Altering the Chain Length Specificity of a Lipase from Pleurotus citrinopileatus for the Application in Cheese Making
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Microorganisms
2.3. Construction of the Protein Models
2.4. Molecular Docking and Prediction of Relevant Positions
2.5. In Silico Analysis of Diffusion and Binding of Triglycerides
2.6. Site-Directed Mutagenesis
2.7. Protein Expression and Purification
2.8. Determination of Hydrolytic Activity
2.9. Biochemical Characterization
2.10. Application in Feta-Type Cheese Production and Sensory Evaluation
2.11. SPME-GC-MS Analysis of vFFA
3. Results and Discussion
3.1. In Silico Analysis of the Relevant Positions
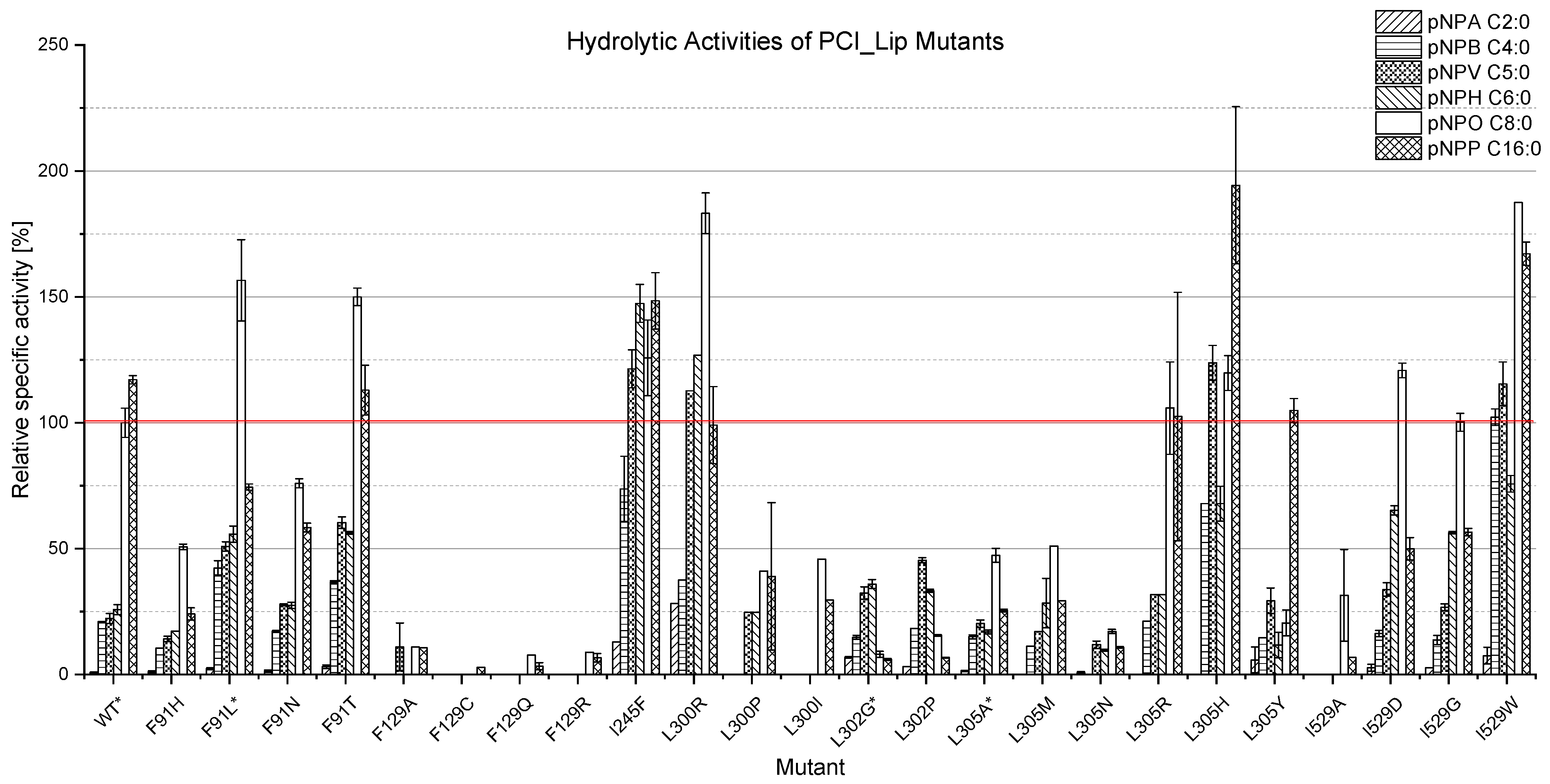
3.2. Expression of the Mutants and Characterisation of Their Hydrolysis Profiles
3.3. Biochemical Characterization of Mutants
3.4. Analysis of Mutants and Molecular Dynamic Simulations
3.5. Application of the Mutants in Cheese Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McSweeney, P.L.H. Biochemistry of cheese ripening. Int. J. Dairy Tech. 2004, 57, 127–144. [Google Scholar] [CrossRef]
- Qian, M.; Reineccius, G. Identification of Aroma Compounds in Parmigiano-Reggiano Cheese by Gas Chromatography/Olfactometry. J. Dairy Sci. 2002, 85, 1362–1369. [Google Scholar] [CrossRef]
- Månsson, H.L. Fatty acids in bovine milk fat. Food Nutr. Res. 2008, 52, 1821. [Google Scholar] [CrossRef]
- Barłowska, J.; Szwajkowska, M.; Litwińczuk, Z.; Król, J. Nutritional Value and Technological Suitability of Milk from Various Animal Species Used for Dairy Production. Compr. Rev. Food Sci. Food Saf. 2011, 10, 291–302. [Google Scholar] [CrossRef]
- Moschopoulou, E. Characteristics of rennet and other enzymes from small ruminants used in cheese production. Small Rumin. Res. 2011, 101, 188–195. [Google Scholar] [CrossRef]
- Richardson, G.H.; Nelson, J.H.; Farnham, M.G. Gastric Lipase Characterization and Utilization in Cheese Manufacture. J. Dairy Sci. 1971, 54, 643–647. [Google Scholar] [CrossRef]
- Nelson, J.H.; Jensen, R.G.; Pitas, R.E. Pregastric Esterase and other Oral Lipases—A Review. J. Dairy Sci. 1977, 60, 327–362. [Google Scholar] [CrossRef]
- Dekker, P. Dairy Enzymes. In Industrial Enzyme Applications; Vogel, A., May, O., Eds.; John Wiley & Sons, Ltd: Chichester, UK, 2019; pp. 143–166. ISBN 9783527813780. [Google Scholar]
- Arnold, R.G.; Shahani, K.M.; Dwivedi, B.K. Application of Lipolytic Enzymes to Flavor Development in Dairy Products. J. Dairy Sci. 1975, 58, 1127–1143. [Google Scholar] [CrossRef]
