Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. PCR and 16S rRNA Gene Sequencing
2.4. Microbial Data Pre-Processing
2.5. Statistical Analysis
3. Results
3.1. Sequence Quality Control
3.2. Alpha and Beta Diversities of Milk Microbiota Based on Months
3.3. Differences in Microbial Composition of Raw Milk between Samples Collected in Different Months
3.4. Correlation Analysis between Dominant Bacterial Genera and Raw Milk Quality Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| RDA | redundancy analysis |
| PCoA | Principal coordinate analysis |
| ASVs | amplicon sequence variants |
| TA | titratable acidity |
| TBC | total bacteria count |
| SCC | somatic cell count |
References
- Skeie, S.B.; Håland, M.; Thorsen, I.M.; Narvhus, J.; Porcellato, D. Bulk tank raw milk microbiota differs within and between farms: A moving goalpost challenging quality control. J. Dairy Sci. 2019, 102, 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Morandi, S.; Ceccarani, C.; Battelli, G.; Castiglioni, B.; Cologna, N.; Goss, A.; Severgnini, M.; Mazzucchi, M.; Partel, E.; et al. Raw Milk Microbiota Modifications as Affected by Chlorine Usage for Cleaning Procedures: The Trentingrana PDO Case. Front. Microbiol. 2020, 11, 564749. [Google Scholar] [CrossRef] [PubMed]
- Berhanu, L.; Gume, B.; Kassa, T.; Dadi, L.S.; Tegegne, D.; Getnet, M.; Bediru, H.; Getaneh, A.; Suleman, S.; Mereta, S.T. Microbial quality of raw cow milk and its predictors along the dairy value chain in Southwest Ethiopia. Int. J. Food Microbiol. 2021, 350, 109228. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.D.; De Block, J.; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef]
- Parente, E.; Ricciardi, A.; Zotta, T. The microbiota of dairy milk: A review. Int. Dairy J. 2020, 107, 104714. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; You, C.; Ren, J.; Chen, W.; Zheng, H.; Liu, Z. Variation in Raw Milk Microbiota Throughout 12 Months and the Impact of Weather Conditions. Sci. Rep. 2018, 8, 2371. [Google Scholar] [CrossRef]
- Naing, Y.W.; Wai, S.S.; Lin, T.N.; Thu, W.P.; Htun, L.L.; Bawm, S.; Myaing, T.T. Bacterial content and associated risk factors influencing the quality of bulk tank milk collected from dairy cattle farms in Mandalay Region. Food Sci. Nutr. 2019, 7, 1063–1071. [Google Scholar] [CrossRef]
- Hornik, B.; Czarny, J.; Staninska-Pięta, J.; Wolko, Ł.; Cyplik, P.; Piotrowska-Cyplik, A. The Raw Milk Microbiota from Semi-Subsistence Farms Characteristics by NGS Analysis Method. Molecules 2021, 26, 5029. [Google Scholar] [CrossRef]
- Breitenwieser, F.; Doll, E.; Clavel, T.; Scherer, S.; Wenning, M. Complementary Use of Cultivation and High-Throughput Amplicon Sequencing Reveals High Biodiversity Within Raw Milk Microbiota. Front. Microbiol. 2020, 11, 1557. [Google Scholar] [CrossRef]
- Joishy, T.K.; Dehingia, M.; Khan, M.R. Bacterial diversity and metabolite profiles of curd prepared by natural fermentation of raw milk and back sloping of boiled milk. World J. Microbiol. Biotechnol. 2019, 35, 102. [Google Scholar] [CrossRef]
