The Changes in Bell Pepper Flesh as a Result of Lacto-Fermentation Evaluated Using Image Features and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
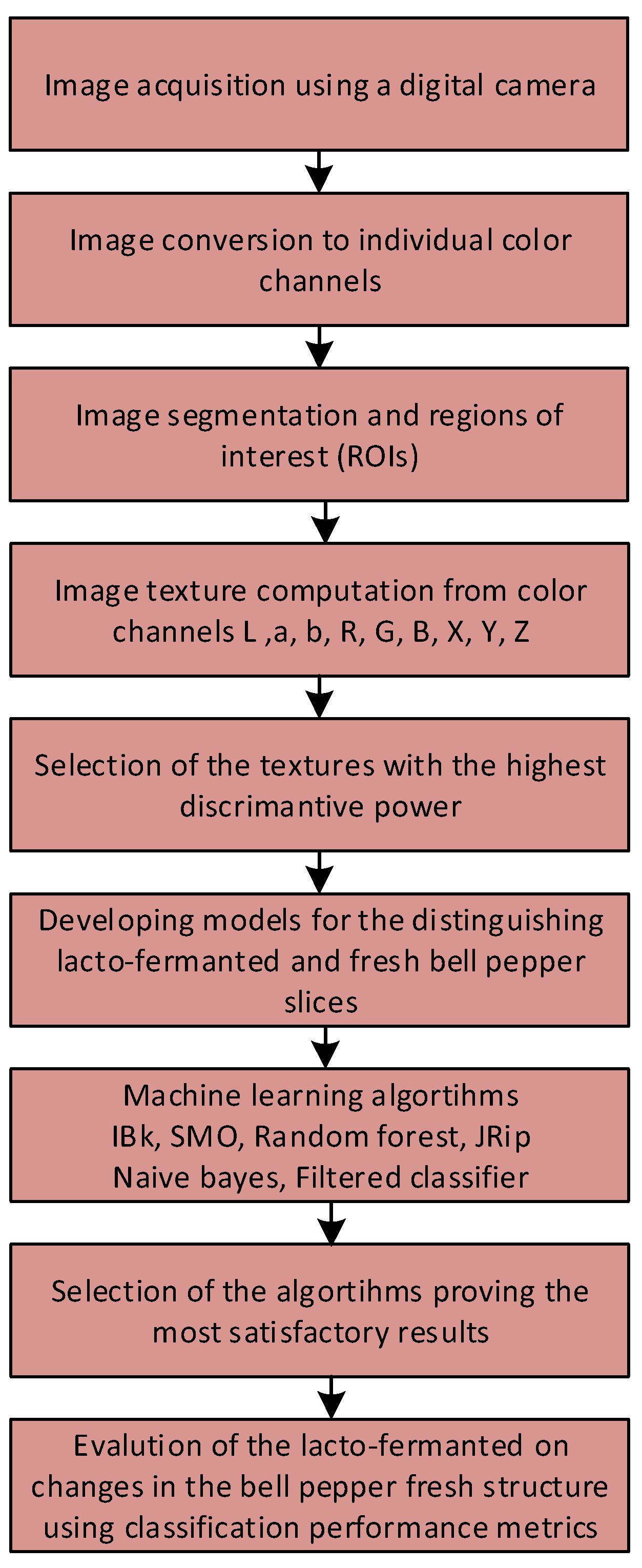
2.2. Image Acquisition and Processing
2.3. Statistical Analysis
3. Results
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Reale, S.; Biancolillo, A.; Gasparrini, C.; Di Martino, L.; Di Cecco, V.; Manzi, A.; Di Santo, M.; D’Archivio, A.A. Geographical discrimination of bell pepper (Capsicum annuum) spices by (HS)-SPME/GC-MS aroma profiling and chemometrics. Molecules 2021, 26, 6177. [Google Scholar] [CrossRef]
- Sutliff, A.K.; Saint-Cyr, M.; Hendricks, A.E.; Chen, S.S.; Doenges, K.A.; Quinn, K.; Westcott, J.; Tang, M.; Borengasser, S.J.; Reisdorph, R.M. Lipidomics-based comparison of molecular compositions of green, yellow, and red bell peppers. Metabolites 2021, 11, 241. [Google Scholar] [CrossRef]
- Martínez-Zamora, L.; Castillejo, N.; Artés-Hernández, F. Postharvest UV-B and photoperiod with blue+ red LEDs as strategies to stimulate carotenogenesis in bell peppers. Appl. Sci. 2021, 11, 3736. [Google Scholar] [CrossRef]
- Chávez-Mendoza, C.; Sanchez, E.; Muñoz-Marquez, E.; Sida-Arreola, J.P.; Flores-Cordova, M.A. Bioactive compounds and antioxidant activity in different grafted varieties of bell pepper. Antioxidants 2015, 4, 427–446. [Google Scholar] [CrossRef]
- Althaus, B.; Blanke, M. Non-destructive, opto-electronic determination of the freshness and shrivel of Bell pepper fruits. J. Imaging 2020, 6, 122. [Google Scholar] [CrossRef]
- Barbosa, C.; Machado, T.B.; Alves, M.R.; Oliveira, M.B.P. Fresh-cut bell peppers in modified atmosphere packaging: Improving shelf life to answer food security concerns. Molecules 2020, 25, 2323. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Sathiyaseelan, A.; Mariadoss, A.V.A.; Chelliah, R.; Hu, X.; Oh, D.H.; Wang, M.-H. Lactobacillus rhamnosus GG and biochemical agents enrich the shelf life of fresh-cut bell pepper (Capsicum annuum L. var. grossum (l.) sendt). Foods 2020, 9, 1252. [Google Scholar] [CrossRef]
- Zahoor, I.; Khan, M.A. Microwave assisted fluidized bed drying of red bell pepper: Drying kinetics and optimization of process conditions using statistical models and response surface methodology. Sci. Hortic. 2021, 286, 110209. [Google Scholar] [CrossRef]