- Sowa, M.A.; Kreuter, N.; Sella, N.; Albuquerque, W.; Manhard, J.; Siegl, A.; Ghezellou, P.; Li, B.; Spengler, B.; Weichhard, E.; et al. Replacement of Pregastric Lipases in Cheese Production: Identification and Heterologous Expression of a Lipase from Pleurotus citrinopileatus. J. Agric. Food Chem. 2022, 70, 2998–3008. [Google Scholar] [CrossRef]
- Lutz, S. Beyond directed evolution-semi-rational protein engineering and design. Curr. Opin. Biotechnol. 2010, 21, 734–743. [Google Scholar] [CrossRef] [Green Version]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef]
- Bauer, T.L.; Buchholz, P.C.F.; Pleiss, J. The modular structure of α/β-hydrolases. FEBS J. 2020, 287, 1035–1053. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef]
- Santarossa, G.; Lafranconi, P.G.; Alquati, C.; DeGioia, L.; Alberghina, L.; Fantucci, P.; Lotti, M. Mutations in the “lid” region affect chain length specificity and thermostability of a Pseudomonas fragi lipase. FEBS Lett. 2005, 579, 2383–2386. [Google Scholar] [CrossRef]
- Schmitt, J.; Brocca, S.; Schmid, R.D.; Pleiss, J. Blocking the tunnel: Engineering of Candida rugosa lipase mutants with short chain length specificity. Protein Eng. 2002, 15, 595–601. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Phillips, J.C.; Zheng, G.; Kumar, S.; Kale, L.V. NAMD: Biomolecular Simulation on Thousands of Processors. In Proceedings of the IEEE/ACM SC2002 Supercomputing Conference, Baltimore, MD, USA, 16–22 November 2002; p. 36, ISBN 0-7695-1524-X. [Google Scholar]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Qi, X.; Yu, W.; Zhang, X.; Zhang, T.; Li, B. Highly efficient biosynthesis of phosphatidylserine by the surface adsorption-catalysis in purely aqueous media and mechanism study by biomolecular simulation. Mol. Catal. 2021, 502, 111397. [Google Scholar] [CrossRef]
- Green, R.; Rogers, E.J. Transformation of chemically competent E. coli. Methods Enzymol. 2013, 529, 329–336. [Google Scholar] [CrossRef]
- Nishihara, K.; Kanemori, M.; Kitagawa, M.; Yanagi, H.; Yura, T. Chaperone coexpression plasmids: Differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 1694–1699. [Google Scholar] [CrossRef]
- Nishihara, K.; Kanemori, M.; Yanagi, H.; Yura, T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 884–889. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Purdy, R.E.; Kolattukudy, P.E. Depolymerization of a hydroxy fatty acid biopolymer, cutin, by an extracellular enzyme from Fusarium solani f. pisi: Isolation and some properties of the enzyme. Arch. Biochem. Biophys. 1973, 159, 61–69. [Google Scholar] [CrossRef]
- Winkler, U.K.; Stuckmann, M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 1979, 138, 663–670. [Google Scholar] [CrossRef]
