Correlation and Difference between Core Micro-Organisms and Volatile Compounds of Suan Rou from Six Regions of China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.2.1. Total Genomic DNA Extraction and PCR Amplification
2.2.2. Illumina Sequencing and Bioinformatics Processing
2.2.3. Data Processing
2.3. Analysis of Volatile Compounds
2.4. Statistical Analysis
3. Results and Discussion
3.1. Microbial Community Characteristics of SR from Different Regions
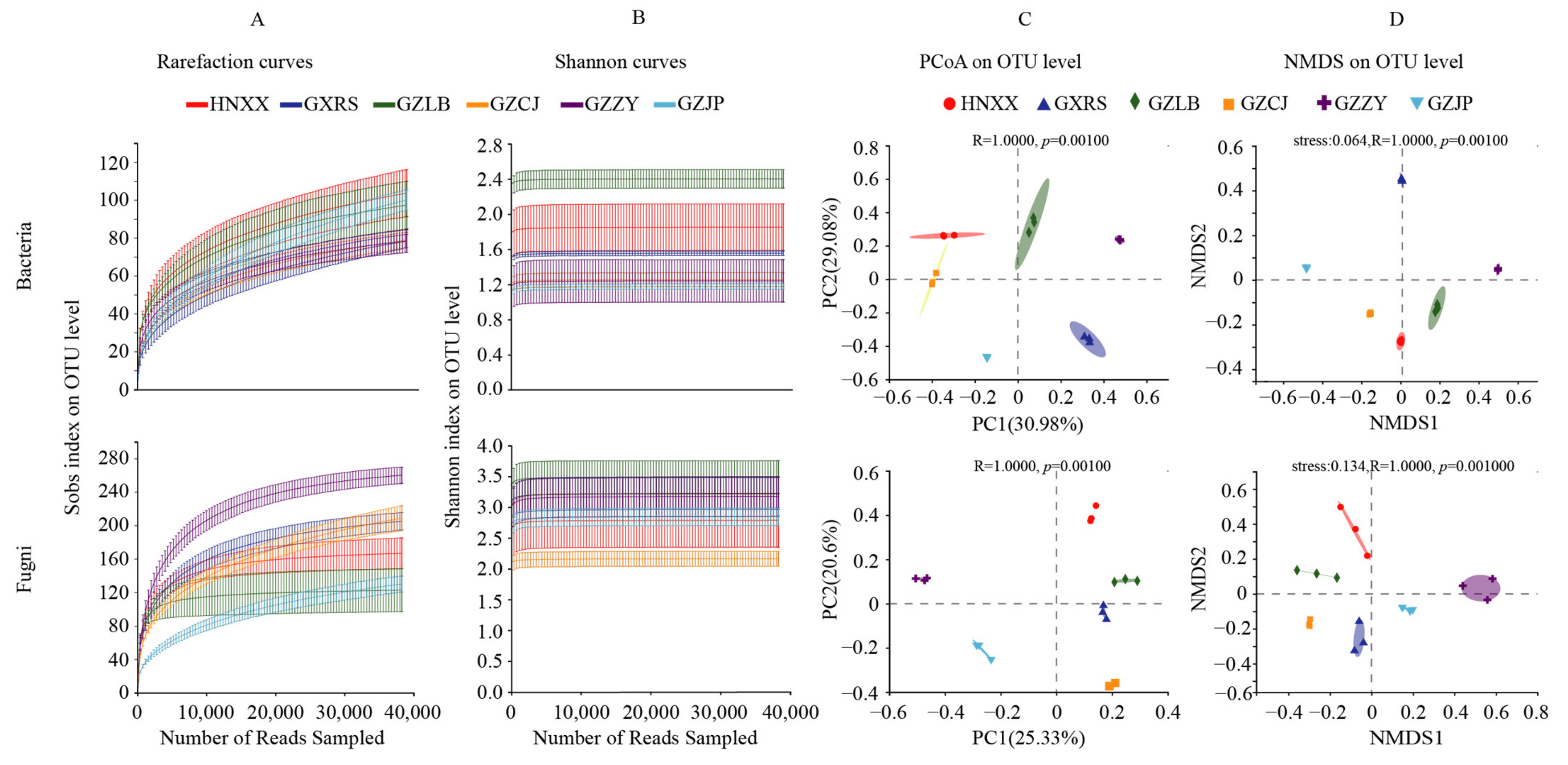
3.1.1. Abundance and Diversity of Bacterial and Fungi
Microbial Composition Analysis
Microbial Difference Analysis
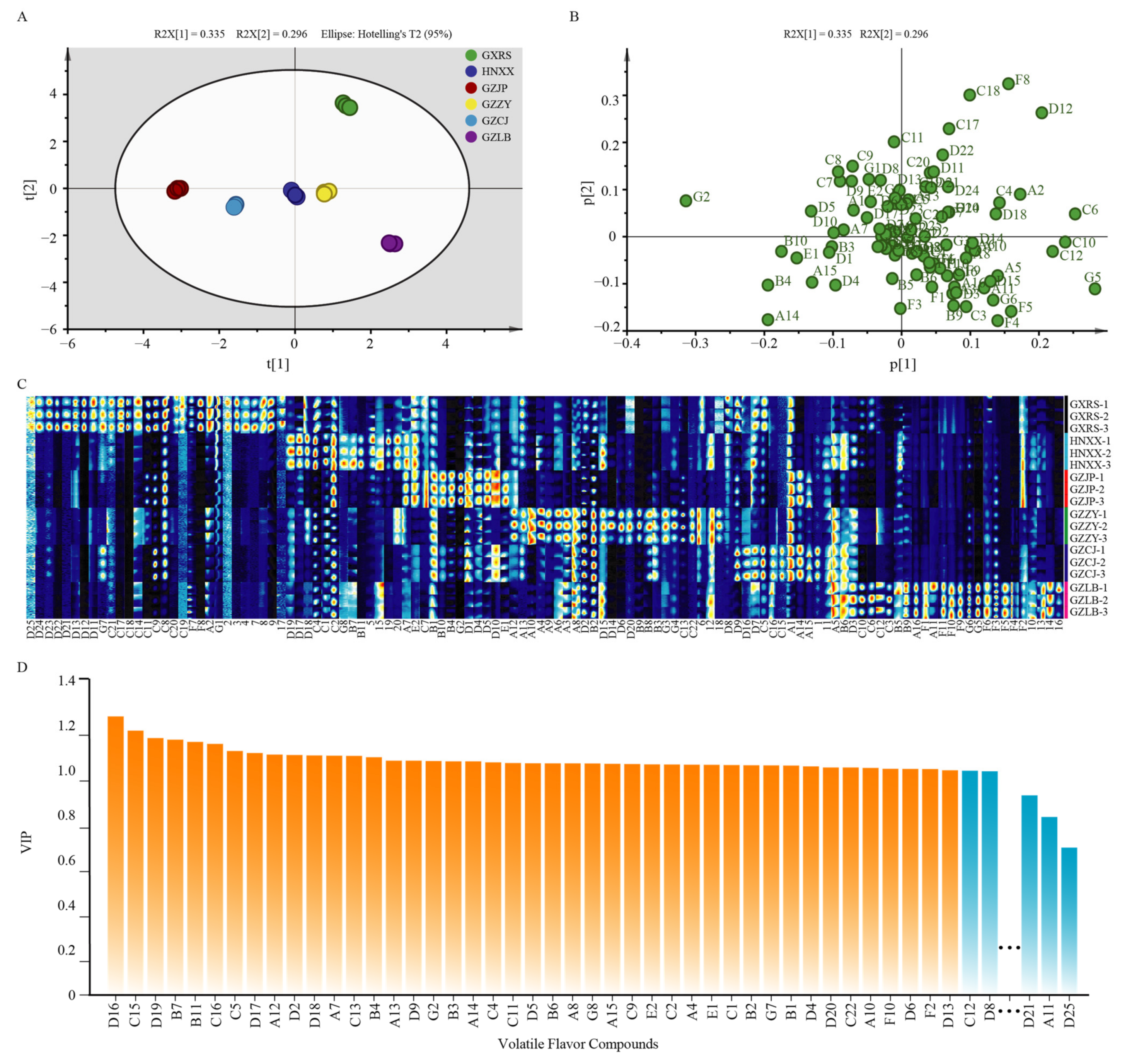
3.2. Flavor Compounds Analysis
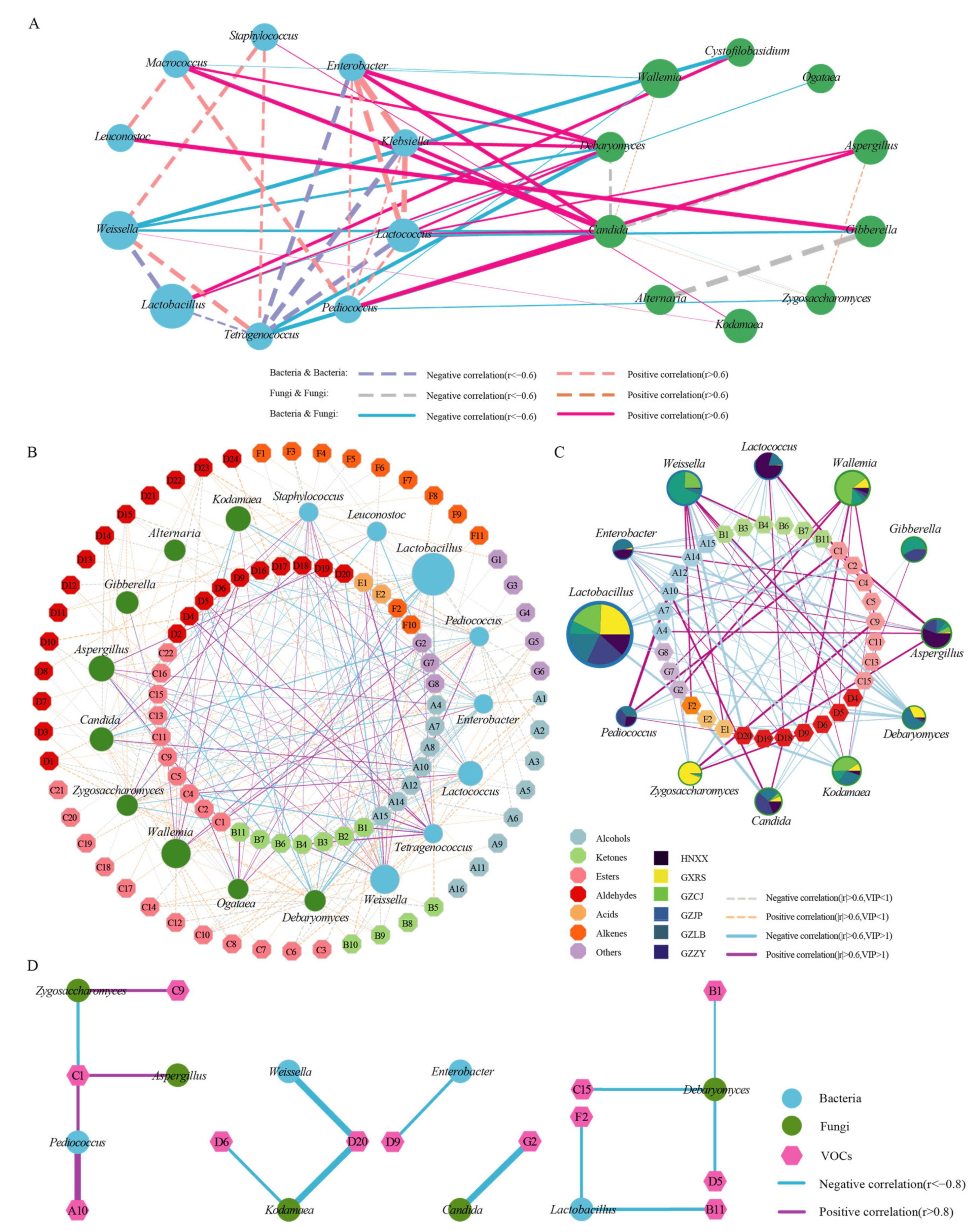
3.3. Co-Occurrence and Exclusion Analyses Revealed the Relationships between Different Microbes
3.4. Correlations between Micro-Organisms and Volatile Organic Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health benefits of fermented foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Voidarou, C.; Antoniadou, Μ.; Rozos, G.; Tzora, A.; Skoufos, I.; Varzakas, T.; Lagiou, A.; Bezirtzoglou, E. Fermentative Foods: Microbiology, Biochemistry, Potential Human Health Benefits and Public Health Issues. Foods 2020, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Yu, Q.L.; Wan, Z.; Li, H.Y.; Liu, J.; Wang, J. Effect of Antioxidant Activity of Starter Cultures on the Quality of Fermented Meat Products: A Review. Food Sci. 2021, 42, 302–312. [Google Scholar] [CrossRef]
- Rul, F.; Béra-Maillet, C.; Champomier-Vergès, M.C.; El-Mecherfi, K.E.; Foligné, B.; Michalski, M.C.; Milenkovic, D.; Savary-Auzeloux, I. Underlying evidence for the health benefits of fermented foods in humans. Food Funct. 2022, 13, 4804–4824. [Google Scholar] [CrossRef]
- Chen, X.; Mi, R.; Qi, B.; Xiong, S.; Li, J.; Qu, C.; Qiao, X.; Chen, W.; Wang, S. Effect of proteolytic starter culture isolated from Chinese Dong fermented pork (Nanx Wudl) on microbiological, biochemical and organoleptic attributes in dry fermented sausages. Food Sci. Hum. Well. 2021, 10, 13–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, P.; Xie, Y.; Wang, X. Co-fermentation with Lactobacillus curvatus LAB26 and Pediococcus pentosaceus SWU73571 for improving quality and safety of sour meat. Meat Sci. 2020, 170, 108240. [Google Scholar] [CrossRef]
- Mariutti, R.B.L.; Bragagnolo, N. Influence of salt on lipid oxidation in meat and seafood products: A review. Food Res. Int. 2017, 94, 90–100. [Google Scholar] [CrossRef]
- Flores, M. Understanding the implications of current health trends on the aroma of wet and dry cured meat products. Meat Sci. 2018, 144, 53–61. [Google Scholar] [CrossRef]
- Gan, X.; Li, H.; Wang, Z.; Emara, A.M.; Zhang, D.; He, Z. Does protein oxidation affect proteolysis in low sodium Chinese traditional bacon processing? Meat Sci. 2019, 150, 14–22. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, P.; Guo, L.; Wang, J.; Han, R.; Sun, J.; Yang, Q. Effects of partial replacement of sodium nitrite with Lactobacillus pentosus inoculation on quality of fermented sausages. J. Food Process Pres. 2019, 43, e13932. [Google Scholar] [CrossRef]
- Wang, Y.; Han, J.; Wang, D.X.; Gao, F.; Zhang, K.P.; Tian, J.J.; Jin, Y. Research Update on the Impact of Lactic Acid Bacteria on the Substance Metabolism, Flavor, and Quality Characteristics of Fermented Meat Products. Foods 2022, 11, 2090. [Google Scholar] [CrossRef]
- Lv, J.; Xu, W.; Ji, C.; Liang, H.; Li, S.; Yang, Z.; Zhang, S.; Lin, X. Relationships between the bacterial diversity and metabolites of a Chinese fermented pork product, sour meat. Int. J. Food Sci. Tech. 2020, 56, 2742–2750. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, X.; Hadiatullah, H.; Zhang, J.; Zhao, G. Determination of microbial diversities and aroma characteristics of Beitang shrimp paste. Food Chem. 2020, 344, 128695. [Google Scholar] [CrossRef]