- Xu, X.; Wu, B.; Zhao, W.; Lao, F.; Chen, F.; Liao, X.; Wu, J. Shifts in autochthonous microbial diversity and volatile metabolites during the fermentation of chili pepper (Capsicum frutescens L.). Food Chem. 2021, 335, 127512. [Google Scholar] [CrossRef]
- Muhialdin, B.J.; Kadum, H.; Zarei, M.; Hussin, A.S.M. Effects of metabolite changes during lacto-fermentation on the biological activity and consumer acceptability for dragon fruit juice. LWT 2020, 121, 108992. [Google Scholar] [CrossRef]
- Frediansyah, A.; Romadhoni, F.; Nurhayati, R.; Wibowo, A.T. Fermentation of Jamaican cherries juice using Lactobacillus plantarum elevates antioxidant potential and inhibitory activity against Type II diabetes-related enzymes. Molecules 2021, 26, 2868. [Google Scholar] [CrossRef]
- López-Salas, D.; Oney-Montalvo, J.E.; Ramírez-Rivera, E.; Ramírez-Sucre, M.O.; Rodríguez-Buenfil, I.M. Fermentation of Habanero Pepper by Two Lactic Acid Bacteria and Its Effect on the Production of Volatile Compounds. Fermentation 2022, 8, 219. [Google Scholar] [CrossRef]
- Villaseñor-Aguilar, M.-J.; Bravo-Sánchez, M.-G.; Padilla-Medina, J.-A.; Vázquez-Vera, J.L.; Guevara-González, R.-G.; García-Rodríguez, F.-J.; Barranco-Gutiérrez, A.-I. A maturity estimation of bell pepper (Capsicum annuum L.) by artificial vision system for quality control. Appl. Sci. 2020, 10, 5097. [Google Scholar] [CrossRef]
- Subeesh, A.; Bhole, S.; Singh, K.; Chandel, N.; Rajwade, Y.; Rao, K.; Kumar, S.; Jat, D. Deep convolutional neural network models for weed detection in polyhouse grown bell peppers. Artif. Intell. Agric. 2022, 6, 47–54. [Google Scholar] [CrossRef]
- Aslan, M.F.; Durdu, A.; Sabanci, K.; Ropelewska, E.; Gültekin, S.S. A Comprehensive Survey of the Recent Studies with UAV for Precision Agriculture in Open Fields and Greenhouses. Appl. Sci. 2022, 12, 1047. [Google Scholar] [CrossRef]
- Kasinathan, T.; Uyyala, S.R. Machine learning ensemble with image processing for pest identification and classification in field crops. Neural Comput. Appl. 2021, 33, 7491–7504. [Google Scholar] [CrossRef]
