Sacha Inchi Oil Press-Cake Protein Hydrolysates Exhibit Anti-Hyperuricemic Activity via Attenuating Renal Damage and Regulating Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation and Characterization of SISH
2.2. Animals and Experimental Design
2.3. Biochemical Determination
2.4. Histology
2.5. 16S rRNA Gene Amplification and Sequencing
2.6. Serum Metabolomic Analysis
2.7. Renal Transcriptome Analysis
2.8. Correlation Analysis and Statistical Analysis
3. Results
3.1. Identification of SISH Composition
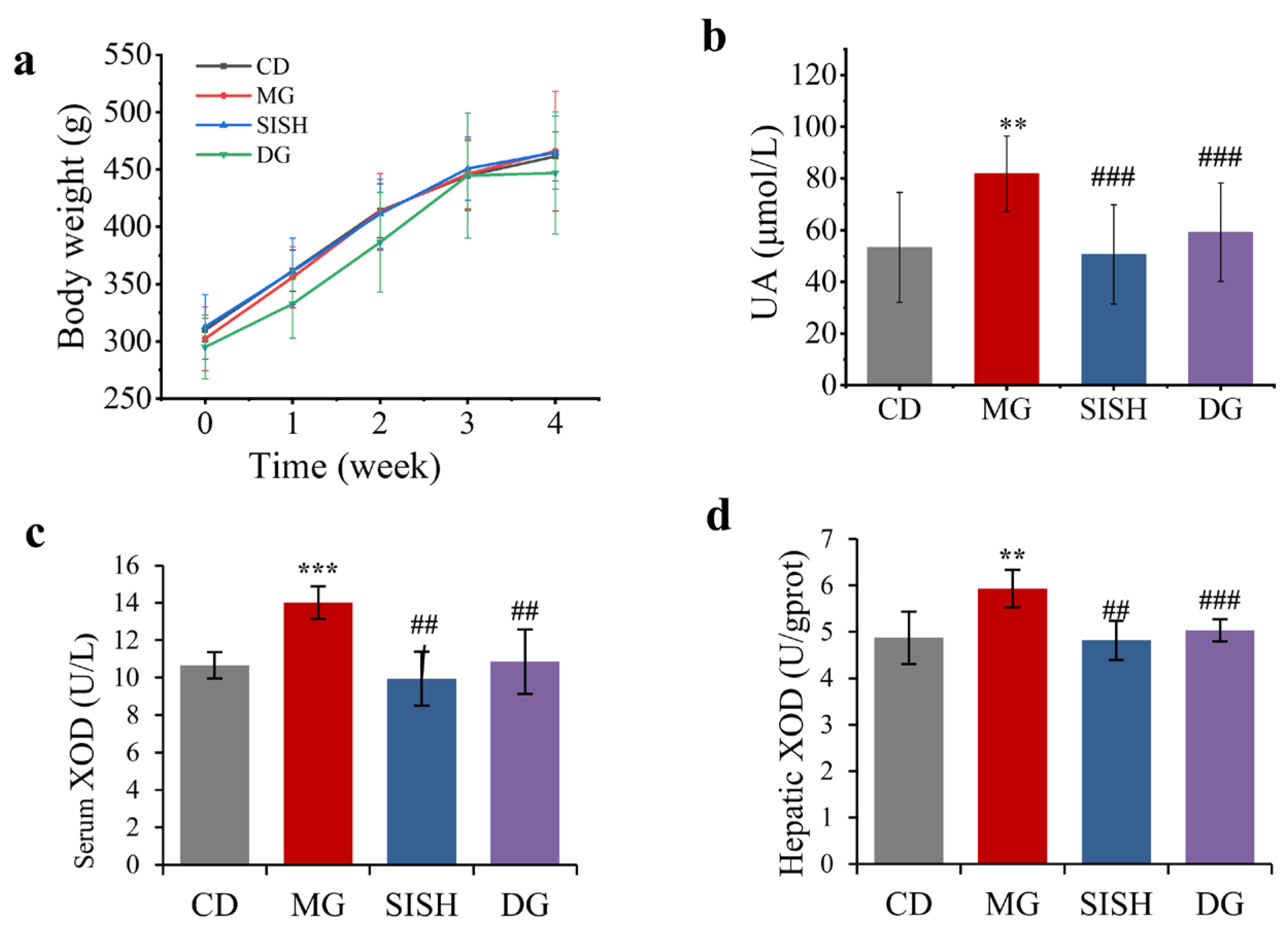
