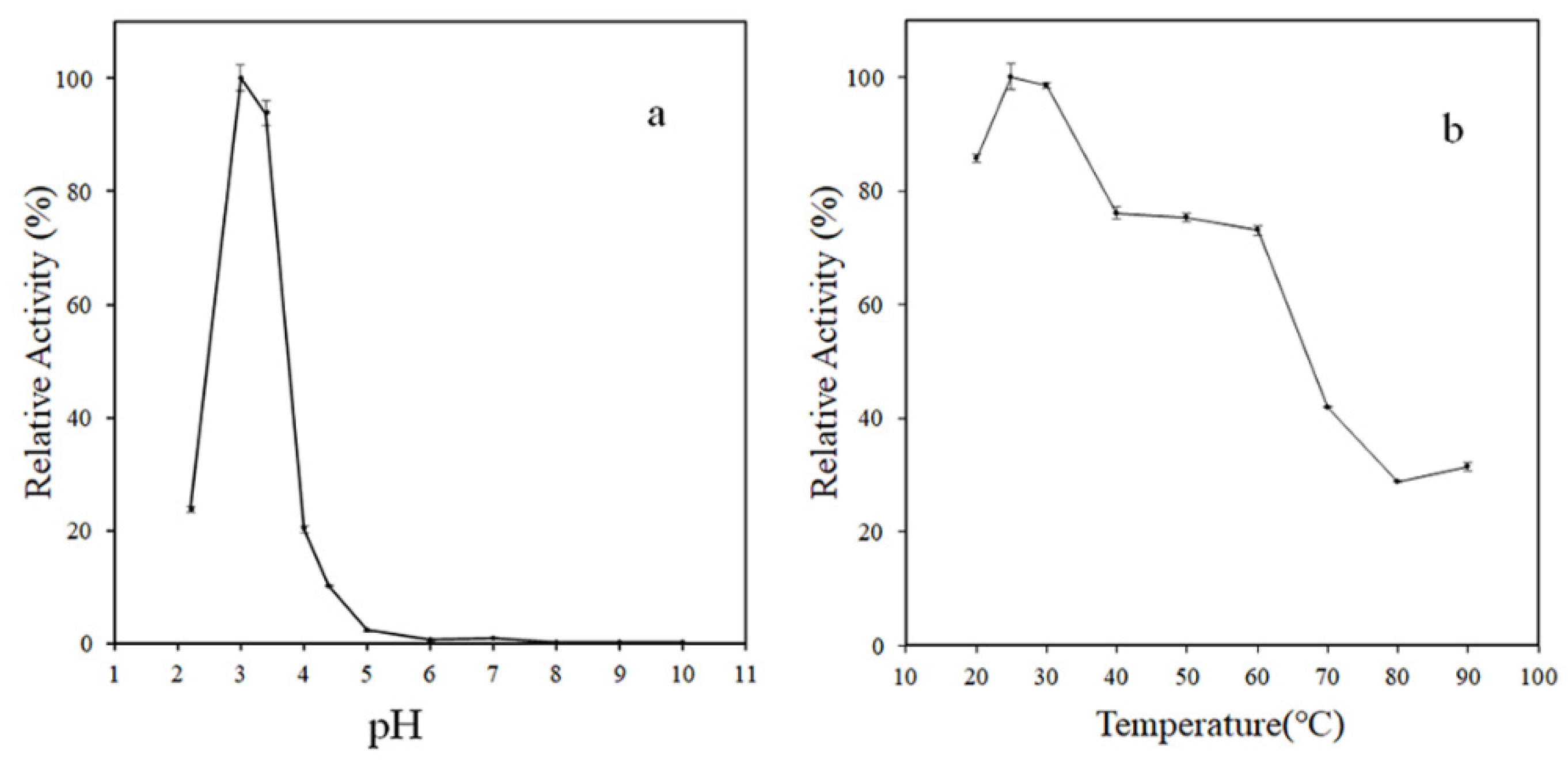
3.2. Enzymatic Characterization of rMCO
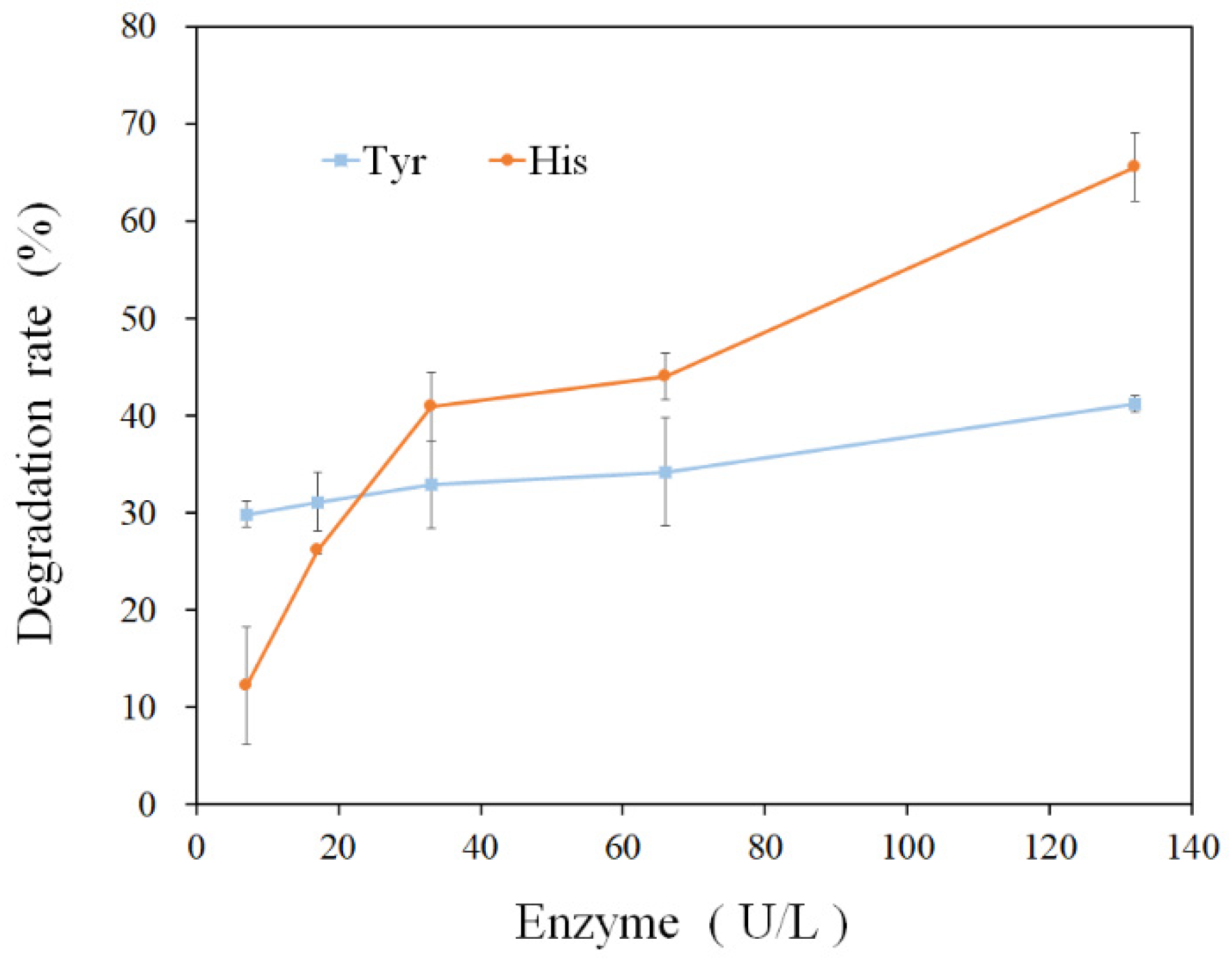
3.3. Amine-Oxidizing Activity of rMCO
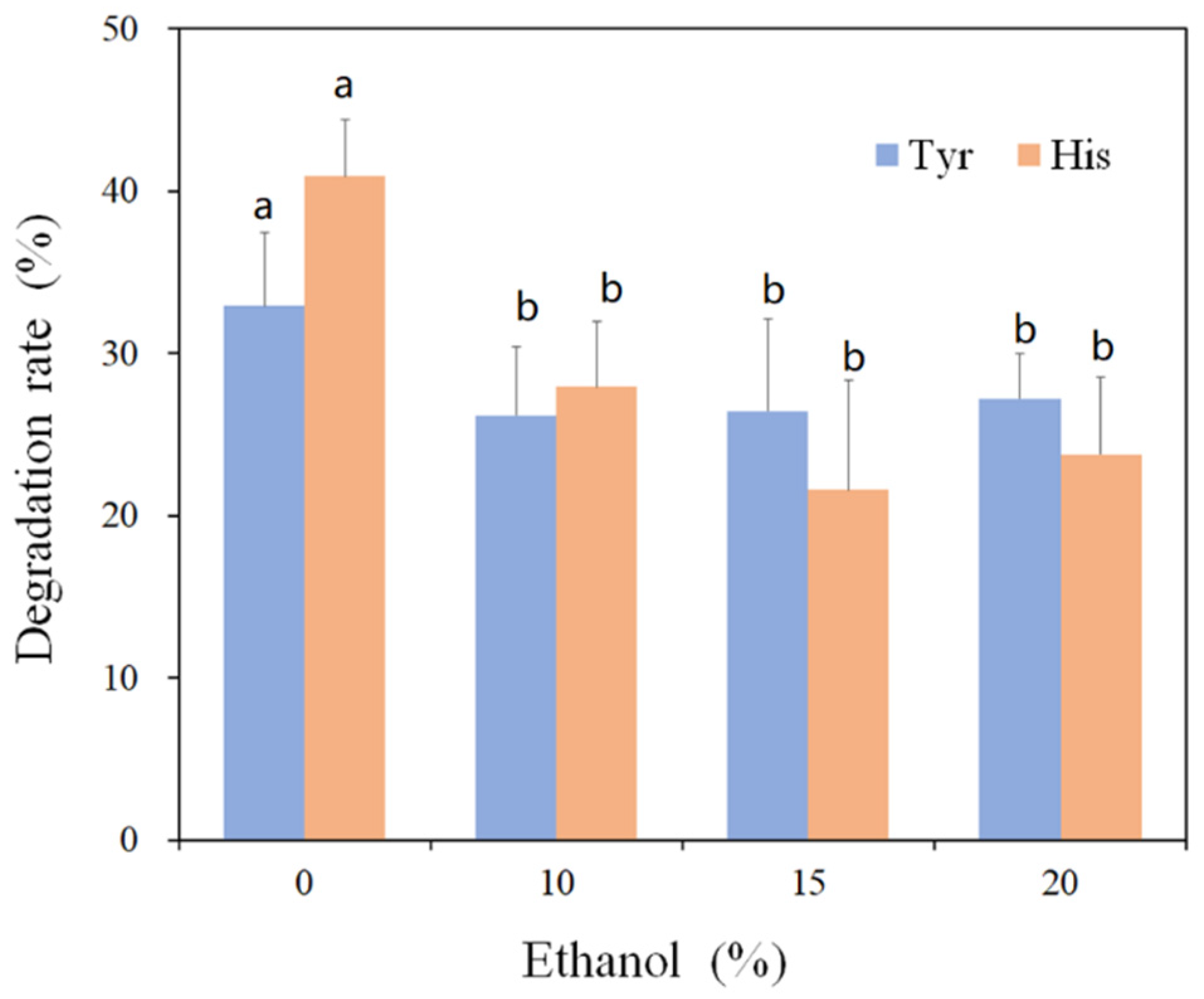
3.4. Effect of Food Matrix on the Amine-Oxidizing Activity of rMCO
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, B.; Redruello, B.; Linares, D.M.; Ladero, V.; Fernandez, M.; Martin, M.C.; Ruas-Madiedo, P.; Alvarez, M.A. The dietary biogenic amines tyramine and histamine show synergistic toxicity towards intestinal cells in culture. Food Chem. 2017, 218, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Chen, J.; Du, G.; Fang, F. Food-grade expression of multicopper oxidase with improved capability in degrading biogenic amines. Syst. Microbiol. Biomanuf. 2021, 2, 285–295. [Google Scholar] [CrossRef]
- Guan, Z.-B.; Luo, Q.; Wang, H.-R.; Chen, Y.; Liao, X.-R. Bacterial laccases: Promising biological green tools for industrial applications. Cell. Mol. Life Sci. 2018, 75, 3569–3592. [Google Scholar] [CrossRef]
- Guarcello, R.; De Angelis, M.; Settanni, L.; Formiglio, S.; Gaglio, R.; Minervini, F.; Moschetti, G.; Gobbetti, M. Selection of Amine-Oxidizing Dairy Lactic Acid Bacteria and Identification of the Enzyme and Gene Involved in the Decrease of Biogenic Amines. Appl. Environ. Microbiol. 2016, 82, 6870–6880. [Google Scholar] [CrossRef]
- Callejón, S.; Sendra, R.; Ferrer, S.; Pardo, I. Cloning and characterization of a new laccase from Lactobacillus plantarum J16 CECT 8944 catalyzing biogenic amines degradation. Appl. Microbiol. Biotechnol. 2016, 100, 3113–3124. [Google Scholar] [CrossRef]