- Bosilj, P.; Duckett, T.; Cielniak, G. Connected attribute morphology for unified vegetation segmentation and classification in precision agriculture. Comput. Ind. 2018, 98, 226–240. [Google Scholar] [CrossRef]
- Liu, H.; Sun, H.; Li, M.; Iida, M. Application of Color Featuring and Deep Learning in Maize Plant Detection. Remote Sens. 2020, 12, 2229. [Google Scholar] [CrossRef]
- Koklu, M.; Unlersen, M.F.; Ozkan, I.A.; Aslan, M.F.; Sabanci, K. A CNN-SVM study based on selected deep features for grapevine leaves classification. Measurement 2022, 188, 110425. [Google Scholar] [CrossRef]
- Sabanci, K.; Aslan, M.F.; Durdu, A. Bread and durum wheat classification using wavelet based image fusion. J. Sci. Food Agric. 2020, 100, 5577–5585. [Google Scholar] [CrossRef]
- Szczypiński, P.M.; Strzelecki, M.; Materka, A.; Klepaczko, A. MaZda—A software package for image texture analysis. Comput. Methods Programs Biomed. 2009, 94, 66–76. [Google Scholar] [CrossRef]
- Ropelewska, E. Distinguishing lacto-fermented and fresh carrot slice images using the Multilayer Perceptron neural network and other machine learning algorithms from the groups of Functions, Meta, Trees, Lazy, Bayes and Rules. Eur. Food Res. Technol. 2022, 248, 2421–2429. [Google Scholar] [CrossRef]
- Ropelewska, E.; Sabanci, K.; Aslan, M.F. Preservation effects evaluated using innovative models developed by machine learning on cucumber flesh. Eur. Food Res. Technol. 2022, 248, 1929–1937. [Google Scholar] [CrossRef]
- Ropelewska, E.; Wrzodak, A.; Sabanci, K.; Aslan, M.F. Effect of lacto-fermentation and freeze-drying on the quality of beetroot evaluated using machine vision and sensory analysis. Eur. Food Res. Technol. 2022, 248, 153–161. [Google Scholar] [CrossRef]
- Ropelewska, E.; Wrzodak, A. The use of image analysis and sensory analysis for the evaluation of cultivar differentiation of freeze-dried and lacto-fermented beetroot (Beta vulgaris L.). Food Anal. Methods 2022, 15, 1026–1041. [Google Scholar] [CrossRef]
- Szczypinski, P.M.; Strzelecki, M.; Materka, A. Mazda-a software for texture analysis. In Proceedings of the 2007 International Symposium on Information Technology Convergence (ISITC 2007), Jeonju, Korea, 23–24 November 2007; pp. 245–249. [Google Scholar]