3.2. SISH Attenuated Hyperuricemia
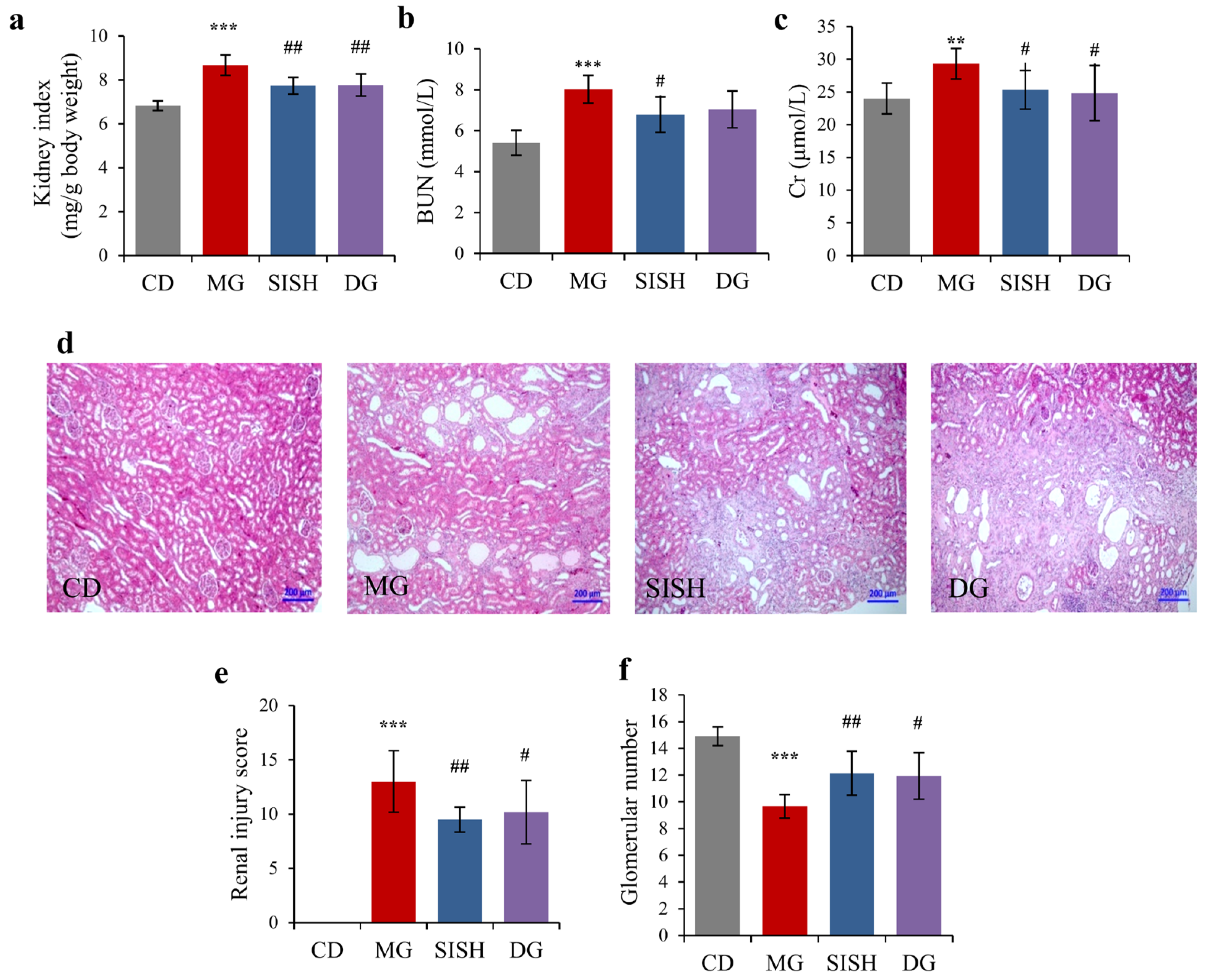
3.3. SISH Attenuated Renal Damage in Hyperuricemic Rats
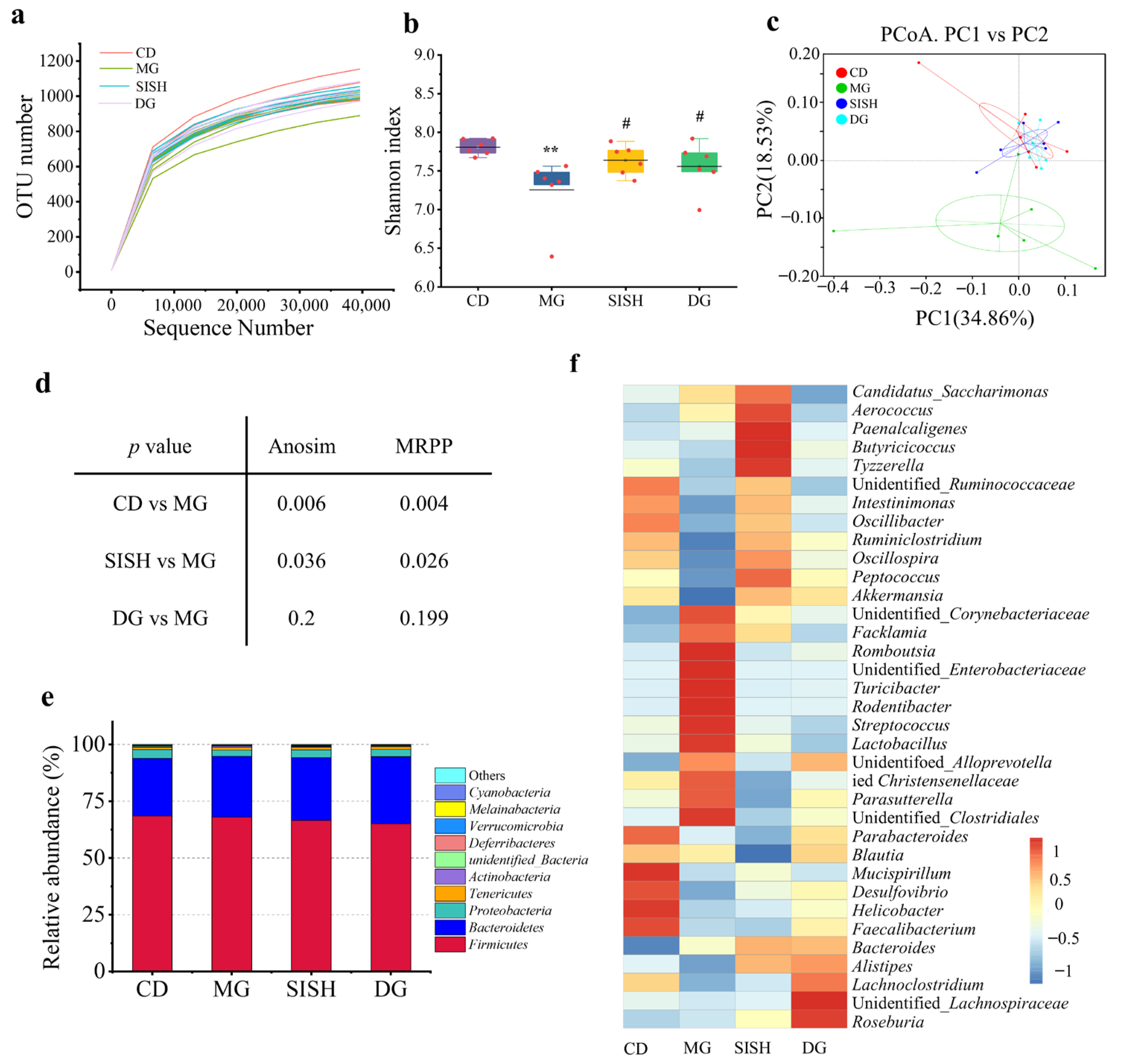
3.4. SISH Reshaped the Gut Microbiota Community in Hyperuricemic Rats
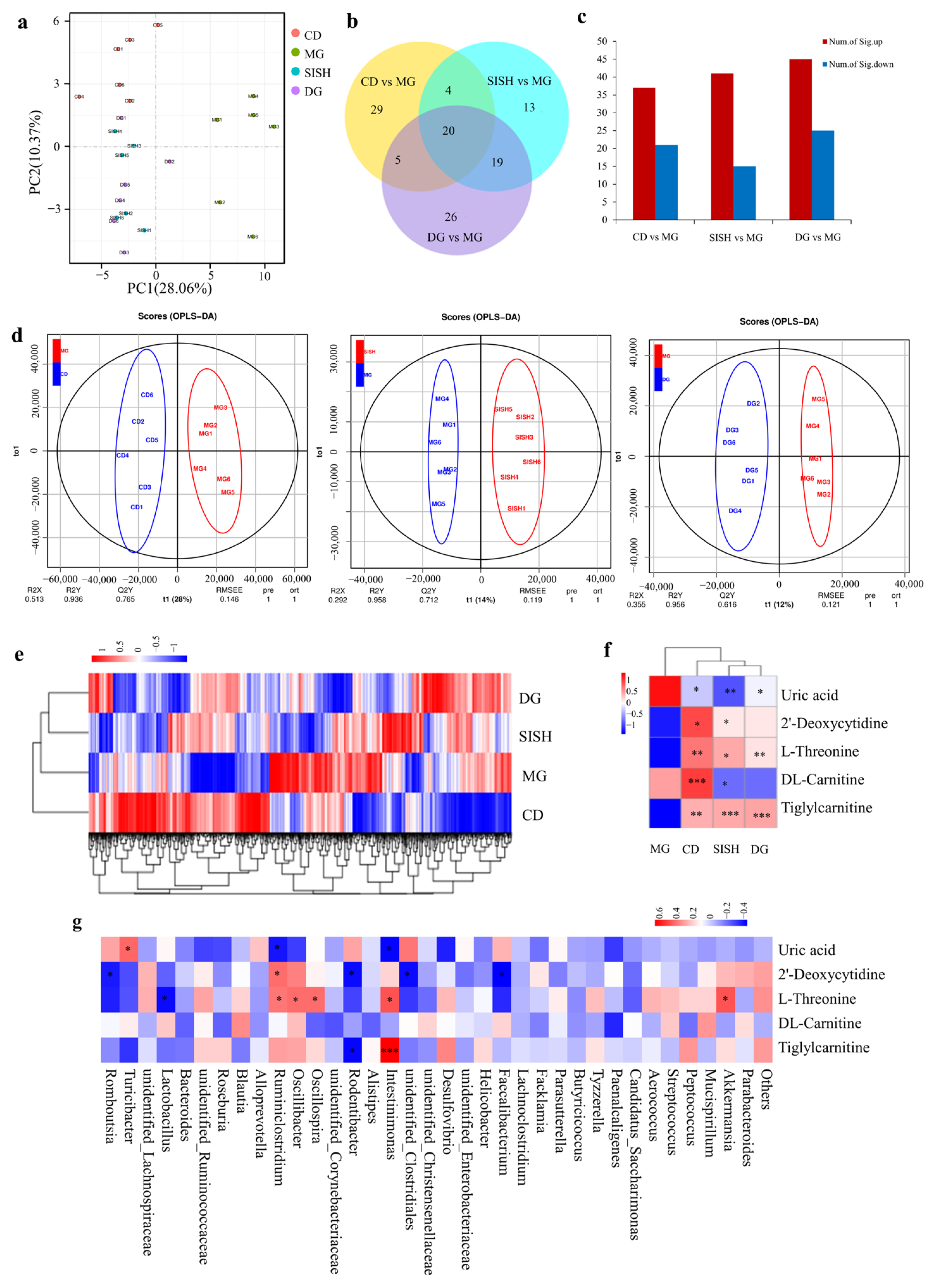
3.5. SISH Intervention Altered Serum Metabolome in Hyperuricemic Rats
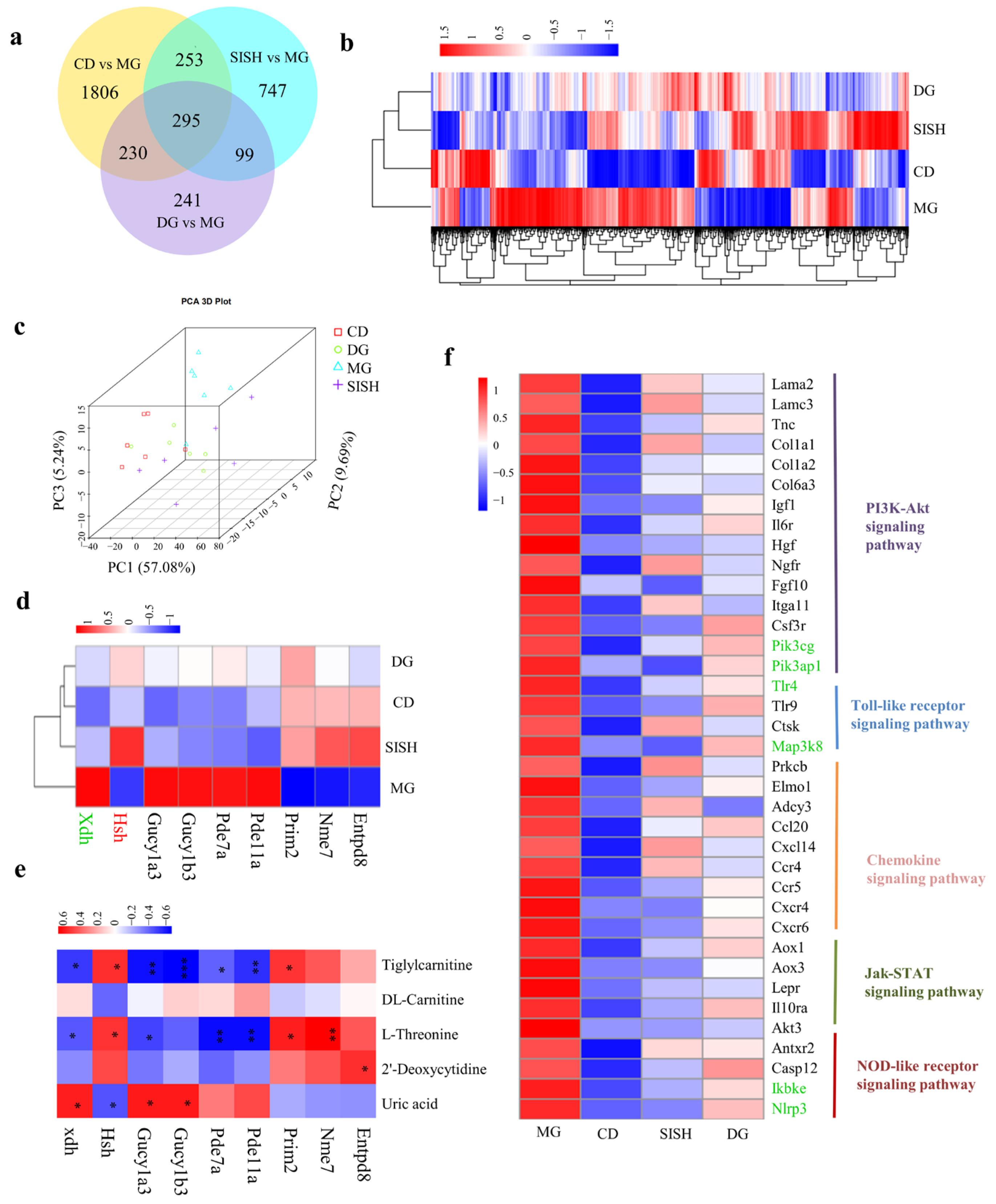
3.6. Identification of the Potential SISH Target Genes and Signaling Pathways Involved in Hyperuricemic