- Cao, H.; Yan, Y.; Wang, L.; Dong, L.; Pang, X.; Tang, S.; Li, A.; Xiang, A.; Zhang, L.; Zheng, B. High-Throughput Sequencing Reveals Bacterial Diversity in Raw Milk Production Environment and Production Chain in Tangshan City of China. Food Sci. Anim. Resour. 2021, 41, 452–467. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Meng, L.; Liu, H.; Zheng, N.; Zhang, Y.; Guo, X.; Zhao, S.; Li, F.; Wang, J. Impacts of Milking and Housing Environment on Milk Microbiota. Animals 2020, 10, 2339. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Chieffi, D.; Fanelli, F.; Logrieco, A.F.; Cho, G.; Kabisch, J.; Böhnlein, C.; Franz, C.M.A.P. Microbial quality and safety of milk and milk products in the 21st century. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2013–2049. [Google Scholar] [CrossRef] [PubMed]
- Gastélum-Barrios, A.; Soto-Zarazúa, G.M.; Escamilla-García, A.; Toledano-Ayala, M.; Macías-Bobadilla, G.; Jauregui-Vazquez, D. Optical Methods Based on Ultraviolet, Visible, and Near-Infrared Spectra to Estimate Fat and Protein in Raw Milk: A Review. Sensors 2020, 20, 3356. [Google Scholar] [CrossRef]
- Goli, M.; Ezzatpanah, H.; Ghavami, M.; Chamani, M.; Aminafshar, M.; Toghiani, M.; Eghbalsaied, S. The effect of multiplex-PCR-assessed major pathogens causing subclinical mastitis on somatic cell profiles. Trop. Anim. Health Prod. 2012, 44, 1673–1680. [Google Scholar] [CrossRef]
- Carloni, E.; Petruzzelli, A.; Amagliani, G.; Brandi, G.; Caverni, F.; Mangili, P.; Tonucci, F. Effect of farm characteristics and practices on hygienic quality of ovine raw milk used for artisan cheese production in central Italy. Anim. Sci. J. 2016, 87, 591–599. [Google Scholar] [CrossRef]
- Ndahetuye, J.B.; Artursson, K.; Båge, R.; Ingabire, A.; Karege, C.; Djangwani, J.; Nyman, A.-K.; Ongol, M.P.; Tukei, M.; Persson, Y. MILK Symposium review: Microbiological quality and safety of milk from farm to milk collection centers in Rwanda. J. Dairy Sci. 2020, 103, 9730–9739. [Google Scholar] [CrossRef]
- Olatoye, O.; Amosun, A.; Ogbu, U.; Okunlade, Y. Bulk tank somatic cell count and associated microbial quality of milk from selected dairy cattle herds in Oyo State, Nigeria. Ital. J. Food Saf. 2018, 7, 7130. [Google Scholar] [CrossRef]
- Zhang, D.; Palmer, J.; Teh, K.H.; Biggs, P.; Flint, S. 16S rDNA high-throughput sequencing and MALDI-TOF MS are complementary when studying psychrotrophic bacterial diversity of raw cows’ milk. Int. Dairy J. 2019, 97, 86–91. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.; Ross, R.; Fitzgerald, G.; Cotter, P. A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J. Appl. Microbiol. 2012, 113, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Watson, M.; McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Chen, H. VennDiagram: Generate High-Resolution Venn and Euler Plots. 2018. Available online: https://CRAN.R-project.org/package=VennDiagram (accessed on 11 September 2019).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 15 February 2020).