- van den Dool, H.; Kratz, P. A generalization of the retention index system including linear temperature programmed gas—liquid partition chromatography. J. Chromatogr. A 1963, 11, 463–471. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Jaeger, K.-E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef]
- Mancheño, J.M.; Pernas, M.A.; Martínez, M.J.; Ochoa, B.; Rúa, M.L.; Hermoso, J.A. Structural insights into the lipase/esterase behavior in the Candida rugosa lipases family: Crystal structure of the lipase 2 isoenzyme at 1.97A resolution. J. Mol. Biol. 2003, 332, 1059–1069. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.-C.; Shaw, J.-F. Protein engineering and applications of Candida rugosa lipase isoforms. Lipids 2004, 39, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Moharana, T.R.; Rao, N.M. Substrate structure and computation guided engineering of a Lipase for Omega-3 fatty acid selectivity. PLoS ONE 2020, 15, e0231177. [Google Scholar] [CrossRef] [PubMed]
- Nobili, A.; Gall, M.G.; Pavlidis, I.V.; Thompson, M.L.; Schmidt, M.; Bornscheuer, U.T. Use of ‘small but smart’ libraries to enhance the enantioselectivity of an esterase from Bacillus stearothermophilus towards tetrahydrofuran-3-yl acetate. FEBS J. 2013, 280, 3084–3093. [Google Scholar] [CrossRef] [PubMed]
- Schrauber, H.; Eisenhaber, F.; Argos, P. Rotamers: To be or not to be? An analysis of amino acid side-chain conformations in globular proteins. J. Mol. Biol. 1993, 230, 592–612. [Google Scholar] [CrossRef]
- Kumari, A.; Gupta, R. Phenylalanine to leucine point mutation in oxyanion hole improved catalytic efficiency of Lip12 from Yarrowia lipolytica. Enzyme Microb. Technol. 2013, 53, 386–390. [Google Scholar] [CrossRef]
- Domínguez de María, P.; Sánchez-Montero, J.M.; Sinisterra, J.V.; Alcántara, A.R. Understanding Candida rugosa lipases: An overview. Biotechnol. Adv. 2006, 24, 180–196. [Google Scholar] [CrossRef]

| Amino Acid | Mutant | Expression | Amino Acid | Mutant | Expression |
|---|---|---|---|---|---|
| F91 | G | - | L300 | I | x |
| H | x | P | x | ||
| L | x | R | x | ||
| N | x | Q | - | ||
| T | x | L302 | F | - | |
| F129 | A | x | G | x | |
| C | x | P | x | ||
| M | - | L305 | A | x | |
| Q | x | H | x | ||
| R | x | M | x | ||
| S163 | H | - | N | x | |
| M | - | R | x | ||
| P | - | Y | x | ||
| V | - | I529 | A | x | |
| Y | - | D | x | ||
| I245 | F | x | E | - | |
| W | - | G | x | ||
| W | x |
| Protein Concentration [µmol L−1] | Substrate | vmax [µmol min−1 L−1] | KM [mmol L−1] | kcat [s−1] | kcat/KM [s−1 mol−1 L] | |
|---|---|---|---|---|---|---|
| WT | 1.96 | pNPB | 50.12 | 0.647 | 0.43 | 658 |
| pNPH | 78.89 | 0.549 | 0.67 | 1222 | ||
| pNPO | 215.82 | 0.718 | 1.83 | 2555 | ||
| pNPP | 103.52 | 0.022 | 0.88 | 40,177 | ||
| F91L | 2.74 | pNPB | 115.89 | 0.786 | 0.71 | 898 |