- Almeida, M.A.D.; Saldaña, E.; Pinto, J.S.D.S.; Palacios, J.; Contreras-Castillo, C.J.; Sentandreu, M.A.; Fadda, S.G. A peptidomic approach of meat protein degradation in a low-sodium fermented sausage model using autochthonous starter cultures. Food Res. Int. 2018, 109, 368–379. [Google Scholar] [CrossRef]
- Wen, R.; Sun, F.; Li, X.; Chen, Q.; Kong, B. The potential correlations between the fungal communities and volatile compounds of traditional dry sausages from Northeast China. Food Microbiol. 2021, 98, 103787. [Google Scholar] [CrossRef]
- Bis-Souza, C.V.; Pateiro, M.; Dominguez, R.; Lorenzo, J.M.; Penna, A.L.B.; Barretto, A.C.D. Volatile profile of fermented sausages with commercial probiotic strains and fructooligosaccharides. J. Food Sci. Tech. Mys. 2019, 56, 5465–5473. [Google Scholar] [CrossRef]
- Zeng, X.; Meng, J.; Zhang, W.; He, L.; Deng, L.; Ye, C. Changes in the microbiological, physicochemical properties of Chinese traditional fermented Suan rou at ripening fermentation. Food Sci. Nutr. 2021, 9, 5899–5913. [Google Scholar] [CrossRef]
- Zhong, A.; Chen, W.; Duan, Y.; Li, K.; Tang, X.; Tian, X.; Wu, Z.; Li, Z.; Wang, Y.; Wang, C. The potential correlation between microbial communities and flavors in traditional fermented sour meat. LWT-Food Sci. Technol. 2021, 149, 111873. [Google Scholar] [CrossRef]
- Lv, J.; Yang, Z.; Xu, W.; Li, S.; Liang, H.; Ji, C.; Yu, C.; Zhu, B.; Lin, X. Relationships between bacterial community and metabolites of Suan Rou at different temperature during the fermentation. Int. J. Food Microbiol. 2019, 307, 108286. [Google Scholar] [CrossRef]
- Emiel, V.R.; Christina, C.; David, V.V.; Wim, B.; Luc, D.V.; Stefan, W.; Frédéric, L. Application of a High-Throughput Amplicon Sequencing Method to Chart the Bacterial Communities that Are Associated with European Fermented Meats from Different Origins. Foods 2020, 9, 1247. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Ji, L.; Zhang, J.; Zhao, Z.; Zhang, R.; Bai, T.; Hou, B.; Zhang, Y.; Liu, D.; et al. A Review: Microbial Diversity and Function of Fermented Meat Products in China. Front. Microbiol. 2021, 12, 645435. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Su, W.; Mu, Y.; Mu, Y.C. Major metabolites and microbial community of fermented black glutinous rice wine with different starters. Front. Microbiol. 2020, 11, 593. [Google Scholar] [CrossRef] [PubMed]
- Buée, M.; Reich, M.; Murat, C.; Morin, E.; Nilsson, R.H.; Uroz, S.; Martin, F. 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009, 184, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Su, W.; Mu, Y.; Zhao, C. Correlation Between Microbial Diversity and Volatile Flavor Compounds of Suan Zuo rou, a Fermented Meat Product From Guizhou, China. Front. Microbiol. 2021, 12, 736525. [Google Scholar] [CrossRef]
- Wang, M.; Wang, C.; Yang, C.; Peng, L.; Xie, Q.; Zheng, R.; Dai, Y.; Liu, S.; Peng, X. Effects of Lactobacillus plantarum C7 and Staphylococcus warneri S6 on flavor quality and bacterial diversity of fermented meat rice, a traditional Chinese food. Food Res. Int. 2021, 150, 110745. [Google Scholar] [CrossRef]
- Laranjo, M.; Potes, M.E.; Elias, M. Role of Starter Cultures on the Safety of Fermented Meat Products. Front. Microbiol. 2019, 10, 853. [Google Scholar] [CrossRef]
- Murgia, M.A.; Marongiu, A.; Aponte, M.; Blaiotta, G.; Deiana, P.; Mangia, N.P. Impact of a selected Debaryomyces hansenii strain’s inoculation on the quality of Sardinian fermented sausages. Food Res. Int. 2019, 121, 144–150. [Google Scholar] [CrossRef]
- Mu, Y.; Su, W.; Mu, Y.; Jiang, L. Combined Application of High-Throughput Sequencing and Metabolomics Reveals Metabolically Active Microorganisms During Panxian Ham Processing. Front. Microbiol. 2019, 10, 3012. [Google Scholar] [CrossRef]