- Strzelecki, M.; Szczypinski, P.; Materka, A.; Klepaczko, A. A software tool for automatic classification and segmentation of 2D/3D medical images. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2013, 702, 137–140. [Google Scholar] [CrossRef]
- Ibraheem, N.A.; Hasan, M.M.; Khan, R.Z.; Mishra, P.K. Understanding color models: A review. ARPN J. Sci. Technol. 2012, 2, 265–275. [Google Scholar]
- Bouckaert, R.R.; Frank, E.; Hall, M.; Kirkby, R.; Reutemann, P.; Seewald, A.; Scuse, D. WEKA Manual for Version 3-9-1; University of Waikato: Hamilton, New Zealand, 2016. [Google Scholar]
- Eibe, F.; Hall, M.A.; Witten, I.H. The WEKA workbench. Online appendix for data mining: Practical machine learning tools and techniques. In Morgan Kaufmann; Morgan Kaufmann Publishers: Burlington, MA, USA, 2016. [Google Scholar]
- Ian, H.W.; Eibe, F. Data Mining: Practical Machine Learning Tools and Techniques, 2nd ed.; Elsevier: San Francisco, CA, USA, 2005. [Google Scholar]
- Ropelewska, E.; Szwejda-Grzybowska, J. A comparative analysis of the discrimination of pepper (Capsicum annuum L.) based on the cross-section and seed textures determined using image processing. J. Food Process Eng. 2021, 44, e13694. [Google Scholar] [CrossRef]
- Sabanci, K.; Aslan, M.F.; Ropelewska, E.; Unlersen, M.F. A convolutional neural network-based comparative study for pepper seed classification: Analysis of selected deep features with support vector machine. J. Food Process Eng. 2022, 45, e13955. [Google Scholar] [CrossRef]
- Ropelewska, E. Classification of the pits of different sour cherry cultivars based on the surface textural features. J. Saudi Soc. Agric. Sci. 2021, 20, 52–57. [Google Scholar] [CrossRef]
- Meshram, V.; Patil, K.; Meshram, V.; Hanchate, D.; Ramkteke, S. Machine learning in agriculture domain: A state-of-art survey. Artif. Intell. Life Sci. 2021, 1, 100010. [Google Scholar] [CrossRef]
- Rasekh, M.; Karami, H.; Fuentes, S.; Kaveh, M.; Rusinek, R.; Gancarz, M. Preliminary study non-destructive sorting techniques for pepper (Capsicum annuum L.) using odor parameter. LWT 2022, 164, 113667. [Google Scholar] [CrossRef]