- Li, B.; Wang, Y.; Xue, L.; Lu, S. Heterologous Expression and Application of Multicopper Oxidases from Enterococcus spp. for Degradation of Biogenic Amines. Protein Pept. Lett. 2021, 28, 183–194. [Google Scholar] [CrossRef]
- Olmeda, I.; Casino, P.; Collins, R.E.; Sendra, R.; Callejón, S.; Huesa, J.; Soares, A.S.; Ferrer, S.; Pardo, I. Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species. Microb. Biotechnol. 2021, 14, 1026–1043. [Google Scholar] [CrossRef]
- Wang, S.; Han, J.; Zhang, J.B.; Lin, X.P.; Liang, H.P.; Li, S.J.; Dong, X.P.; Ji, C.F. Effects of Temperature on Bacterial Biodiversity and Qualities of Fermented Yucha Products. J. Aquat. Food Prod. Technol. 2020, 29, 43–54. [Google Scholar] [CrossRef]
- Li, T.; Huang, L.; Li, Y.; Xu, Z.; Ge, X.; Zhang, Y.; Wang, N.; Wang, S.; Yang, W.; Lu, F.; et al. The heterologous expression, characterization, and application of a novel laccase from Bacillus velezensis. Sci. Total Environ. 2020, 713, 136713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, J.; Wang, S.; Lin, X.; Liang, H.; Li, S.; Yu, C.; Dong, X.; Ji, C. Developing and Validating a UPLC-MS Method with a StageTip-Based Extraction for the Biogenic Amines Analysis in Fish. J. Food Sci. 2019, 84, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Liu, H.; Zeng, X. Structural and functional study on cysteine 495, coordinating ligand to T1Cu site in multicopper oxidase CopA. Chemosphere 2021, 281, 130807. [Google Scholar] [CrossRef] [PubMed]
