Fed-Batch Fermentation of Saccharomyces pastorianus with High Ribonucleic Acid Yield
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Medium
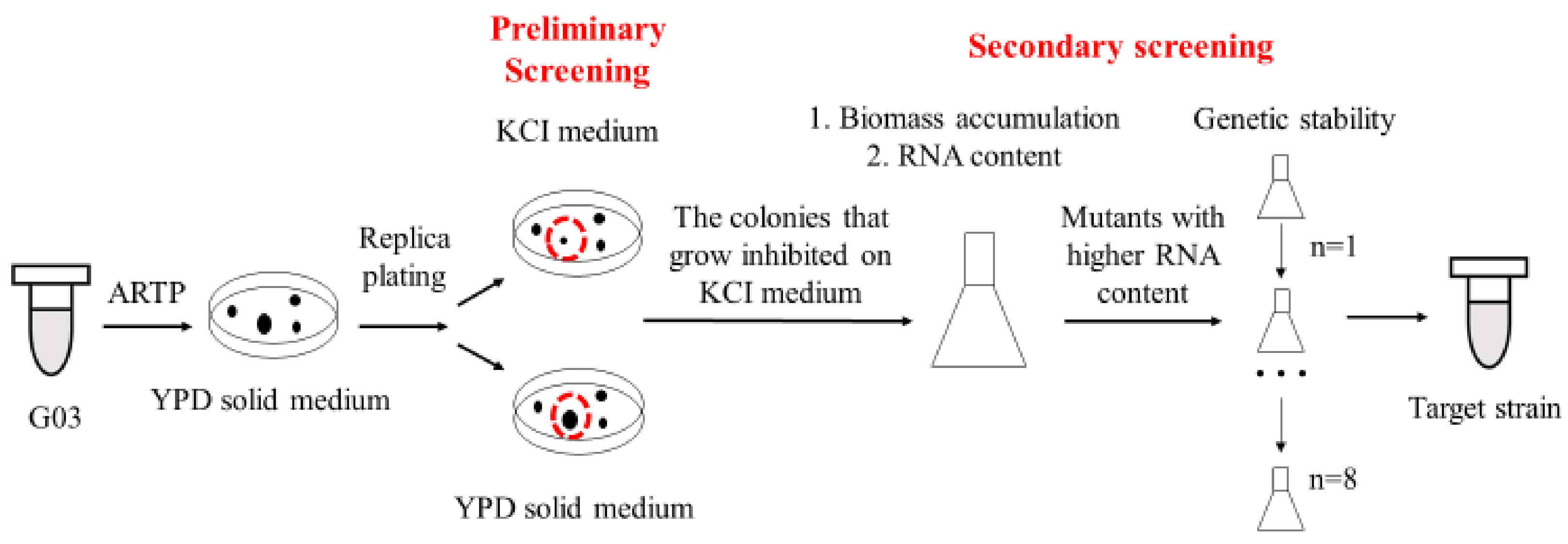
2.2. Screening of Mutants
2.3. Physiological Characteristics Analysis
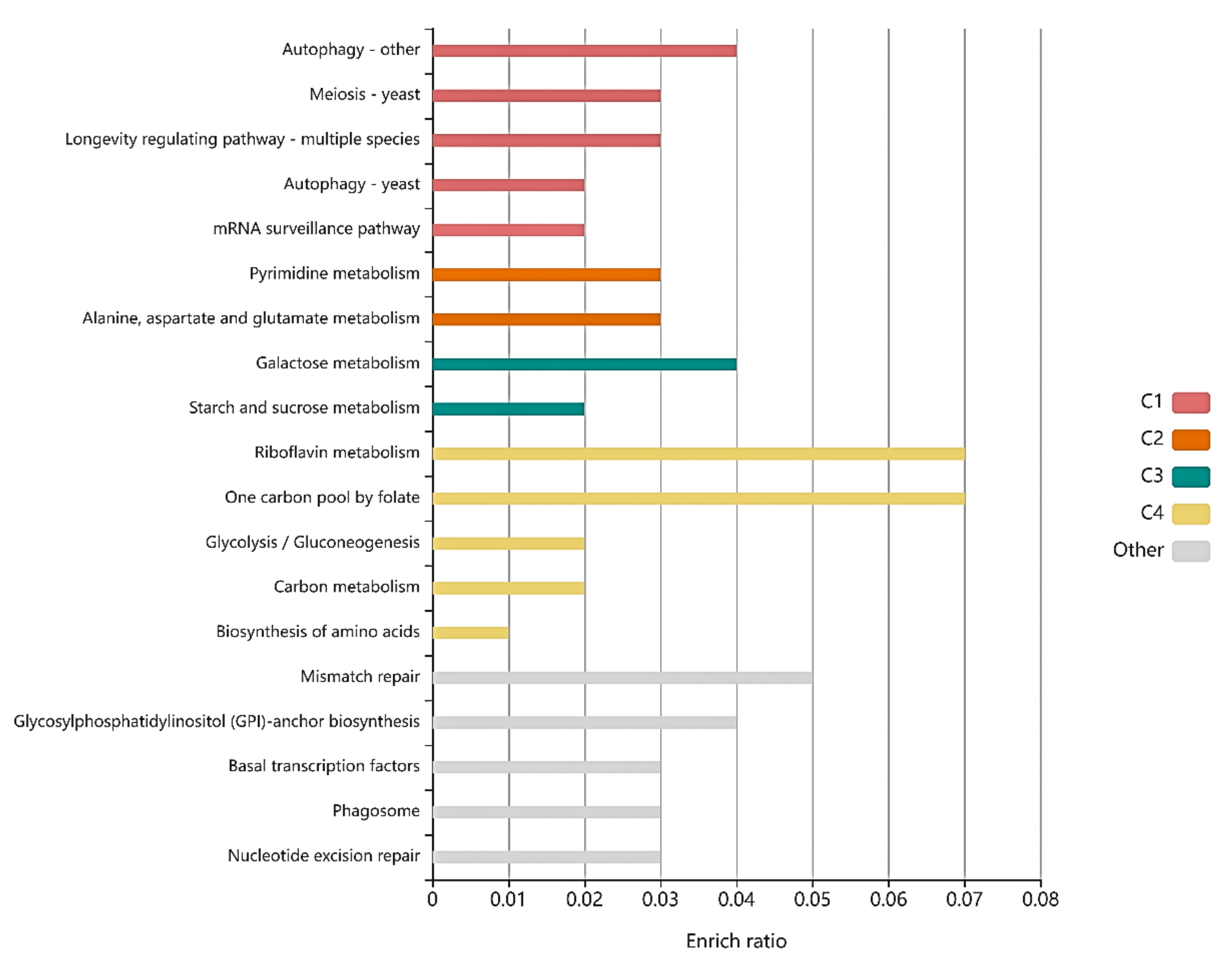
2.4. Genome Sequencing and Enrichment Analysis
2.5. High-Cell-Density Fermentation
2.6. Measurement of Reducing Sugar Content
2.7. Statistical Analysis
3. Results
3.1. Screening of High RNA Producing Yeast
3.2. Genomic Sequencing of Mutant Strain
3.3. High-Density Fermentation Strategy
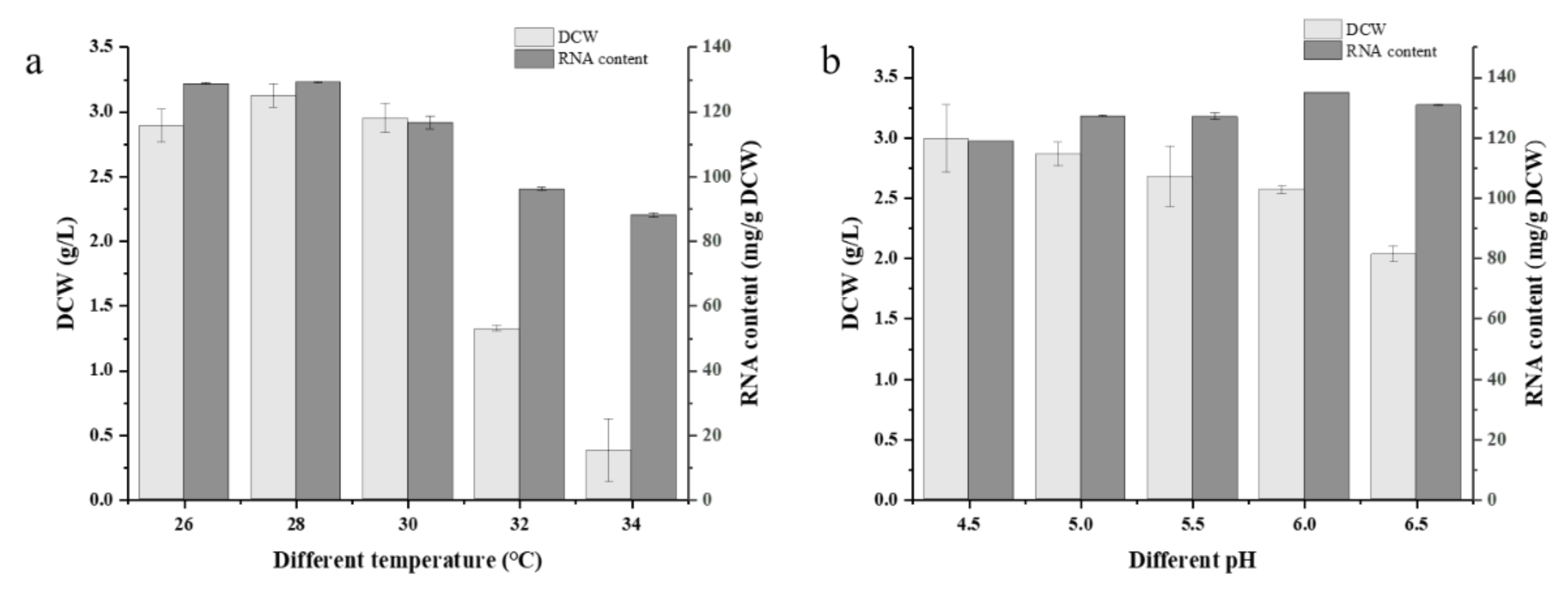
3.3.1. Optimization of Culture Conditions
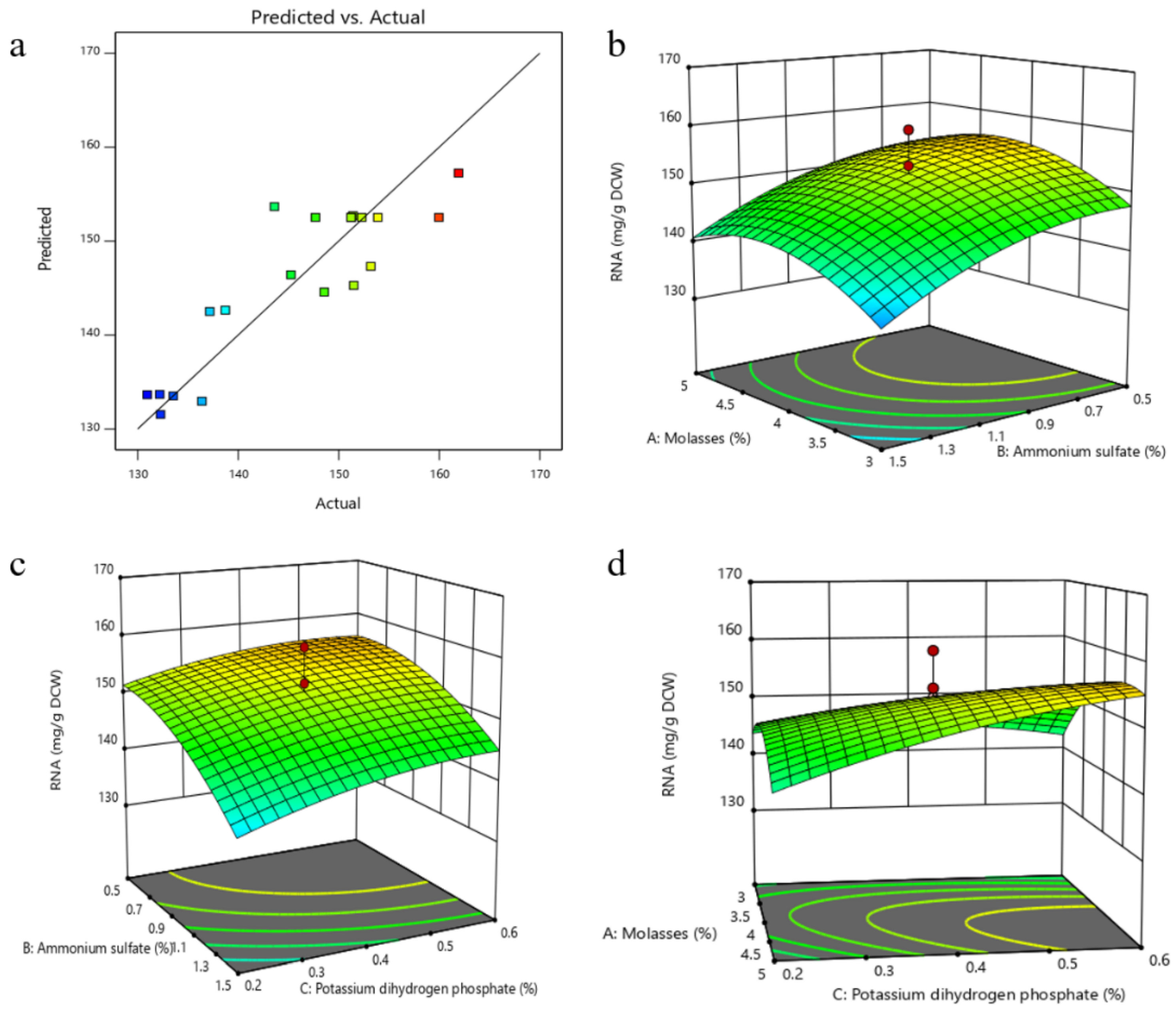
3.3.2. Optimization of Fermentation Medium
3.3.3. Fed-Batch Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slobodianik, N.H. Dietary ribonucleotides and health. Nutrition 2003, 19, 68–69. [Google Scholar] [CrossRef]
- Burmeister, G.; Rainsford, K.D.J.I. Discriminating effects of a nucleotide-rich yeast extract, Probioticum R, as an immunomodulator contrasted with actions in chronic immuno-inflammatory disease (adjuvant-induced arthritis) in rodents. Inflammopharmacology 1991, 1, 161–183. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Olmedo, F.; Iturbe, F.; Gomezhernandez, J.; Lopezmunguia, A. Continuous Production of 5′-Ribonucleotides from Yeast RNA by Hydrolysis with Immobilized 5′-Phosphodiesterase and 5′-Adenylate Deaminase. World J. Microbiol. Biotechnol. 1994, 10, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Li, B.B.; Liu, Y.; Wang, L.Z.; Hong, J.; Chen, Y.; Ying, H.J. RNA accumulation in Candida tropicalis based on cofactor engineering. FEMS Yeast Res. 2019, 19, foz028. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Chen, X.; Cao, J.; Li, L.; Bai, J.; Chen, Y.; Xiong, J.; Ying, H. Determination of optimal conditions for ribonucleic acid production by Candida tropicalis no. 121. Korean J. Chem. Eng. 2011, 28, 1721–1726. [Google Scholar] [CrossRef]
- Holzschu, D.L.; Chandler, F.W.; Ajello, L.; Ahearn, D.G.J.S. Evaluation of industrial yeasts for pathogenicity. Sabouraudia J. Med. Vet. Mycol. 1979, 17, 71–78. [Google Scholar] [CrossRef]
- Guo, X.; Zhao, B.; Zhou, X.; Lu, D.; Wang, Y.; Chen, Y.; Xiao, D. Analysis of the molecular basis of Saccharomyces cerevisiae mutant with high nucleic acid content by comparative transcriptomics. Food Res. Int. 2021, 142, 110188. [Google Scholar] [CrossRef]
- Xiaokun, L.; Wang, W.; Ying, L.; Shuli, L. Screening of high-yield nucleic acid Saccharomyces cerevisiae strain by atmospheric and room-temperature plasma (ARTP) technique. Mod. Food Sci. Technol. 2018, 34, 137–144. [Google Scholar] [CrossRef]
- Khatun, F.; Kurata, K.; Chuwattanakul, V.; Sugiyama, M.; Kaneko, Y.; Harashima, S. Increased transcription of RPL40A and RPL40B is important for the improvement of RNA production in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2013, 116, 423–432. [Google Scholar] [CrossRef]
- Pei, Y.; Lehman, K.; Tian, L.G.; Shuman, S. Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 2000, 28, 1885–1892. [Google Scholar] [CrossRef][Green Version]
- Sasano, Y.; Kariya, T.; Usugi, S.; Sugiyama, M.; Harashima, S. Molecular breeding of Saccharomyces cerevisiae with high RNA content by harnessing essential ribosomal RNA transcription regulator. AMB Express 2017, 7, 32. [Google Scholar] [CrossRef]