- Corral, S.; Belloch, C.; López-Díez, J.J.; Flores, M. Lipolysis and aroma generation as mechanisms involved in masking boar taint in sodium reduced fermented sausages inoculated with Debaryomyces hansenii yeast. J. Sci. Food Agr. 2018, 98, 2121–2130. [Google Scholar] [CrossRef]
- Li, X.; Eu, A.; Liu, S. Effect of co-fermentation and sequential fermentation of Candida versatilis and Lactococcus lactis subsp. cremoris on unsalted pork hydrolysates components. Int. J. Food Sci. Tech. 2021, 56, 6451–6462. [Google Scholar] [CrossRef]
- Mi, R.; Chen, X.; Xiong, S.; Qi, B.; Li, J.; Qiao, X.; Chen, W.; Qu, C.; Wang, S. Predominant yeasts in Chinese Dong fermented pork (Nanx Wudl) and their aroma-producing properties in fermented sausage condition. Food Sci. Hum. Well. 2021, 10, 231–240. [Google Scholar] [CrossRef]
- Peromingo, B.; Andrade, M.J.; Delgado, J.; Sánchez-Montero, L.; Núñez, F. Biocontrol of aflatoxigenic Aspergillus parasiticus by native Debaryomyces hansenii in dry-cured meat products. Food Microbiol. 2019, 82, 269–276. [Google Scholar] [CrossRef]
- He, W.; Ren, F.; Wang, Y.; Gao, X.; Wang, X.; Dai, X.; Song, J. Application of GC-IMS in Detection of Food Flavor Substances. Proceedings of 2020 International Conference on Green Chemical and 2020 International Conference on Green Chemical and Environmental Science (ICGCES 2020), Qing Dao, China, 12 June 2020. [Google Scholar]
- Giannetti, V.; Mariani, M.B.; Mannino, P.; Marini, F. Volatile fraction analysis by HS-SPME/GC-MS and chemometric modeling for traceability of apples cultivated in the Northeast Italy. Food Control 2017, 78, 215–221. [Google Scholar] [CrossRef]
- Lin, H.; Yu, X.; Fang, J.; Lu, Y.; Liu, P.; Xing, Y.; Wang, Q.; Che, Z.; He, Q. Flavor compounds in Pixian broad-bean paste: Non-volatile organic acids and amino acids. Molecules 2018, 23, 1299. [Google Scholar] [CrossRef]
- Olivares, A.; Navarro, J.L.; Flores, M. Effect of fat content on aroma generation during processing of dry fermented sausages. Meat Sci. 2011, 87, 264–273. [Google Scholar] [CrossRef]
- Shi, Y.; Li, X.; Huang, A. A metabolomics-based approach investigates volatile flavor formation and characteristic compounds of the Dahe black pig dry-cured ham. Meat Sci. 2019, 158, 107904. [Google Scholar] [CrossRef]
- Sidira, M.; Kandylis, P.; Kanellaki, M.; Kourkoutas, Y. Effect of immobilized Lactobacillus casei on the evolution of flavor compounds in probiotic dry-fermented sausages during ripening. Meat Sci. 2016, 100, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Castelán, C.; García-Cano, I.; Escobar-Zepeda, A.; Azaola-Espinosa, A.; Álvarez-Cisneros, Y.; Ponce-Alquicira, E. Evaluation of the bacterial diversity of Spanish-type chorizo during the ripening process using high-throughput sequencing and physicochemical characterization. Meat Sci. 2018, 150, 7–13. [Google Scholar] [CrossRef]
- Iacumin, L.; Osualdini, M.; Bovolenta, S.; Boscolo, D.; Chiesa, L.; Panseri, S.; Comi, G. Microbial, chemico-physical and volatile aromatic compounds characterization of Pitina PGI, a peculiar sausage-like product of North East Italy. Meat Sci. 2020, 163, 108081. [Google Scholar] [CrossRef]
- Cruxen, C.E.S.; Funck, G.D.; Dannenberg, G.S.; Haubert, L.; Marques, J.L.; Kroning, I.S.; Chaves, F.C.; Silva, W.P.; Fiorentini, Â.M. Characterization of Staphylococcus xylosus LQ3 and its application in dried cured sausage. LWT-Food Sci. Technol. 2017, 86, 538–543. [Google Scholar] [CrossRef]