- Huy, N.D.; My Le, N.T.; Chew, K.W.; Park, S.-M.; Show, P.L. Characterization of a recombinant laccase from Fusarium oxysporum HUIB02 for biochemical application on dyes removal. Biochem. Eng. J. 2021, 168, 107958. [Google Scholar] [CrossRef]
- Wu, Y.-R.; Luo, Z.-H.; Kwok-Kei Chow, R.; Vrijmoed, L.L.P. Purification and characterization of an extracellular laccase from the anthracene-degrading fungus Fusarium solani MAS2. Bioresour. Technol. 2010, 101, 9772–9777. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Zhu, Q.; Wu, Y.; Lin, X. Oxidation of polycyclic aromatic hydrocarbons using Bacillus subtilis CotA with high laccase activity and copper independence. Chemosphere 2016, 148, 1–7. [Google Scholar] [CrossRef]
- Dewey-Mattia, D.; Manikonda, K.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1–11. [Google Scholar] [CrossRef]
- Reiss, R.; Ihssen, J.; Richter, M.; Eichhorn, E.; Schilling, B.; Thöny-Meyer, L. Laccase versus laccase-like multi-copper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PLoS ONE 2013, 8, e65633. [Google Scholar] [CrossRef]
- Wan, Y.-Y.; Lu, R.; Xiao, L.; Du, Y.-M.; Miyakoshi, T.; Chen, C.-L.; Knill, C.J.; Kennedy, J.F. Effects of organic solvents on the activity of free and immobilised laccase from Rhus vernicifera. Int. J. Biol. Macromol. 2010, 47, 488–495. [Google Scholar] [CrossRef]