- Wang, Q.; Feng, L.-R.; Wei, L.; Li, H.-G.; Wang, L.; Zhou, Y.; Yu, X.-B. Mutation Breeding of Lycopene-Producing Strain Blakeslea Trispora by a Novel Atmospheric and Room Temperature Plasma (ARTP). Appl. Biochem. Biotechnol. 2014, 174, 452–460. [Google Scholar]
- Zhang, X.; Zhang, C.; Zhou, Q.-Q.; Zhang, X.-F.; Wang, L.-Y.; Chang, H.-B.; Li, H.-P.; Oda, Y.; Xing, X.-H. Quantitative evaluation of DNA damage and mutation rate by atmospheric and room-temperature plasma (ARTP) and conventional mutagenesis. Appl. Microbiol. Biotechnol. 2015, 99, 5639–5646. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Xu, G.; Zhang, X.; Shi, J.; Xu, Z. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve L-serine yield in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018, 102, 5939–5951. [Google Scholar] [CrossRef]
- Wen, S.H.; Zhang, T.; Tan, T.W. Maximizing production of glutathione by amino acid modulation and hih-cell-densit fed-batch culture of Saccharomyces cerevisiae. Process Biochem. 2006, 41, 2424–2428. [Google Scholar] [CrossRef]
- Brethauer, S.; Wyman, C.E. Review: Continuous hydrolysis and fermentation for cellulosic ethanol production. Bioresour. Technol. 2010, 101, 4862–4874. [Google Scholar] [CrossRef]
- Rodrigues, D.; Pillaca-Pullo, O.; Torres-Obreque, K.; Flores-Santos, J.; Sanchez-Moguel, I.; Pimenta, M.V.; Basi, T.; Converti, A.; Lopes, A.M.; Monteiro, G.; et al. Fed-Batch Production of Saccharomyces cerevisiae L-Asparaginase II by Recombinant Pichia pastoris MUTs Strain. Front. Bioeng. Biotechnol. 2019, 7, 16. [Google Scholar] [CrossRef]
- Schmacht, M.; Lorenz, E.; Stahl, U.; Senz, M. Medium optimization based on yeast’s elemental composition for glutathione production in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2017, 123, 555–561. [Google Scholar] [CrossRef]