- Yap, M.; Gleeson, D.; O’Toole, P.W.; O’Sullivan, O.; Cotter, P.D. Seasonality and Geography Have a Greater Influence than the Use of Chlorine-Based Cleaning Agents on the Microbiota of Bulk Tank Raw Milk. Appl. Environ. Microbiol. 2021, 87, e0108121. [Google Scholar] [CrossRef]
- Kable, M.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The Core and Seasonal Microbiota of Raw Bovine Milk in Tanker Trucks and the Impact of Transfer to a Milk Processing Facility. mBio 2016, 7, e00836-16. [Google Scholar] [CrossRef]
- Vithanage, N.R.; Dissanayake, M.; Bolge, G.; Palombo, E.A.; Yeager, T.R.; Datta, N. Biodiversity of culturable psychrotrophic microbiota in raw milk attributable to refrigeration conditions, seasonality and their spoilage potential. Int. Dairy J. 2016, 57, 80–90. [Google Scholar] [CrossRef]
- Liang, L.; Wang, P.; Zhao, X.; He, L.; Qu, T.; Chen, Y. Single-molecule real-time sequencing reveals differences in bacterial diversity in raw milk in different regions and seasons in China. J. Dairy Sci. 2022, 105, 5669–5684. [Google Scholar] [CrossRef]
- Ryu, S.; Park, W.S.; Yun, B.; Shin, M.; Go, G.-W.; Kim, J.N.; Oh, S.; Kim, Y. Diversity and characteristics of raw milk microbiota from Korean dairy farms using metagenomic and culturomic analysis. Food Control 2021, 127, 108160. [Google Scholar] [CrossRef]
- Lage, O.M.; van Niftrik, L.; Jogler, C.; Devos, D.P. Planctomycetes. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar] [CrossRef]