- Cretin, B.N.; Dubourdieu, D.; Marchal, A. Influence of ethanol content on sweetness and bitterness perception in dry wines. LWT 2018, 87, 61–66. [Google Scholar]
- Xu, F. Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 1997, 272, 924–928. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, S.; Xie, Y.; Fang, Z.; Xiao, Y.; Fang, W.; Zhang, X. Mechanism of the salt activation of laccase Lac15. Biochem. Biophys. Res. Commun. 2020, 521, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Steffensen, C.L.; Stensballe, A.; Kidmose, U.; Degn, P.E.; Andersen, M.L.; Nielsen, J.H. Modifications of amino acids during ferulic acid-mediated, laccase-catalysed cross-linking of peptides. Free Radic. Res. 2009, 43, 1167–1178. [Google Scholar] [CrossRef]
- Baiano, A. Phenolic compounds and antioxidant activity of experimental and industrial table grape juices. J. Food Process. Preserv. 2020, 44, e14655. [Google Scholar] [CrossRef]
- Mohamedshah, Z.; Chadwick-Corbin, S.; Wightman, J.D.; Ferruzzi, M.G. Comparative assessment of phenolic bioaccessibility from 100% grape juice and whole grapes. Food Funct. 2020, 11, 6433–6445. [Google Scholar] [CrossRef]
- Calcaterra, A.; Galli, C.; Gentili, P. Phenolic compounds as likely natural mediators of laccase: A mechanistic assessment. J. Mol. Catal. B Enzym. 2008, 51, 118–120. [Google Scholar] [CrossRef]
- Terrón, M.C.; López-Fernández, M.; Carbajo, J.M.; Junca, H.; Téllez, A.; Yagüe, S.; Arana-Cuenca, A.; González, T.; González, A.E. Tannic acid interferes with the commonly used laccase-detection assay based on ABTS as the substrate. Biochimie 2004, 86, 519–522. [Google Scholar] [CrossRef]