- Can, Y.U.; Zheng, G.; Yao, J.; Ku, L.I.; Tang, G.; Jiangbo, L.V.; Wang, Z.; Chen, X.; Yeast, A.J.C.B. Breeding and high density fermentation technology of Saccharomyces cerevisiae with high RNA content. China Brew. 2016, 35, 66–71. (In Chinese) [Google Scholar]
- Li, B.; Chen, X.; Ren, H.; Li, L.; Xiong, J.; Bai, J.; Chen, Y.; Wu, J.; Ying, H. Kinetic models of ribonucleic acid fermentation and continuous culture by Candida tropicalis no.121. Bioprocess Biosyst. Eng. 2012, 35, 415–422. [Google Scholar] [CrossRef]
- Tyurin, B.K.; Biktashev, R.U.; Zharkov, V.I.; Anisin, S.D.; Avrutskaya, I.A.; Tyurina, I.B. Production of a Yeast Biomass. U.S. Patent US3909352A, 30 September 1975. [Google Scholar]
- Liu, K.; Fang, H.; Cui, F.; Nyabako, B.A.; Tao, T.; Zan, X.; Chen, H.; Sun, W. ARTP mutation and adaptive laboratory evolution improve probiotic performance of Bacillus coagulans. Appl. Microbiol. Biotechnol. 2020, 104, 6363–6373. [Google Scholar] [CrossRef]
- Liye, W.; Haocheng, W.; Shan, M.; Weibing, L.; Zehan, L.; Jingjing, S.; Huafeng, Y.; Ying, Z.; Yun, W. Breeding of Saccharomyces cerevisiae high-yield of alcohol and acid by atmospheric room temperature plasma. Food Mach. 2019, 35, 26–31. (In Chinese) [Google Scholar] [CrossRef]
- Charpentier, C.; Aussenac, J.; Charpentier, M.; Prome, J.C.; Duteurtre, B.; Feuillat, M. Release of nucleotides and nucleosides during yeast autolysis: Kinetics and potential impact on flavor. J. Agric. Food Chem. 2005, 53, 3000–3007. [Google Scholar] [CrossRef]
- Li, X.; Jia, O.; Xu, Y.; Chen, M.; Song, X.; Yong, Q.; Yu, S. Optimization of culture conditions for production of yeast biomass using bamboo wastewater by response surface methodology. Bioresour. Technol. 2009, 100, 3613–3617. [Google Scholar] [CrossRef]