- Perin, L.M.; Pereira, J.G.; Bersot, L.S.; Nero, L.A. The Microbiology of Raw Milk. In Raw Milk; Academic press: Cambridge, MA, USA, 2019; pp. 45–64. [Google Scholar] [CrossRef]
- Rodrigues, M.; Lima, S.; Higgins, C.; Canniatti-Brazaca, S.; Bicalho, R. The Lactococcus genus as a potential emerging mastitis pathogen group: A report on an outbreak investigation. J. Dairy Sci. 2016, 99, 9864–9874. [Google Scholar] [CrossRef] [PubMed]
- Vithanage, N.R.; Dissanayake, M.; Bolge, G.; Palombo, E.A.; Yeager, T.R.; Datta, N. Microbiological quality of raw milk attributable to prolonged refrigeration conditions. J. Dairy Res. 2017, 84, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Renye, J.A.; Feng, L.; Zeng, Q.; Tang, Y.; Huang, L.; Ren, D.; Yang, P. Characterization of the indigenous microflora in raw and pasteurized buffalo milk during storage at refrigeration temperature by high-throughput sequencing. J. Dairy Sci. 2016, 99, 7016–7024. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Sadiq, F.A.; Liu, T.-J.; Li, Y.; Gu, J.-S.; Yang, H.-Y.; He, G.-Q. Spoilage potential of psychrotrophic bacteria isolated from raw milk and the thermo-stability of their enzymes. J. Zhejiang Univ. Sci. B 2018, 19, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Sadiq, F.A.; Liu, T.; Flint, S.; Chen, J.; Yang, H.; Gu, J.; Zhang, G.; He, G. Psychrotrophic bacterial populations in Chinese raw dairy milk. LWT 2017, 84, 409–418. [Google Scholar] [CrossRef]