- Ai, M.; Qiu, X.; Huang, J.; Wu, C.; Jin, Y.; Zhou, R. Characterizing the microbial diversity and major metabolites of Sichuan bran vinegar augmented by Monascus purpureus. Int. J. Food Microbiol. 2019, 292, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Belleggia, L.; Ferrocino, I.; Reale, A.; Corvaglia, M.R.; Milanović, V.; Cesaro, C.; Boscaino, F.; Di, R.T.; Garofalo, C.; Cardinali, F.; et al. Unfolding microbiota and volatile organic compounds of Portuguese Painho de Porco Preto fermented sausages. Food Res. Int. 2022, 155, 111063. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, J.M.; Franco, D.; Carballo, J. Effect of the inclusion of chestnut in the finishing diet on volatile compounds during the manufacture of dry-cured “Lacon” from Celta pig breed. Meat Sci. 2014, 96, 211–233. [Google Scholar] [CrossRef] [PubMed]


| Sample | Producers | Production Location | Raw Material | Fermented Conditions |
|---|---|---|---|---|
| HNXX | Local farmer 1 | Xiangxi Tujia and Miao Autonomous Prefecture, Hunan Province | Pork belly, Salt, Rice, Pepper | A sealed fermented LNEC |
| GXRS | Local farmer 2 | Rongshui Miao Autonomous County, Liuzhou City, Guangxi Zhuang Autonomous Region | Pork belly, Salt, Chili | A sealed fermented LNEC |
| GZZY | Local farmer 3 | Zunyi City, Guizhou Province | Pork belly, Salt, Rice | A sealed fermented LNEC |
| GZLB | Local farmer 4 | Libo County, Qiannan Buyi and Miao Autonomous Prefecture, Guizhou Province | Pork belly, Salt, Millet, Pepper | A sealed fermented LNEC |
| GZCJ | Local farmer 5 | Congjiang County, Southeast Guizhou Miao and Dong Autonomous Prefecture, Guizhou Province | Pork belly, Salt, Millet, Chili, Pepper | A sealed fermented LNEC |
| GZJP | Local farmer 6 | Jinping County, Southeast Guizhou Miao and Dong Autonomous Prefecture, Guizhou Province | Pork belly, Salt, Chili, Pepper | A sealed fermented LNEC |
| Bacteria | Fungi | |
|---|---|---|
| Template DNA | 10.0 ng | 10.0 ng |
| Primer F (5.0 μM) | 1.0 μL | 0.80 μL |
| Primer R (5.0 μM) | 1.0 μL | 0.80 μL |
| dNTPs (2.5 mM) | 2.0 μL | 2.0 μL |
| Fast Pfu polymerase | 0.50 μL | 0.20 μL |
| 5× Fast Pfu buffer | 4.0 μL | 2.0 μL |
| ddH2O | To a final volume of 20.0 µL | |
| Simple ID | Shannon | Simpson | ACE | Chao1 | ||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | |
| HNXX | 2.08 ± 0.45 a | 3.50 ± 0.21 a | 0.23 ± 0.08 a | 0.06 ± 0.02 e | 135.91 ± 21.93 a | 136.11 ± 18.66 c | 136.15 ± 6.39 a | 137.83 ± 25.86 c |
| GXRS | 1.78 ± 0.39 a | 1.30 ± 0.36 c | 0.29 ± 0.09 a | 0.37 ± 0.09 c | 187.52 ± 43.89 a | 138.74 ± 20.28 c | 153.54 ± 19.70 a | 129.51 ± 25.23 c |
| GZCJ | 1.71 ± 0.48 a | 0.28 ± 0.03 d | 0.36 ± 0.16 a | 0.89 ± 0.00 a | 135.44 ± 32.43 a | 68.06 ± 12.70 d | 125.69 ± 27.57 a | 45.67 ± 17.33 d |
| GZLB | 1.94 ± 0.40 a | 3.22 ± 0.21 a,b | 0.25 ± 0.08 a | 0.09 ± 0.03 d,e | 148.85 ± 15.66 a | 217.38 ± 12.42 b | 145.66 ± 1.19 a | 220.91 ± 16.50 b |
| GZZY | 2.12 ± 0.27 a | 3.18 ± 0.27 a,b | 0.25 ± 0.11 a | 0.10 ± 0.03 d,e | 147.49 ± 10.41 a | 276.27 ± 13.84 a | 150.16 ± 18.67 a | 277.17 ± 12.55 a |
| GZJP | 1.57 ± 0.07 a | 0.82 ± 0.26 e | 0.41 ± 0.07 a | 0.69 ± 0.11 b | 134.76 ± 22.91 a | 89.12 ± 22.06 d | 130.16 ± 14.58 a,b | 78.33 ± 15.93 d |
| NO. | Compound Name | Formula | CAS | Relative Content (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| GZRS | HNXX | GZJP | GZZY | GZCJ | GZLB | ||||
| Alcohols (16) | 11.81 ± 0.07 f | 17.97 ± 0.16 d | 19.9 ± 0.34 c | 29.38 ± 0.24 a | 25.1 ± 0.08 b | 16.52 ± 0.43 e | |||
| A1 | Ethanol | C64175 | C2H6O | 4.96 ± 0.1987 e | 5.70 ± 0.1975 d | 10.29 ± 0.1351 a | 8.24 ± 0.1165 c | 9.21 ± 0.0862 b | 2.35 ± 0.1598 f |
| A2 | Propanol | C71238 | C3H8O | 2.81 ± 0.0364 a | 0.90 ± 0.0358 e | 0.28 ± 0.0220 f | 2.59 ± 0.0773 b | 1.06 ± 0.0414 d | 1.49 ± 0.0473 c |
| A3 | Isobutanol | C78831 | C4H10O | 0.10 ± 0.0013 e | 0.21 ± 0.0169 d | 0.17 ± 0.0051 d | 0.94 ± 0.0399 a | 0.56 ± 0.0225 c | 0.66 ± 0.0528 b |
| A4 | 1-penten-3-ol | C616251 | C5H10O | 0.72 ± 0.0137 f | 2.87 ± 0.1810 b | 1.65 ± 0.0484 d | 4.03 ± 0.0317 a | 1.91 ± 0.0611 c | 0.95 ± 0.0507 e |
| A5 | Isopentanol | C123513 | C5H12O | 0.84 ± 0.0378 c | 2.91 ± 0.0411 b | 0.34 ± 0.0361 d | 2.94 ± 0.1447 b | 2.96 ± 0.0426 b | 3.53 ± 0.0784 a |