- Malairuang, K.; Krajang, M.; Sukna, J.; Chamsart, S.J.P. High-Cell-Density Cultivation of Saccharomyces cerevisiae with Intensive Multiple Sequential Batches Together with a New Technique of Fed-Batch at Cell Level. Processes 2020, 8, 1321. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Woolford, J.L.; Baserga, S.J. Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Morris, R.J.; Kragler, F. Long-Distance Transported RNAs: From Identity to Function. Annu. Rev. Plant Biol. 2022, 73, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Soto, D.; Ruiz-Herrera, J. Functional analysis of the MAPK pathways in fungi. Rev. Iberoam. Micol. 2017, 34, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Mirisola, M.G.; Longo, V.D. Yeast Chronological Lifespan: Longevity Regulatory Genes and Mechanisms. Cells 2022, 11, 1714. [Google Scholar] [CrossRef]
- Camargos, C.V.; Moraes, V.D.; de Oliveira, L.M.; Guidini, C.Z.; Ribeiro, E.J.; Santos, L.D. High Gravity and Very High Gravity Fermentation of Sugarcane Molasses by Flocculating Saccharomyces cerevisiae: Experimental Investigation and Kinetic Modeling. Appl. Biochem. Biotechnol. 2021, 193, 807–821. [Google Scholar] [CrossRef]
- Xing, W.; Wang, X.; Yin, M.; Xiao, Z.; Ma, C.; Lin, Z.; Wang, P.G.; Ping, X. Production of uridine 5′-monophosphate by Corynebacterium ammoniagenes ATCC 6872 using a statistically improved biocatalytic process. Appl. Microbiol. Biotechnol. 2007, 76, 321–328. [Google Scholar]
- Yoshio, N.; Isao, B.; Mitsuzo, K.J.U. Process for Preparing Yeast Cells Containing an Enhanced Amount of Ribonucleic Acid. U.S. Patent No. 3411989, 19 November 1968. [Google Scholar]