- Mohi-Alden, K.; Omid, M.; Firouz, M.S.; Nasiri, A. A Machine Vision-Intelligent Modelling Based Technique for In-Line Bell Pepper Sorting. Inf. Process. Agric. 2022, in press. [Google Scholar] [CrossRef]
- Harel, B.; Parmet, Y.; Edan, Y. Maturity classification of sweet peppers using image datasets acquired in different times. Comput. Ind. 2020, 121, 103274. [Google Scholar] [CrossRef]
- Bazinas, C.; Vrochidou, E.; Kalampokas, T.; Karampatea, A.; Kaburlasos, V.G. A Non-Destructive Method for Grape Ripeness Estimation Using Intervals’ Numbers (INs) Techniques. Agronomy 2022, 12, 1564. [Google Scholar] [CrossRef]
- Palumbo, M.; Cozzolino, R.; Laurino, C.; Malorni, L.; Picariello, G.; Siano, F.; Stocchero, M.; Cefola, M.; Corvino, A.; Romaniello, R. Rapid and Non-Destructive Techniques for the Discrimination of Ripening Stages in Candonga Strawberries. Foods 2022, 11, 1534. [Google Scholar] [CrossRef]
- Karadağ, K.; Tenekeci, M.E.; Taşaltın, R.; Bilgili, A. Detection of pepper fusarium disease using machine learning algorithms based on spectral reflectance. Sustain. Comput. Inform. Syst. 2020, 28, 100299. [Google Scholar] [CrossRef]

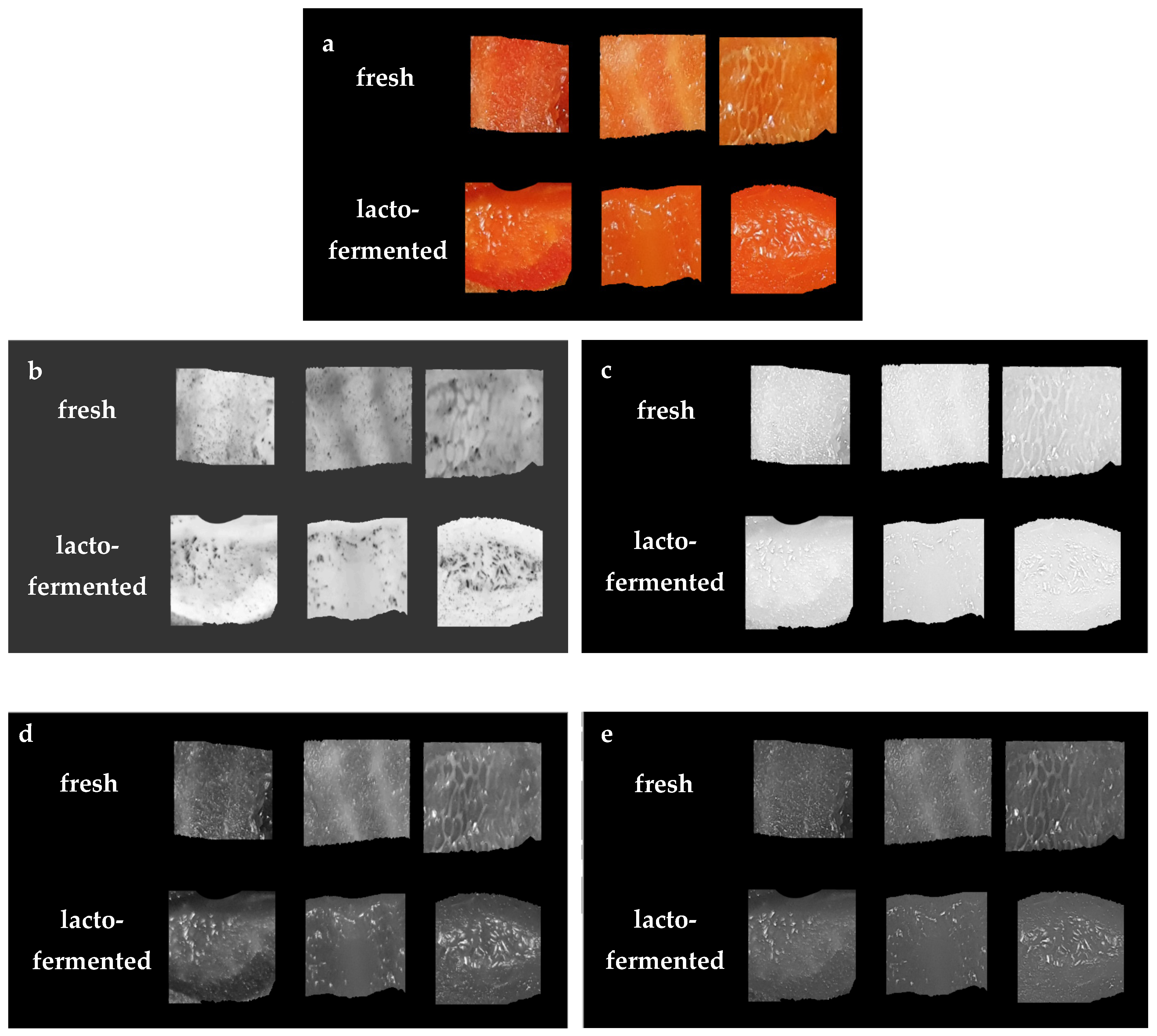
| Color Space Lab | Color Channel L | Color Space RGB | Color Channel R | Color Space XYZ | Color Channel X |
|---|---|---|---|---|---|
| LHMean LHPerc01 LHPerc10 LHMaxm10 LS5SH1SumEntrp LS5SH3Correlat LS5SV5AngScMom LS4RZGLevNonU aS5SZ1Correlat aS4RNRLNonUni bHDomn01 bS5SN1AngScMom bS4RHShrtREmp bS4RNLngREmph | LHMean LHPerc01 LHPerc10 LHPerc99 LHMaxm10 LS5SH1SumEntrp LS5SH3Correlat LS5SV5AngScMom LS4RZGLevNonU LS4RNRLNonUni | RHPerc10 RHMaxm01 RHMaxm10 RSGVariance RS5SH1Correlat RS5SH3Correlat RS5SH3SumVarnc RS5SH5Correlat RS5SV5Correlat RS4RZGLevNonU RATeta1 GHKurtosis GHPerc01 GHPerc50 GHMaxm10 GS5SH1SumEntrp GS5SH3SumVarnc GS5SN5AngScMom GS4RNRLNonUni BHDomn10 | RHPerc10 RHMaxm01 RHMaxm10 RS5SH1Correlat RS5SZ1DifVarnc RS5SH3Correlat RS5SN3AngScMom RS5SH5Correlat RS5SV5Correlat RS4RHGLevNonU RATeta1 | XHPerc10 XHMaxm01 XHMaxm10 XS5SH1Correlat XS5SV1SumAverg XS5SH3SumEntrp XS5SZ3SumEntrp XS5SH5Correlat XS4RVGLevNonU XASigma YHDomn01 YHMaxm10 YS5SZ1SumEntrp YS4RNRLNonUni ZSGNonZeros | XHPerc10 XHMaxm01 XHMaxm10 XS5SH1Correlat XS5SV1SumAverg XS5SH3SumEntrp XS5SZ3SumEntrp XS5SH5Correlat XS4RVGLevNonU XS4RNGLevNonU XASigma |