| A6 | 2-methyl-1-butanol | C137326 | C5H12O | 0.19 ± 0.0068 e | 0.29 ± 0.0401 c | 0.14 ± 0.0062 f | 0.60 ± 0.0083 a | 0.24 ± 0.0023 d | 0.41 ± 0.0084 b |
| A7 | (Z)-2-pentenol | C1576950 | C5H10O | 0.13 ± 0.0055 d | 0.58 ± 0.0721 b | 0.72 ± 0.0275 a | 0.30 ± 0.0380 c | 0.19 ± 0.0019 d | 0.12 ± 0.0118 d |
| A8 | 1-pentanol | C71410 | C5H12O | 0.86 ± 0.0172 d | 1.70 ± 0.0471 c | 0.48 ± 0.0358 e | 2.70 ± 0.0389 a | 2.59 ± 0.0216 a | 2.06 ± 0.1608 b |
| A9 | 3-methyl-1-pentanol | C589355 | C6H14O | 0.11 ± 0.0071 c | 0.18 ± 0.0213 b,c | 0.22 ± 0.0011 b | 0.58 ± 0.1136 a | 0.22 ± 0.0045 b | 0.10 ± 0.0078 c |
| A10 | 1-hexanol | C111273 | C6H14O | 0.29 ± 0.0272 c | 0.58 ± 0.083 b | 0.30 ± 0.0452 c | 3.92 ± 0.1271 a | 0.33 ± 0.0303 c | 0.62 ± 0.0304 b |
| A11 | (Z)-3-hexen-1-ol | C928961 | C6H12O | 0.06 ± 0.0073 b | 0.13 ± 0.0043 b | 0.05 ± 0.0009 b | 0.07 ± 0.0269 b | 0.17 ± 0.0096 b | 0.56 ± 0.3009 a |
| A12 | 1-octen-3-ol | C3391864 | C8H16O | 0.08 ± 0.0026 e | 0.21 ± 0.0050 b | 0.18 ± 0.0149 c | 0.30 ± 0.0165 a | 0.10 ± 0.0079 d | 0.08 ± 0.0054 d,e |
| A13 | 1-octanol | C111875 | C8H18O | 0.20 ± 0.0044 b | 0.25 ± 0.0148 a | 0.10 ± 0.0125 c | 0.26 ± 0.0540 a | 0.29 ± 0.0061 a | 0.07 ± 0.0055 c |
| A14 | Linalool | C78706 | C10H18O | 0.07 ± 0.0032 d | 0.47 ± 0.0336 c | 3.33 ± 0.3002 a | 1.18 ± 0.0309 b | 3.57 ± 0.0733 a | 0.53 ± 0.0190 c |
| A15 | cis-p-menth-2-en-1-ol | C29803825 | C10H18O | 0.08 ± 0.0028 c | 0.23 ± 0.0050 b | 1.07 ± 0.1186 a | 0.34 ± 0.0178 b | 1.04 ± 0.0186 a | 0.29 ± 0.0070 b |
| A16 | alpha-terpineol | C98555 | C10H18O | 0.32 ± 0.0329 c | 0.75 ± 0.0402 b | 0.61 ± 0.0463 b | 0.40 ± 0.0346 c | 0.68 ± 0.0337 b | 2.69 ± 0.1609 a |
| Ketones (11) | 4.24 ± 0.14 e | 9.05 ± 0.29 d | 13.1 ± 0.38 b | 16.6 ± 0.35 a | 13.68 ± 0.47 b | 10.28 ± 0.46 c | |||
| B1 | 2-propanone | C67641 | C3H6O | 1.02 ± 0.0118 f | 1.46 ± 0.0938 d | 2.36 ± 0.0673 a | 1.93 ± 0.0232 c | 2.11 ± 0.0360 b | 1.24 ± 0.0584 e |
| B2 | 2-butanone | C78933 | C4H8O | 2.29 ± 0.0134 e | 2.30 ± 0.1082 e | 3.33 ± 0.2072 d | 7.75 ± 0.1029 a | 6.74 ± 0.1524 b | 4.43 ± 0.2777 c |
| B3 | 2-pentanone | C107879 | C5H10O | 0.14 ± 0.0070 c | 0.22 ± 0.0060 c | 0.62 ± 0.0371 b | 1.28 ± 0.0470 a | 1.31 ± 0.0982 a | 0.14 ± 0.0226 c |
| B4 | 3-hydroxy-2-butanone | C513860 | C4H8O2 | 0.09 ± 0.0052 e,f | 0.18 ± 0.0136 f | 4.31 ± 0.1561 a | 0.71 ± 0.1472 c | 1.10 ± 0.0550 b | 0.37 ± 0.0581 d |
| B5 | 4-methyl-3-penten-2-one | C141797 | C6H10O | 0.06 ± 0.0036 e | 0.43 ± 0.0666 a | 0.26 ± 0.0093 b,c | 0.12 ± 0.005 d,e | 0.18 ± 0.0012 c,d | 0.27 ± 0.0659 b |
| B6 | 2-hexanone | C591786 | C6H12O | 0.08 ± 0.0020 f | 0.30 ± 0.0126 c | 0.16 ± 0.0058 d | 0.12 ± 0.0168 e | 0.32 ± 0.0113b c | 0.34 ± 0.0244 a |
| B7 | Dihydro-2-methyl-3(2H)furanone | C3188009 | C5H8O2 | 0.03 ± 0.0006 d | 0.11 ± 0.0127 a | 0.04 ± 0.0034 c | 0.05 ± 0.002 c,d | 0.07 ± 0.0040 b | 0.04 ± 0.0046 c,d |
| B8 | 2-heptanone | C110430 | C7H14O | 0.05 ± 0.0045 c | 0.05 ± 0.0039 c | 0.08 ± 0.0106 c | 0.80 ± 0.0629 a | 0.45 ± 0.0807 b | 0.07 ± 0.0156 c |
| B9 | 3-octanone | C106683 | C8H16O | 0.28 ± 0.1059 d | 1.15 ± 0.1785 c | 0.99 ± 0.0878 c | 3.58 ± 0.2010 a | 0.89 ± 0.0387 c | 3.06 ± 0.3374 b |
| B10 | 6-methyl-5-hepten-2-one | C110930 | C8H14O | 0.04 ± 0.0052 d | 0.12 ± 0.0061 c | 0.51 ± 0.0172 a | 0.04 ± 0.004 d | 0.25 ± 0.0125 b | 0.06 ± 0.0048 d |
| B11 | Coumarin | C91645 | C9H6O2 | 0.16 ± 0.0208 b | 2.76 ± 0.3605 a | 0.46 ± 0.1660 b | 0.24 ± 0.0190 b | 0.26 ± 0.0116 b | 0.25 ± 0.0331 b |
| Esters (22) | 46.37 ± 0.09 a | 44.95 ± 0.69 a | 35.62 ± 0.58 b | 24.38 ± 0.64 c | 32.21 ± 0.65 d | 27.53 ± 1.46 e | |||
| C1 | Methyl acetate | C79209 | C3H6O2 | 0.88 ± 0.0017 d | 3.10 ± 0.4606 a | 1.37 ± 0.0792 c | 2.26 ± 0.0315 b | 2.00 ± 0.0138 b | 2.05 ± 0.1902 b |
| C2 | Ethyl acetate | C141786 | C4H8O2 | 9.56 ± 0.1527 d | 23.09 ± 0.1598 a | 19.99 ± 0.276 b | 6.22 ± 0.4258 e | 12.95 ± 0.3376 c | 12.51 ± 0.2807 c |