- Vignault, A.; Pascual, O.; Jourdes, M.; Moine, V.; Fermaud, M.; Roudet, J.; Canals, J.-M.; Teissedre, P.-L.; Zamora, F. Impact of enological tannins on laccase activity. OENO One 2019, 53, 27–38. [Google Scholar] [CrossRef]
- Ying, F.; Lin, S.; Li, J.; Zhang, X.; Chen, G. Identification of monoamine oxidases inhibitory peptides from soybean protein hydrolysate through ultrafiltration purification and in silico studies. Food Biosci. 2021, 44, 101355. [Google Scholar] [CrossRef]
- Arii, Y.; Takenaka, Y. Initiation of protein association in tofu formation by metal ions. Biosci. Biotechnol. Biochem. 2014, 78, 86–91. [Google Scholar] [CrossRef] [PubMed]




| Metal Ions /Inhibitors /Organic Reagents | Concentration (mM/%) | Relative Activity (%) |
|---|---|---|
| None | - | 100 ± 2.4 |
| CuCl2 | 0.5 | 95.7 ± 2.4 |
| 5 | 68.1 ± 1.7 | |
| MgCl2 | 0.5 | 101.5 ± 3.4 |
| 5 | 75.9 ± 2.0 | |
| FeCl2 | 0.5 | 11.1 ± 0.8 |
| 5 | 2.2 ± 0.1 | |
| MnSO4 | 0.5 | 98.9 ± 2.7 |
| 5 | 103.1 ± 1.9 | |
| ZnCl2 | 0.5 | 95.9 ± 2.3 |
| 5 | 80.1 ± 1.5 | |
| KCl | 0.5 | 101.2 ± 2.0 |
| 5 | 85.4 ± 2.1 | |
| CaCl2 | 0.5 | 98.8 ± 2.9 |
| 5 | 72.5 ± 1.8 | |
| FeCl3 | 0.5 | 96.6 ± 2.0 |
| 5 | 51.0 ± 1.2 | |
| NaCl | 0.5 | 96.5 ± 2.9 |
| 5 | 79.3 ± 2.1 | |
| SDS | 0.5 | 1.4 ± 0.1 |
| 5 | 0.3 ± 0.1 | |
| EDTA | 0.5 | 100.0 ± 2.6 |
| 5 | 96.8 ± 2.9 | |
| L-Cysteine | 0.5 | 22.2 ± 0.7 |
| 5 | 0.4 ± 0.1 | |