| Algorithm (Group) | Predicted Class (%) | Actual Class | Average Accuracy (%) | Precision | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|
| Fresh | Lacto-Fermented | ||||||
| IBk (Lazy) | 98 | 2 | Fresh | 99 | 1.000 | 0.990 | 0.980 |
| 0 | 100 | Lacto-fermented | 0.980 | 0.990 | 0.980 | ||
| SMO (Functions) | 98 | 2 | Fresh | 98 | 0.980 | 0.980 | 0.960 |
| 2 | 98 | Lacto-fermented | 0.980 | 0.980 | 0.960 | ||
| Random forest (Trees) | 97 | 3 | Fresh | 98 | 0.990 | 0.980 | 0.960 |
| 1 | 99 | Lacto-fermented | 0.971 | 0.980 | 0.960 | ||
| Naïve Bayes (Bayes) | 99 | 1 | Fresh | 98 | 0.971 | 0.980 | 0.960 |
| 3 | 97 | Lacto-fermented | 0.990 | 0.980 | 0.960 | ||
| Filtered classifier (Meta) | 97 | 3 | Fresh | 97 | 0.970 | 0.970 | 0.940 |
| 3 | 97 | Lacto-fermented | 0.970 | 0.970 | 0.940 | ||
| JRip (Rules) | 96 | 4 | Fresh | 96.5 | 0.970 | 0.965 | 0.930 |
| 3 | 97 | Lacto-fermented | 0.960 | 0.965 | 0.930 | ||
| Algorithm (Group) | Predicted Class (%) | Actual Class | Average Accuracy (%) | Precision | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|
| Fresh | Lacto-Fermented | ||||||
| IBk (Lazy) | 98 | 2 | Fresh | 98 | 0.980 | 0.980 | 0.960 |
| 2 | 98 | Lacto-fermented | 0.980 | 0.980 | 0.960 | ||
| SMO (Functions) | 98 | 2 | Fresh | 98 | 0.980 | 0.980 | 0.960 |
| 2 | 98 | Lacto-fermented | 0.980 | 0.980 | 0.960 | ||
| Random forest (Trees) | 98 | 2 | Fresh | 98.5 | 0.990 | 0.985 | 0.970 |
| 1 | 99 | Lacto-fermented | 0.980 | 0.985 | 0.970 | ||
| Naïve Bayes (Bayes) | 97 | 3 | Fresh | 97 | 0.970 | 0.970 | 0.940 |
| 3 | 97 | Lacto-fermented | 0.970 | 0.970 | 0.940 | ||
| Filtered classifier (Meta) | 97 | 3 | Fresh | 97 | 0.970 | 0.970 | 0.940 |
| 3 | 97 | Lacto-fermented | 0.970 | 0.970 | 0.940 | ||
| JRip (Rules) | 97 | 3 | Fresh | 96.5 | 0.960 | 0.965 | 0.930 |
| 4 | 96 | Lacto-fermented | 0.970 | 0.965 | 0.930 | ||
| Algorithm (Group) | Predicted Class (%) | Actual Class | Average Accuracy (%) | Precision | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|
| Fresh | Lacto-Fermented | ||||||
| IBk (Lazy) | 98 | 2 | Fresh | 98.5 | 0.990 | 0.985 | 0.970 |
| 1 | 99 | Lacto-fermented | 0.980 | 0.985 | 0.970 | ||