- Carafa, I.; Navarro, I.C.; Bittante, G.; Tagliapietra, F.; Gallo, L.; Tuohy, K.; Franciosi, E. Shift in the cow milk microbiota during alpine pasture as analyzed by culture dependent and high-throughput sequencing techniques. Food Microbiol. 2020, 91, 103504. [Google Scholar] [CrossRef]
- Porcellato, D.; Smistad, M.; Bombelli, A.; Abdelghani, A.; Jørgensen, H.J.; Skeie, S.B. Longitudinal Study of the Bulk Tank Milk Microbiota Reveals Major Temporal Shifts in Composition. Front. Microbiol. 2021, 12, 616429. [Google Scholar] [CrossRef]
- Kaczorowski, Ł.; Powierska-Czarny, J.; Wolko, Ł.; Piotrowska-Cyplik, A.; Cyplik, P.; Czarny, J. The Influence of Bacteria Causing Subclinical Mastitis on the Structure of the Cow’s Milk Microbiome. Molecules 2022, 27, 1829. [Google Scholar] [CrossRef]
- Murphy, S.C.; Martin, N.H.; Barbano, D.M.; Wiedmann, M. Influence of raw milk quality on processed dairy products: How do raw milk quality test results relate to product quality and yield? J. Dairy Sci. 2016, 99, 10128–10149. [Google Scholar] [CrossRef]
- Celano, G.; Calasso, M.; Costantino, G.; Vacca, M.; Ressa, A.; Nikoloudaki, O.; De Palo, P.; Calabrese, F.M.; Gobbetti, M.; De Angelis, M. Effect of Seasonality on Microbiological Variability of Raw Cow Milk from Apulian Dairy Farms in Italy. Microbiol. Spectr. 2022, 10, e0051422. [Google Scholar] [CrossRef]
- Guo, J.; Mu, R.; Li, S.; Zhang, N.; Fu, Y.; Hu, X. Characterization of the Bacterial Community of Rumen in Dairy Cows with Laminitis. Genes 2021, 12, 1996. [Google Scholar] [CrossRef] [PubMed]