| pNPH | 119.80 | 0.504 | 0.73 | 1448 | ||
| pNPO | 655.00 | 1.328 | 3.99 | 3005 | ||
| pNPP | 136.65 | 0.042 | 0.83 | 20,024 | ||
| L302G | 3.82 | pNPB | 77.53 | 0.756 | 0.34 | 447 |
| pNPH | 223.05 | 1.214 | 0.97 | 801 | ||
| pNPO | 42.82 | 0.706 | 0.19 | 264 | ||
| pNPP | 13.87 | 0.013 | 0.06 | 4733 | ||
| L305A | 4.60 | pNPB | 77.12 | 2.299 | 0.28 | 122 |
| pNPH | 45.51 | 0.645 | 0.16 | 256 | ||
| pNPO | 97.42 | 0.735 | 0.35 | 481 | ||
| pNPP | 33.27 | 0.022 | 0.12 | 5598 |
| No Lipase | Opti-Zym z10uc | WT | F91L | L302G | L305A | |
|---|---|---|---|---|---|---|
| Appearance | Typically cream-colored, crumbly | |||||
| Texture | Firm, dry | Firm, dry | Softer/creamier than reference | Softer/creamier than reference | Firm, dry | Firm, dry |
| Smell | Neutral, milky | Typical for goat lipase, intense, aromatic, goaty, strong, comparable to parmesan | Comparable to opti-zym | Comparable to opti-zym; slightly off | Comparable to opti-zym; slightly off | Comparable to opti-zym; most pleasant and intense smell within this trial |
| Taste | Neutral, slightly sour | Typical for goat lipase, intense, aromatic, piquant, goaty, persistent aftertaste | Comparable to opti-zym | At first comparable to opti-zym but less intense; slightly off, acid taste differing from WT | At first comparable to opti-zym but less intense; slightly off, acid taste differing from WT, slightly rancid/bitter | Comparable to opti-zym, more intense then wildtype, pleasant acid profile, best taste within this trial |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broel, N.; Sowa, M.A.; Manhard, J.; Siegl, A.; Weichhard, E.; Zorn, H.; Li, B.; Gand, M. Altering the Chain Length Specificity of a Lipase from Pleurotus citrinopileatus for the Application in Cheese Making. Foods 2022, 11, 2608. https://doi.org/10.3390/foods11172608
Broel N, Sowa MA, Manhard J, Siegl A, Weichhard E, Zorn H, Li B, Gand M. Altering the Chain Length Specificity of a Lipase from Pleurotus citrinopileatus for the Application in Cheese Making. Foods. 2022; 11(17):2608. https://doi.org/10.3390/foods11172608
Chicago/Turabian StyleBroel, Niklas, Miriam A. Sowa, Julia Manhard, Alexander Siegl, Edgar Weichhard, Holger Zorn, Binglin Li, and Martin Gand. 2022. "Altering the Chain Length Specificity of a Lipase from Pleurotus citrinopileatus for the Application in Cheese Making" Foods 11, no. 17: 2608. https://doi.org/10.3390/foods11172608
APA StyleBroel, N., Sowa, M. A., Manhard, J., Siegl, A., Weichhard, E., Zorn, H., Li, B., & Gand, M. (2022). Altering the Chain Length Specificity of a Lipase from Pleurotus citrinopileatus for the Application in Cheese Making. Foods, 11(17), 2608. https://doi.org/10.3390/foods11172608