| Methanol | 1% | 94.4 ± 2.0 |
| 10% | 80.6 ± 2.2 | |
| Ethanol | 1% | 97.2 ± 1.6 |
| 10% | 70.6 ± 1.9 | |
| Isopropanol | 1% | 98.2 ± 2.3 |
| 10% | 61.8 ± 2.5 | |
| Acetonitrile | 1% | 85.3 ± 1.9 |
| 10% | 24.9 ± 0.6 | |
| Acetone | 1% | 101.9 ± 2.9 |
| 10% | 41.2 ± 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhao, Y.; Zhang, S.; Lin, X.; Liang, H.; Chen, Y.; Ji, C. Heterologous Expression of the Lactobacillus sakei Multiple Copper Oxidase to Degrade Histamine and Tyramine at Different Environmental Conditions. Foods 2022, 11, 3306. https://doi.org/10.3390/foods11203306
Wang X, Zhao Y, Zhang S, Lin X, Liang H, Chen Y, Ji C. Heterologous Expression of the Lactobacillus sakei Multiple Copper Oxidase to Degrade Histamine and Tyramine at Different Environmental Conditions. Foods. 2022; 11(20):3306. https://doi.org/10.3390/foods11203306
Chicago/Turabian StyleWang, Xiaofu, Yunsong Zhao, Sufang Zhang, Xinping Lin, Huipeng Liang, Yingxi Chen, and Chaofan Ji. 2022. "Heterologous Expression of the Lactobacillus sakei Multiple Copper Oxidase to Degrade Histamine and Tyramine at Different Environmental Conditions" Foods 11, no. 20: 3306. https://doi.org/10.3390/foods11203306
APA StyleWang, X., Zhao, Y., Zhang, S., Lin, X., Liang, H., Chen, Y., & Ji, C. (2022). Heterologous Expression of the Lactobacillus sakei Multiple Copper Oxidase to Degrade Histamine and Tyramine at Different Environmental Conditions. Foods, 11(20), 3306. https://doi.org/10.3390/foods11203306