- Zeng, D.W.; Qiu, C.X.; Shen, Y.; Hou, J.; Li, Z.L.; Zhang, J.X.; Liu, S.; Shang, J.L.; Qin, W.S.; Xu, L.L.; et al. An innovative protein expression system using RNA polymerase I for large-scale screening of high-nucleic-acid contentSaccharomyces cerevisiaestrains. Microb. Biotechnol. 2020, 13, 2008–2019. [Google Scholar] [CrossRef]
- Jetti, K.D.; Reddy, G.; Garlapab, D.; Nammi, S.K. Improved ethanol productivity and ethanol tolerance through genome shuffling of Saccharomyces cerevisiae and Pichia stipitis. Int. Microbiol. 2019, 22, 247–254. [Google Scholar] [CrossRef]
- Wess, J.; Brinek, M.; Boles, E. Improving isobutanol production with the yeast Saccharomyces cerevisiae by successively blocking competing metabolic pathways as well as ethanol and glycerol formation. Biotechnol. Biofuels 2019, 12, 173. [Google Scholar] [CrossRef]
- Posada-Uribe, L.F.; Romero-Tabarez, M.; Villegas-Escobar, V. Effect of medium components and culture conditions in Bacillus subtilis EA-CB0575 spore production. Bioprocess Biosyst. Eng. 2015, 38, 1879–1888. [Google Scholar] [CrossRef]
- Jallouli, W.; Jaoua, S.; Zouari, N. Improvement of Photorhabdus temperata strain K122 bioinsecticide production by batch and fed-batch fermentations optimization. Bioprocess Biosyst. Eng. 2012, 35, 1505–1513. [Google Scholar] [CrossRef]
- Salehmin, M.N.I.; Annuar, M.S.M.; Chisti, Y. High cell density fed-batch fermentations for lipase production: Feeding strategies and oxygen transfer. Bioprocess Biosyst. Eng. 2013, 36, 1527–1543. [Google Scholar] [CrossRef]
- Sohoni, S.V.; Nelapati, D.; Sathe, S.; Javadekar-Subhedar, V.; Gaikaiwari, R.P.; Wangikar, P.P. Optimization of high cell density fermentation process for recombinant nitrilase production in E. coli. Bioresour. Technol. 2015, 188, 202–208. [Google Scholar] [CrossRef]