| C3 | Isopropyl acetate | C108214 | C5H10O2 | 0.03 ± 0.0010 b | 0.07 ± 0.0032 b | 0.06 ± 0.0050 b | 0.09 ± 0.0120 b | 0.08 ± 0.0094 b | 0.36 ± 0.0702 a |
| C4 | Propyl acetate | C109604 | C5H10O2 | 7.23 ± 0.1196 b | 10.54 ± 0.0565 a | 1.02 ± 0.0262 e | 3.97 ± 0.244 d | 3.29 ± 0.0682 f | 4.43 ± 0.3286 c |
| C5 | Ethyl 2-methylpropanoate | C97621 | C6H12O2 | 2.15 ± 0.0096 b | 1.10 ± 0.1287 c | 1.03 ± 0.0846 c | 0.49 ± 0.038 d | 2.82 ± 0.0523 a | 1.10 ± 0.0878 c |
| C6 | 2-methylpropyl acetate | C110190 | C6H12O2 | 0.51 ± 0.0207 b | 0.16 ± 0.0470 c | 0.05 ± 0.0012 c | 0.13 ± 0.0045 c | 0.06 ± 0.0055 c | 0.96 ± 0.2729 a |
| C7 | Methyl 3-methylbutanoate | C556241 | C6H12O2 | 0.28 ± 0.0254 b | 0.32 ± 0.0250 b | 0.58 ± 0.0571 a | 0.09 ± 0.006 c,d | 0.15 ± 0.0065 c | 0.07 ± 0.0050 d |
| C8 | Ethyl butanoate | C105544 | C6H12O2 | 3.54 ± 0.0363 b | 2.72 ± 0.0295 d | 4.85 ± 0.0927 a | 3.22 ± 0.0397 c | 3.28 ± 0.0316 c | 0.51 ± 0.0297 e |
| C9 | Ethyl 3-methylbutanoate | C108645 | C7H14O2 | 3.68 ± 0.0484 b | 0.38 ± 0.0524 f | 2.34 ± 0.0673 c | 0.74 ± 0.044 d | 4.06 ± 0.0619 a | 0.56 ± 0.0662 e |
| C10 | Isoamyl acetate | C123922 | C7H14O2 | 0.61 ± 0.0442 b | 0.60 ± 0.2229 b | 0.13 ± 0.0069 c | 0.65 ± 0.0099 c | 0.15 ± 0.0351 b | 2.07 ± 0.3881 a |
| C11 | Propyl butanoate | C105668 | C7H14O2 | 1.15 ± 0.0852 b | 0.17 ± 0.0107 d | 0.96 ± 0.0165 c | 1.55 ± 0.0362 a | 0.15 ± 0.0082 d | 0.11 ± 0.0065 d |
| C12 | Methyl hexanoate | C106707 | C7H14O2 | 0.08 ± 0.0013 c | 0.04 ± 0.0000 d | — | 0.26 ± 0.0144 b | — | 0.37 ± 0.0256 a |
| C13 | Ethyl hexanoate | C123660 | C8H16O2 | 0.69 ± 0.0250 c | 0.36 ± 0.0298 d | 1.11 ± 0.0056 b | 1.93 ± 0.1395 a | 0.51 ± 0.0208 d | 0.37 ± 0.0930 d |
| C14 | Methyl heptanoate | C106730 | C8H16O2 | 0.11 ± 0.0106 a | 0.07 ± 0.0033 c,d | 0.06 ± 0.0055 d | 0.09 ± 0.002 b,c | 0.07 ± 0.0041 d | 0.09 ± 0.0097 b |
| C15 | Ethyl 2-hydroxy-4-methylpentanoate-D | C10348477 | C8H16O3 | 0.07 ± 0.0084 d | 0.11 ± 0.0094 b,c | 0.14 ± 0.0022 b | 0.11 ± 0.017 b,c | 0.24 ± 0.0216 a | 0.10 ± 0.0060 c |
| C16 | Ethyl 2-hydroxy-4-methylpentanoate-M | C10348477 | C8H16O3 | 0.15 ± 0.0465 d | 0.21 ± 0.0244 c,d | 0.13 ± 0.0103 d | 0.30 ± 0.0075 c | 0.80 ± 0.0183 a | 0.59 ± 0.0908 b |
| C17 | Ethyl 3-hydroxyhexanoate-M | C2305251 | C8H16O3 | 4.06 ± 0.0775 a | 0.71 ± 0.1820 b | 0.48 ± 0.0454 c | 0.35 ± 0.0308 c | 0.38 ± 0.0354 c | 0.33 ± 0.0180 c |
| C18 | Ethyl 3-hydroxyhexanoate-D | C2305251 | C8H16O3 | 10.59 ± 0.382 a | 0.46 ± 0.0281 b | 0.48 ± 0.0129 b | 0.42 ± 0.0286 b | 0.45 ± 0.0378 b | 0.37 ± 0.0105 b |
| C19 | Ethyl heptanoate-D | C106309 | C9H18O2 | 0.22 ± 0.0113 a | 0.09 ± 0.0048 c,d | 0.10 ± 0.0042 b | 0.09 ± 0.007 c,d | 0.10 ± 0.0036 c | 0.08 ± 0.0066 d |
| C20 | Ethyl heptanoate-M | C106309 | C9H18O2 | 0.41 ± 0.0751 a | 0.16 ± 0.0633 c | 0.14 ± 0.0106 c | 0.30 ± 0.0205 b | 0.15 ± 0.0201 c | 0.09 ± 0.0110 c |
| C21 | (Z)-3-hexenyl butanoate | C16491364 | C10H18O2 | 0.17 ± 0.0059 b,c | 0.19 ± 0.0232 a,b | 0.17 ± 0.0099 b,c | 0.21 ± 0.0127 a | 0.18 ± 0.0227 a,b | 0.14 ± 0.0044 c |
| C22 | Geranyl acetate | C105873 | C12H20O2 | 0.23 ± 0.0248 d | 0.33 ± 0.0256 c | 0.44 ± 0.0350 b | 0.94 ± 0.0546 a | 0.34 ± 0.0289 c | 0.29 ± 0.0071 c,d |
| Aldehydes (25) | 16.16 ± 0.11 c | 15.78 ± 0.65 c | 18.79 ± 0.76 a | 17.55 ± 0.69 b | 19.13 ± 0.16 a | 10 ± 0.04 d | |||
| D1 | Propanal | C123386 | C3H6O | 0.82 ± 0.0626 f | 2.47 ± 0.1159 c | 5.36 ± 0.1381 a | 2.21 ± 0.011 d | 4.03 ± 0.0787 b | 1.25 ± 0.0483 e |
| D2 | Butanal | C123728 | C4H8O | 2.00 ± 0.0091 c | 1.25 ± 0.0391 d | 1.99 ± 0.1079 c | 3.67 ± 0.0515 a | 3.78 ± 0.0348 a | 2.75 ± 0.2000 b |
| D3 | 3-methylbutanal | C590863 | C5H10O | 0.15 ± 0.0067 c | 0.84 ± 0.0302 b | 0.21 ± 0.0166 c | 0.96 ± 0.049 a,b | 0.86 ± 0.0406 a,b | 0.98 ± 0.1069 a |
| D4 | 2-methylbutanal | C96173 | C5H10O | 0.09 ± 0.0026 d | 0.40 ± 0.0081 c | 1.21 ± 0.0397 a | 0.67 ± 0.0614 b | 0.70 ± 0.0233 b | 0.38 ± 0.0114 c |