- Shao, T.; Shao, L.; Li, H.; Xie, Z.; He, Z.; Wen, C. Combined Signature of the Fecal Microbiome and Metabolome in Patients with Gout. Front. Microbiol. 2017, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Tian, S.; Wang, J.; Zhu, W. Galacto-oligosaccharides improve barrier function and relieve colonic inflammation via modulating mucosa-associated microbiota composition in lipopolysaccharides-challenged piglets. J. Anim. Sci. Biotechnol. 2021, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhao, Q.; Yousaf, L.; Khan, J.; Xue, Y.; Shen, Q. Consumption of mung bean (Vigna radiata L.) attenuates obesity, ameliorates lipid metabolic disorders and modifies the gut microbiota composition in mice fed a high-fat diet. J. Funct. Foods 2020, 64, 103687. [Google Scholar] [CrossRef]
- Sun, Z.; Yu, Z.; Wang, B. Perilla frutescens Leaf Alters the Rumen Microbial Community of Lactating Dairy Cows. Microorganisms 2019, 7, 562. [Google Scholar] [CrossRef]
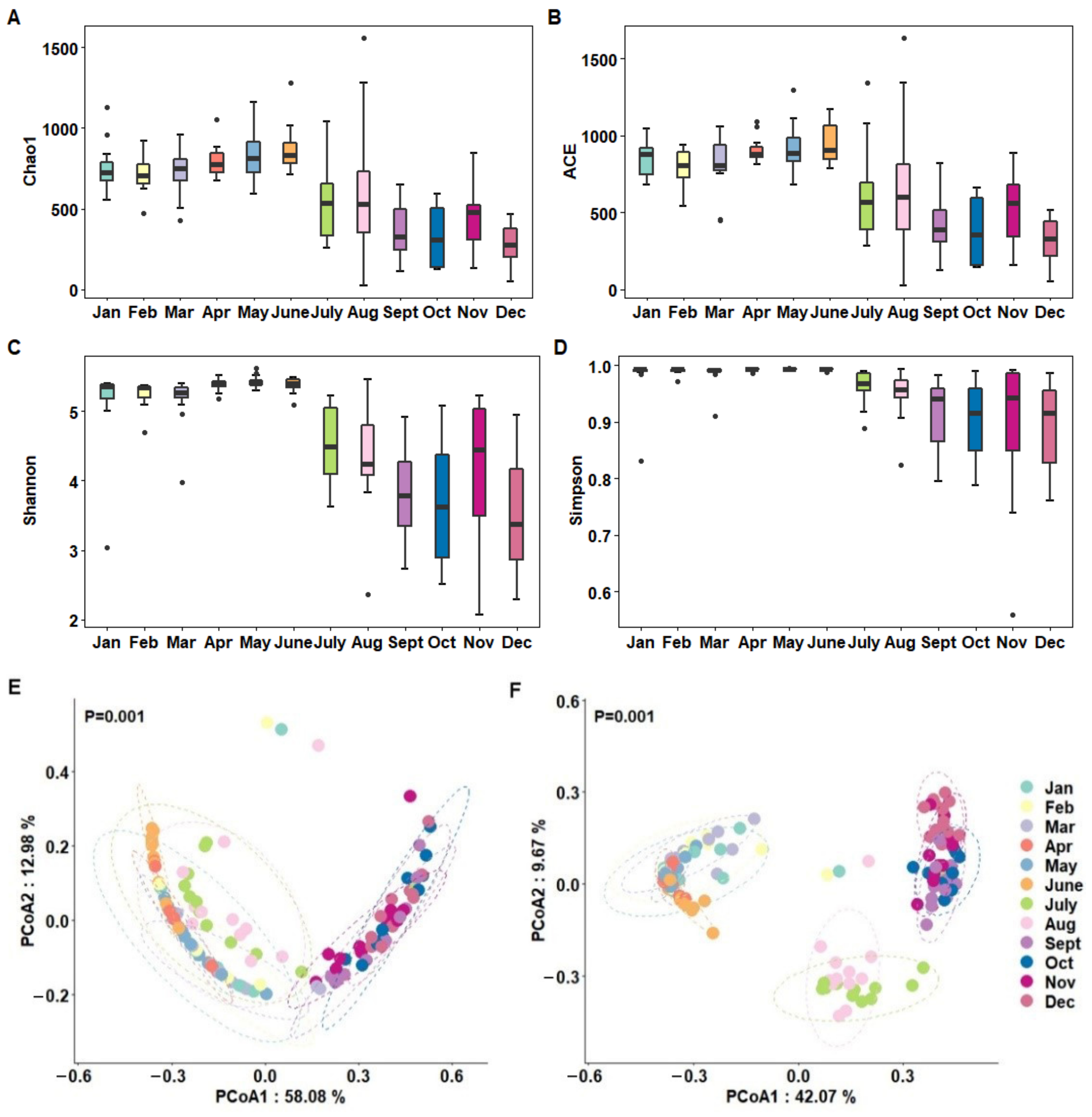
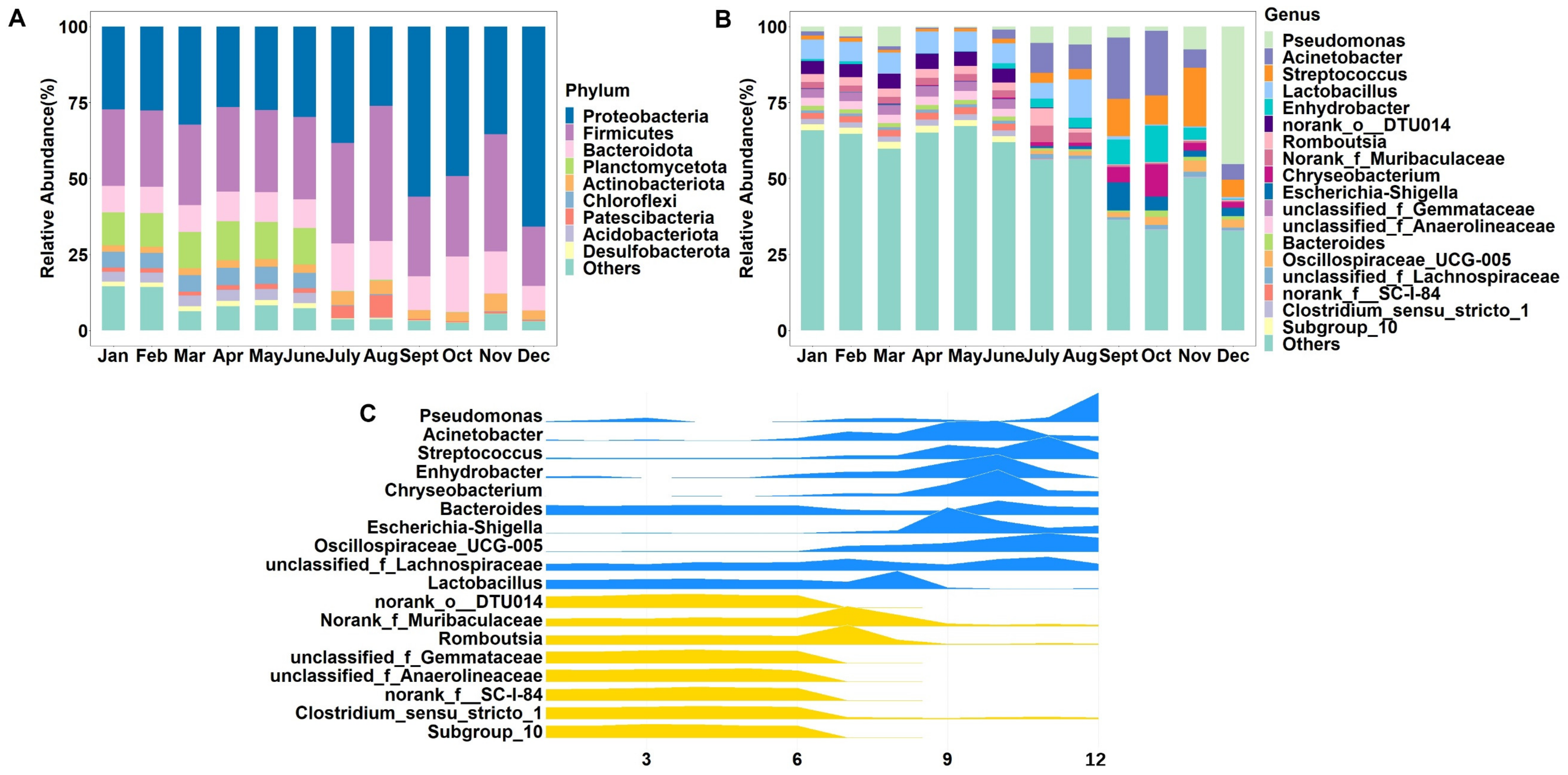
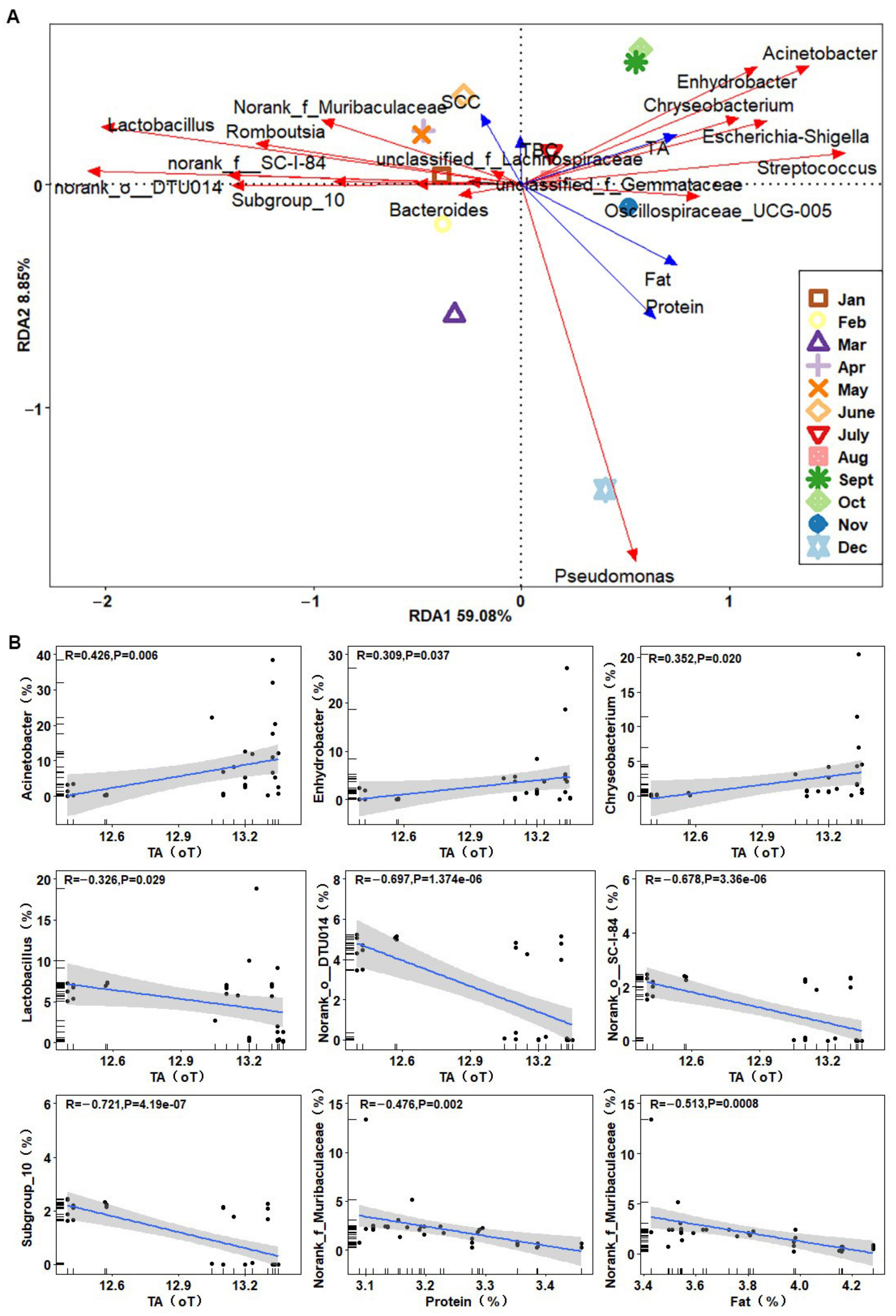
| Month | Chao 1 | ACE | Shannon | Simpson |
|---|---|---|---|---|
| Jan | 755.69 ± 45.64 ab | 846.90 ± 34.25 ab | 5.12 ± 0.19 b | 0.98 ± 0.01 b |
| Feb | 708.80 ± 32.51 ab | 797.71 ± 33.77 ab | 5.24 ± 0.06 ab | 0.99 ± 0.002 ab |
| Mar | 728.85 ± 44.85 ab | 809.75 ± 56.20 ab | 5.16 ± 0.11 b | 0.98 ± 0.01 b |
| Apr | 794.71 ± 30.32 a | 906.28 ± 25.24 a | 5.39 ± 0.03 ab | 0.99 ± 0.00 ab |