- Tashiro, Y.; Takeda, K.; Kobayashi, G.; Sonomoto, K. High production of acetone-butanol-ethanol with high cell density culture by cell-recycling and bleeding. J. Biotechnol. 2005, 120, 197–206. [Google Scholar] [CrossRef]
- Chang, H.N.; Kim, N.J.; Kang, J.; Jeong, C.M.; Choi, J.D.R.; Fei, Q.; Kim, B.J.; Kwon, S.; Lee, S.Y.; Kim, J. Multi-stage high cell continuous fermentation for high productivity and titer. Bioprocess Biosyst. Eng. 2011, 34, 419–431. [Google Scholar] [CrossRef]


| Factor | Level | |
|---|---|---|
| −1 | 1 | |
| Molasses/% | 2.00 | 4.00 |
| (NH4)2SO4/% | 1.00 | 3.00 |
| MgSO4·7H2O/% | 0.10 | 0.30 |
| KH2PO4/% | 0.10 | 0.30 |
| FeSO4·7H2O/(g/L) | 0.02 | 0.10 |
| ZnSO4·7H2O/(g/L) | 0.02 | 0.10 |
| Yeast extract/% | 0.50 | 1.50 |
| Factor | Level | ||||
|---|---|---|---|---|---|
| −1.68 | −1 | 0 | 1 | 1.68 | |
| Molasses/% | 2.32 | 3.0 | 4.0 | 5.0 | 5.68 |
| (NH4)2SO4/% | 0.16 | 0.5 | 1.0 | 1.5 | 1.84 |
| KH2PO4/% | 0.06 | 0.2 | 0.4 | 0.6 | 0.74 |
| Regression Equation | R2 | Retention Time/Min | Peak Area/mAU | DCW/mg | GMP + IMP/µg | ||||
|---|---|---|---|---|---|---|---|---|---|
| G03 | G03H8 | G03 | G03H8 | G03 | G03H8 | ||||
| GMP | y = 121,634x − 22,559 | 0.9997 | 6.16 | 2,001,359.50 | 2,752,181.00 | 2.51 | 3.14 | 88.53 | 178.46 |
| IMP | y = 129,215x + 56,214 | 0.9992 | 7.26 | 3,626,890.00 | 8639122.50 | ||||
| Item | Coefficient | T-Value | p-Value |
|---|---|---|---|
| Constant | 14.7740 | 357.41 | 0.000 * |
| Molasses/% | 0.2191 | 5.30 | 0.006 * |
| (NH4)2SO4/% | −0.1424 | −3.44 | 0.026 * |
| MgSO4·7H2O/% | −0.1015 | −2.46 | 0.070 |
| KH2PO4/% | 0.1835 | 4.44 | 0.011 * |
| FeSO4·7H2O/(g/L) | −0.0757 | −1.83 | 0.141 |
| ZnSO4·7H2O/(g/L) | −0.0598 | −1.45 | 0.222 |