| D5 | Pentanal | C110623 | C5H10O | 0.25 ± 0.0066 d | 0.32 ± 0.0087 c | 1.32 ± 0.0241 a | 0.28 ± 0.072 c,d | 0.42 ± 0.0124 b | 0.13 ± 0.0129 e |
| D6 | (E)-2-pentenal | C1576870 | C5H8O | 0.07 ± 0.0026 b | 0.15 ± 0.0053 b | 0.16 ± 0.0242 b | 0.67 ± 0.1905 a | 0.06 ± 0.0013 b | 0.03 ± 0.0005 b |
| D7 | 3-methyl-2-butenal | C107868 | C5H8O | 0.11 ± 0.0099 b,c | 0.20 ± 0.0185 a | 0.19 ± 0.0149 a | 0.07 ± 0.0138 c | 0.19 ± 0.0109 a | 0.12 ± 0.0354 b |
| D8 | Hexanal-M | C66251 | C6H12O | 1.31 ± 0.1736 b | 1.94 ± 0.1741 a | 1.34 ± 0.1765 b | 1.10 ± 0.0302 b | 0.66 ± 0.0293 c | 0.30 ± 0.0246 d |
| D9 | Hexanal-D | C66251 | C6H12O | 3.50 ± 0.0904 b | 1.61 ± 0.0508 e | 3.18 ± 0.1823 c | 2.07 ± 0.102 d | 5.13 ± 0.1578 a | 0.69 ± 0.0736 f |
| D10 | Methional | C3268493 | C4H8OS | 0.05 ± 0.0536 d | 0.16 ± 0.1638 b | 0.24 ± 0.2373 a | — | 0.11 ± 0.1112 c | 0.06 ± 0.0037 d |
| D11 | Heptanal | C111717 | C7H14O | 0.81 ± 0.1290 a | 0.43 ± 0.0297 b | 0.21 ± 0.0168 c | 0.25 ± 0.0164 c | 0.30 ± 0.0096 c | 0.18 ± 0.0095 c |
| D12 | (Z)-4-heptenal-D | C6728310 | C7H12O | 2.47 ± 0.0436 a | 0.07 ± 0.0048 d | 0.07 ± 0.0092 d | 0.40 ± 0.0130 b | 0.07 ± 0.0099 d | 0.18 ± 0.0114 c |
| D13 | (Z)-4-heptenal-M | C6728310 | C7H12O | 0.80 ± 0.0167 b | 0.57 ± 0.0944 c | 0.98 ± 0.0551 a | 0.28 ± 0.090 d | 0.24 ± 0.0264 d | 0.39 ± 0.0540 d |
| D14 | (E)-2-heptenal-D | C18829555 | C7H12O | 0.04 ± 0.0036 b | 0.29 ± 0.0331 b | 0.04 ± 0.0000 b | 1.20 ± 0.4321 a | 0.04 ± 0.0000 b | 0.05 ± 0.0048 b |
| D15 | (E)-2-heptenal-M | C18829555 | C7H12O | 0.12 ± 0.0079 d | 0.56 ± 0.0612 b | 0.11 ± 0.0222 d | 0.49 ± 0.0395 b | 0.21 ± 0.0017 c | 0.71 ± 0.0382 a |
| D16 | Benzaldehyde | C100527 | C7H6O | 0.03 ± 0.0057 d | 0.08 ± 0.0083 b | 0.05 ± 0.0000 c,d | 0.05 ± 0.0045 c | 0.15 ± 0.0154 a | 0.05 ± 0.0026 c,d |
| D17 | 2,4-heptadienal | C5910850 | C7H10O | 0.29 ± 0.0824 c | 1.50 ± 0.1896 a | 0.73 ± 0.0547 b | 0.37 ± 0.0155 c | 0.28 ± 0.0199 c | 0.18 ± 0.0164 c |
| D18 | Octanal | C124130 | C8H16O | 0.55 ± 0.0617 c | 1.10 ± 0.0750 a | 0.15 ± 0.0076 d | 0.71 ± 0.0154 b | 0.22 ± 0.0048 d | 0.48 ± 0.0188 c |
| D19 | Benzeneacetaldehyde | C122781 | C8H8O | 0.11 ± 0.0027 d | 0.69 ± 0.0465 a | 0.14 ± 0.0151 c,d | 0.40 ± 0.0358 b | 0.40 ± 0.0174 b | 0.18 ± 0.0178 c |
| D20 | Nonanal | C124196 | C9H18O | 0.15 ± 0.0079 b | 0.13 ± 0.0024 b,c | 0.1 ± 0.0070 d | 0.30 ± 0.0324 a | 0.08 ± 0.0059 d | 0.11 ± 0.0067 c,d |
| D21 | (Z)-4-decenal-D | C21662099 | C10H18O | 0.15 ± 0.0370 a | 0.07 ± 0.0080 b | 0.07 ± 0.0023 b | 0.05 ± 0.0011 b | 0.06 ± 0.0074 b | 0.06 ± 0.0038 b |
| D22 | (Z)-4-decenal-M | C21662099 | C10H18O | 1.39 ± 0.1362 a | 0.31 ± 0.0131 c,d | 0.28 ± 0.0506 c,d | 0.62 ± 0.0394 b | 0.35 ± 0.0241 c | 0.19 ± 0.0083 d |
| D23 | 2-decenal | C3913711 | C10H18O | 0.40 ± 0.0201 a | 0.24 ± 0.0291 c,d | 0.34 ± 0.0028 b | 0.28 ± 0.0077 c | 0.40 ± 0.0174 a | 0.22 ± 0.0137 d |
| D24 | (E,E)-2,4-nonadienal | C5910872 | C9H14O | 0.30 ± 0.0269 a | 0.13 ± 0.0119 c | 0.11 ± 0.0093 c | 0.17 ± 0.0051 b | 0.11 ± 0.0072 c | 0.12 ± 0.0072 c |
| D25 | (E,E)-2,4-decadienal | C25152845 | C10H16O | 0.19 ± 0.0210 a | 0.26 ± 0.0257 a | 0.24 ± 0.0150 a | 0.25 ± 0.0267 a | 0.26 ± 0.0352 a | 0.22 ± 0.0420 a |
| Acids (2) | 0.16 ± 0.01 c | 0.26 ± 0.01 b | 0.51 ± 0.02 a | 0.09 ± 0.01 d | 0.17 ± 0.01 c | 0.09 ± 0.01 d | |||
| E1 | Propanoic acid | C79094 | C3H6O2 | — | — | 0.19 ± 0.0163 a | 0.04 ± 0.0018 b | 0.09 ± 0.0043 c | — |
| E2 | 3-methylbutanoic acid | C503742 | C5H10O2 | 0.16 ± 0.0063 c | 0.26 ± 0.0098 b | 0.32 ± 0.0071 a | 0.05 ± 0.0010 e | 0.08 ± 0.0037 d | 0.09 ± 0.0096 d |
| Alkenes (11) | 17.38 ± 0.09 a | 4.79 ± 0.23 c | 6.61 ± 0.47 b | 5.31 ± 0.17 c | 5.44 ± 0.17 c | 17.44 ± 0.44 a | |||
| F1 | Tricyclene | C508327 | C10H16 | 0.08 ± 0.0029 e | 0.21 ± 0.0399 c | 0.27 ± 0.0139 b | 0.07 ± 0.0091 e | 0.12 ± 0.0047 d | 0.69 ± 0.0070 a |