| SMO (Functions) | 100 | 0 | Fresh | 99 | 0.980 | 0.990 | 0.980 |
| 2 | 98 | Lacto-fermented | 1.000 | 0.990 | 0.980 | ||
| Random forest (Trees) | 96 | 4 | Fresh | 96.5 | 0.970 | 0.965 | 0.930 |
| 3 | 97 | Lacto-fermented | 0.960 | 0.965 | 0.930 | ||
| Naïve Bayes (Bayes) | 99 | 1 | Fresh | 98.5 | 0.980 | 0.985 | 0.970 |
| 2 | 98 | Lacto-fermented | 0.990 | 0.985 | 0.970 | ||
| Filtered classifier (Meta) | 94 | 6 | Fresh | 93 | 0.922 | 0.931 | 0.860 |
| 8 | 92 | Lacto-fermented | 0.939 | 0.929 | 0.860 | ||
| JRip (Rules) | 91 | 9 | Fresh | 93.5 | 0.958 | 0.933 | 0.871 |
| 4 | 96 | Lacto-fermented | 0.914 | 0.937 | 0.871 | ||
| Algorithm (Group) | Predicted Class (%) | Actual Class | Average Accuracy (%) | Precision | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|
| Fresh | Lacto-Fermented | ||||||
| IBk (Lazy) | 98 | 2 | Fresh | 97.5 | 0.970 | 0.975 | 0.950 |
| 3 | 97 | Lacto-fermented | 0.980 | 0.975 | 0.950 | ||
| SMO (Functions) | 99 | 1 | Fresh | 98.5 | 0.980 | 0.985 | 0.970 |
| 2 | 98 | Lacto-fermented | 0.990 | 0.985 | 0.970 | ||
| Random forest (Trees) | 96 | 4 | Fresh | 97 | 0.980 | 0.970 | 0.940 |
| 2 | 98 | Lacto-fermented | 0.961 | 0.970 | 0.940 | ||
| Naïve Bayes (Bayes) | 95 | 5 | Fresh | 96 | 0.969 | 0.960 | 0.920 |
| 3 | 97 | Lacto-fermented | 0.951 | 0.960 | 0.920 | ||
| Filtered classifier (Meta) | 94 | 6 | Fresh | 91.5 | 0.895 | 0.917 | 0.831 |
| 11 | 89 | Lacto-fermented | 0.937 | 0.913 | 0.831 | ||
| JRip (Rules) | 89 | 11 | Fresh | 90.5 | 0.918 | 0.904 | 0.810 |
| 8 | 92 | Lacto-fermented | 0.893 | 0.906 | 0.810 | ||
| Algorithm (Group) | Predicted Class (%) | Actual Class | Average Accuracy (%) | Precision | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|
| Fresh | Lacto-Fermented | ||||||
| IBk (Lazy) | 99 | 1 | Fresh | 99 | 0.990 | 0.990 | 0.980 |
| 1 | 99 | Lacto-fermented | 0.990 | 0.990 | 0.980 | ||
| SMO (Functions) | 100 | 0 | Fresh | 99 | 0.980 | 0.990 | 0.980 |
| 2 | 98 | Lacto-fermented | 1.000 | 0.990 | 0.980 | ||
| Random forest (Trees) | 97 | 3 | Fresh | 98 | 0.990 | 0.980 | 0.960 |
| 1 | 99 | Lacto-fermented | 0.971 | 0.980 | 0.960 | ||
| Naïve Bayes (Bayes) | 98 | 2 | Fresh | 98 | 0.980 | 0.980 | 0.960 |
| 2 | 98 | Lacto-fermented | 0.980 | 0.980 | 0.960 | ||