| Yeast extract/% | −0.0770 | −1.86 | 0.136 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Model | 13.4900 | 9 | 1.5000 | 4.1100 | 0.0190 |
| A-Molasses | 0.9769 | 1 | 0.9769 | 2.6800 | 0.1328 |
| B-(NH4)2SO4 | 5.1700 | 1 | 5.1700 | 14.1800 | 0.0037 |
| C-KH2PO3 | 1.2500 | 1 | 1.2500 | 3.4400 | 0.0934 |
| AB | 0.0000 | 1 | 0.0000 | 0.0001 | 0.9942 |
| AC | 1.0800 | 1 | 1.0800 | 2.9600 | 0.1163 |
| BC | 0.0425 | 1 | 0.0425 | 0.1164 | 0.7400 |
| A2 | 3.7200 | 1 | 3.7200 | 10.1900 | 0.0096 |
| B2 | 1.5300 | 1 | 1.5300 | 4.1800 | 0.0680 |
| C2 | 0.4349 | 1 | 0.4349 | 1.1900 | 0.3005 |
| Fermentation Method | x (g/L) | µ (h−1) | Yx/s (g/g) | rx (g/L/h) | rs (g/L/h) | Ef (%) |
|---|---|---|---|---|---|---|
| Batch | 6.17 | 0.13 | 0.19 | 0.26 | 1.12 | 33.33 |
| Fed-batch | 60.58 | 0.12 | 0.43 | 1.58 | 3.20 | 75.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Wang, J.; Li, Q.; Xu, X.; Niu, C.; Zheng, F.; Liu, C. Fed-Batch Fermentation of Saccharomyces pastorianus with High Ribonucleic Acid Yield. Foods 2022, 11, 2742. https://doi.org/10.3390/foods11182742
Chen H, Wang J, Li Q, Xu X, Niu C, Zheng F, Liu C. Fed-Batch Fermentation of Saccharomyces pastorianus with High Ribonucleic Acid Yield. Foods. 2022; 11(18):2742. https://doi.org/10.3390/foods11182742
Chicago/Turabian StyleChen, Hao, Jinjing Wang, Qi Li, Xin Xu, Chengtuo Niu, Feiyun Zheng, and Chunfeng Liu. 2022. "Fed-Batch Fermentation of Saccharomyces pastorianus with High Ribonucleic Acid Yield" Foods 11, no. 18: 2742. https://doi.org/10.3390/foods11182742
APA StyleChen, H., Wang, J., Li, Q., Xu, X., Niu, C., Zheng, F., & Liu, C. (2022). Fed-Batch Fermentation of Saccharomyces pastorianus with High Ribonucleic Acid Yield. Foods, 11(18), 2742. https://doi.org/10.3390/foods11182742