| May | 831.98 ± 46.90 a | 916.97 ± 50.83 a | 5.43 ± 0.03 a | 0.99 ± 0.00 a |
| June | 870.34 ± 44.58 a | 942.46 ± 37.77 a | 5.38 ± 0.03 ab | 0.99 ± 0.00 ab |
| July | 525.98 ± 67.74 bc | 617.68 ± 92.42 bc | 4.49 ± 0.16 c | 0.96 ± 0.01 c |
| Aug | 631.03 ± 129.00 b | 692.62 ± 134.51 b | 4.29 ± 0.22 c | 0.95 ± 0.01 c |
| Sept | 354.40 ± 52.52 bc | 424.13 ± 66.44 bc | 3.82 ± 0.22 c | 0.91 ± 0.19 c |
| Oct | 323.12 ± 58.78 c | 375.38 ± 68.90 c | 3.67 ± 0.28 c | 0.90 ± 0.02 c |
| Nov | 444.40 ± 53.90 bc | 523.92 ± 65.27 bc | 4.18 ± 0.30 c | 0.90 ± 0.04 c |
| Dec | 268.13 ± 37.29 c | 313.85 ± 43.82 c | 3.54 ± 0.26 c | 0.90 ± 0.22 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, H.; Han, S.; Zhang, S.; Xue, Y.; Zhang, Y.; Lu, H.; Wang, S. Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters. Foods 2022, 11, 3077. https://doi.org/10.3390/foods11193077
Yuan H, Han S, Zhang S, Xue Y, Zhang Y, Lu H, Wang S. Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters. Foods. 2022; 11(19):3077. https://doi.org/10.3390/foods11193077
Chicago/Turabian StyleYuan, Huizhi, Sufang Han, Shufei Zhang, Yuling Xue, Yaoguang Zhang, Han Lu, and Shijie Wang. 2022. "Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters" Foods 11, no. 19: 3077. https://doi.org/10.3390/foods11193077
APA StyleYuan, H., Han, S., Zhang, S., Xue, Y., Zhang, Y., Lu, H., & Wang, S. (2022). Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters. Foods, 11(19), 3077. https://doi.org/10.3390/foods11193077