| Filtered classifier (Meta) | 95 | 5 | Fresh | 97 | 0.990 | 0.969 | 0.941 |
| 1 | 99 | Lacto-fermented | 0.952 | 0.971 | 0.941 | ||
| JRip (Rules) | 95 | 5 | Fresh | 96.5 | 0.979 | 0.964 | 0.930 |
| 2 | 98 | Lacto-fermented | 0.951 | 0.966 | 0.930 | ||
| Algorithm (Group) | Predicted Class (%) | Actual Class | Average Accuracy (%) | Precision | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|
| Fresh | Lacto-Fermented | ||||||
| IBk (Lazy) | 99 | 1 | Fresh | 99 | 0.990 | 0.990 | 0.980 |
| 1 | 99 | Lacto-fermented | 0.990 | 0.990 | 0.980 | ||
| SMO (Functions) | 100 | 0 | Fresh | 99 | 0.980 | 0.990 | 0.980 |
| 2 | 98 | Lacto-fermented | 1.000 | 0.990 | 0.980 | ||
| Random forest (Trees) | 96 | 4 | Fresh | 97 | 0.980 | 0.970 | 0.940 |
| 2 | 98 | Lacto-fermented | 0.961 | 0.970 | 0.940 | ||
| Naïve Bayes (Bayes) | 99 | 1 | Fresh | 98.5 | 0.980 | 0.985 | 0.970 |
| 2 | 98 | Lacto-fermented | 0.990 | 0.985 | 0.970 | ||
| Filtered classifier (Meta) | 93 | 7 | Fresh | 95.5 | 0.979 | 0.954 | 0.911 |
| 2 | 98 | Lacto-fermented | 0.933 | 0.956 | 0.911 | ||
| JRip (Rules) | 94 | 6 | Fresh | 93.5 | 0.931 | 0.935 | 0.870 |
| 7 | 93 | Lacto-fermented | 0.939 | 0.935 | 0.870 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ropelewska, E.; Sabanci, K.; Aslan, M.F. The Changes in Bell Pepper Flesh as a Result of Lacto-Fermentation Evaluated Using Image Features and Machine Learning. Foods 2022, 11, 2956. https://doi.org/10.3390/foods11192956
Ropelewska E, Sabanci K, Aslan MF. The Changes in Bell Pepper Flesh as a Result of Lacto-Fermentation Evaluated Using Image Features and Machine Learning. Foods. 2022; 11(19):2956. https://doi.org/10.3390/foods11192956
Chicago/Turabian StyleRopelewska, Ewa, Kadir Sabanci, and Muhammet Fatih Aslan. 2022. "The Changes in Bell Pepper Flesh as a Result of Lacto-Fermentation Evaluated Using Image Features and Machine Learning" Foods 11, no. 19: 2956. https://doi.org/10.3390/foods11192956
APA StyleRopelewska, E., Sabanci, K., & Aslan, M. F. (2022). The Changes in Bell Pepper Flesh as a Result of Lacto-Fermentation Evaluated Using Image Features and Machine Learning. Foods, 11(19), 2956. https://doi.org/10.3390/foods11192956