| F2 | Alpha-pinene | C80568 | C10H16 | 0.13 ± 0.0052 d | 0.29 ± 0.0284 b | 0.46 ± 0.0332 a | 0.10 ± 0.009 d | 0.21 ± 0.0213 c | 0.27 ± 0.0218 b |
| F3 | Beta-pinene-M | C127913 | C10H16 | 0.30 ± 0.0090 d | 0.98 ± 0.0938 c | 2.04 ± 0.2142 b | 1.23 ± 0.1524 c | 1.89 ± 0.0840 b | 3.57 ± 0.0228 a |
| F4 | Beta-pinene-D | C127913 | C10H16 | 0.18 ± 0.0091 d | 0.27 ± 0.0058 d | 0.42 ± 0.0375 c | 0.93 ± 0.0477 b | 0.51 ± 0.0340 c | 4.90 ± 0.1473 a |
| F5 | Beta-pinene-T | C127913 | C10H16 | 0.05 ± 0.0114 b | 0.07 ± 0.0043 b | 0.09 ± 0.0153 b | 0.08 ± 0.0116 b | 0.08 ± 0.0047 b | 1.40 ± 0.0648 a |
| F6 | Myrcene | C123353 | C10H16 | 0.15 ± 0.0159 c | 0.20 ± 0.0038 c | 0.31 ± 0.0129 b | 0.29 ± 0.0596 b | 0.23 ± 0.0078 b,c | 0.58 ± 0.0646 a |
| F7 | Alpha-phellandrene-M | C99832 | C10H16 | 2.33 ± 0.0553 a | 1.44 ± 0.2083 b | 1.54 ± 0.1210 b | 1.07 ± 0.1216 c | 1.39 ± 0.0468 b | 2.40 ± 0.1704 a |
| F8 | Alpha-phellandrene-D | C99832 | C10H16 | 13.61 ± 0.040 a | 0.45 ± 0.1576 c | 0.37 ± 0.0304 c | 1.06 ± 0.1179 b | 0.29 ± 0.0135 c | 0.39 ± 0.0560 c |
| F9 | Beta-ocimene | C13877913 | C10H16 | 0.27 ± 0.0141 c | 0.46 ± 0.0313 b | 0.44 ± 0.0324 b | 0.19 ± 0.019 d | 0.33 ± 0.0126 c | 1.85 ± 0.0619 a |
| F10 | Gamma-terpinene | C99854 | C10H16 | 0.14 ± 0.0053 c | 0.17 ± 0.0235 c | 0.34 ± 0.0266 b | 0.17 ± 0.0107 c | 0.16 ± 0.0248 c | 0.72 ± 0.0114 a |
| F11 | Terpinolene | C586629 | C10H16 | 0.14 ± 0.0108 d | 0.24 ± 0.0081 c | 0.32 ± 0.0206 b | 0.12 ± 0.014 d | 0.22 ± 0.0178 c | 0.67 ± 0.0404 a |
| Others (8) | 3.87 ± 0.02 e | 7.18 ± 0.27 b | 5.47 ± 0.15 d | 6.68 ± 0.01 c | 4.28 ± 0.05 e | 18.11 ± 0.42 a | |||
| G1 | Dimethylamine | C124403 | C2H7N | 2.11 ± 0.2938 b | 2.70 ± 0.1530 a | 2.19 ± 0.1176 b | 1.72 ± 0.0640 c | 1.36 ± 0.0208 d | 0.43 ± 0.0143 e |
| G2 | Dimethyl disulfide | C624920 | C2H6S2 | 0.06 ± 0.0025 c | 0.04 ± 0.0001 c | 1.60 ± 0.0556 a | — | 0.34 ± 0.0119 b | — |
| G3 | 2-acetylfuran | C1192627 | C6H6O2 | 0.04 ± 0.0048 c | 0.05 ± 0.0073 c | 0.04 ± 0.0001 c | 0.18 ± 0.0128 a | 0.08 ± 0.0036 b | 0.07 ± 0.0137 b |
| G4 | 2-pentylfuran | C3777693 | C9H14O | 0.10 ± 0.0195 d | 0.35 ± 0.0210 b | 0.19 ± 0.0133 c | 0.55 ± 0.0777 a | 0.22 ± 0.0032 c | 0.27 ± 0.0493 b,c |
| G5 | 1,8-cineole-D | C470826 | C10H18O | 0.48 ± 0.0311 b | 0.35 ± 0.0242 b,c | 0.15 ± 0.0072 c | 0.47 ± 0.0178 b | 0.19 ± 0.0176 c | 8.57 ± 0.2251 a |
| G6 | 1,8-cineole-M | C470826 | C10H18O | 0.76 ± 0.2324 d | 3.21 ± 0.2589 b | 1.02 ± 0.0743 d | 3.54 ± 0.1134 b | 1.69 ± 0.0605 c | 8.49 ± 0.2093 a |
| G7 | 2,3-diethyl-5-methylpyrazine | C18138040 | C9H14N2 | 0.23 ± 0.0039 a,b | 0.20 ± 0.0477 b | 0.14 ± 0.0264 c | 0.10 ± 0.0103 c | 0.28 ± 0.0186 a | 0.09 ± 0.0053 c |
| G8 | Methyl chavicol | C140670 | C10H12O | 0.08 ± 0.0036 d | 0.28 ± 0.0330 a | 0.14 ± 0.0193 b,c | 0.10 ± 0.010 c,d | 0.13 ± 0.0188 c | 0.18 ± 0.0162 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, K.; Wang, X.; Wan, J.; Zhou, Y.; Li, H.; Zhu, Q. Correlation and Difference between Core Micro-Organisms and Volatile Compounds of Suan Rou from Six Regions of China. Foods 2022, 11, 2708. https://doi.org/10.3390/foods11172708
Lu K, Wang X, Wan J, Zhou Y, Li H, Zhu Q. Correlation and Difference between Core Micro-Organisms and Volatile Compounds of Suan Rou from Six Regions of China. Foods. 2022; 11(17):2708. https://doi.org/10.3390/foods11172708
Chicago/Turabian StyleLu, Kuan, Xueya Wang, Jing Wan, Ying Zhou, Hongying Li, and Qiujin Zhu. 2022. "Correlation and Difference between Core Micro-Organisms and Volatile Compounds of Suan Rou from Six Regions of China" Foods 11, no. 17: 2708. https://doi.org/10.3390/foods11172708
APA StyleLu, K., Wang, X., Wan, J., Zhou, Y., Li, H., & Zhu, Q. (2022). Correlation and Difference between Core Micro-Organisms and Volatile Compounds of Suan Rou from Six Regions of China. Foods, 11(17), 2708. https://doi.org/10.3390/foods11172708