Rats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, Z.; Zhang, J.C.; Wang, Z.L.; Ang, K.Y.; Huang, S.; Hou, Q.C.; Su, X.Q.; Qiao, J.M.; Zheng, Y.; Wang, L.F.; et al. Intestinal Microbiota Distinguish Gout Patients from Healthy Humans. Sci. Rep. 2016, 6, 20602. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Zhan, S.Y.; Li, S.Y.; Zhu, Z.Z.; He, J.R.; Lorenzo, J.M.; Barba, F.J. Anti-hyperuricemic and nephroprotective effects of extracts from Chaenomeles sinensis (Thouin) Koehne in hyperuricemic mice. Food Funct. 2018, 9, 5778–5790. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Gao, X.X.; Zhang, H.; Liu, Y.Y.; Sarker, M.M.R.; Wu, Y.J.; Chen, X.H.; Zhao, C. The anti-hyperuricemic effects of green alga Enteromorpha prolifera polysaccharide via regulation of the uric acid transporters in vivo. Food Chem. Toxicol. 2021, 158, 112630. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Zhao, L.; Ishaq, M.; Xin, W.; Zhao, L.; Wang, C.T.; Hossen, I.; Zhang, H.M.; Lian, Y.H.; Xu, M.L. Anti-hyperuricemic potential of stevia (Stevia rebaudiana Bertoni) residue extract in hyperuricemic mice. Food Funct. 2020, 11, 6387–6406. [Google Scholar] [CrossRef] [PubMed]
- Abeles, A.M. Hyperuricemia, gout, and cardiovascular disease: An update. Curr. Rheumatol. Rep. 2015, 17, 13. [Google Scholar] [CrossRef]
- Ma, Z.M.; Wang, Y.F.; Xu, C.N.; Ai, F.L.; Huang, L.; Wang, J.P.; Peng, J.; Zhou, Y.M.; Yin, M.H.; Zhang, S.; et al. Obesity-Related Genetic Variants and Hyperuricemia Risk in Chinese Men. Front. Endocrinol. 2019, 10, 230. [Google Scholar] [CrossRef]
- Grassi, D.; Desideri, G.; Di Giacomantonio, A.V.; Di Giosia, P.; Ferri, C. Hyperuricemia and cardiovascular risk. High Blood Press. Cardiovasc. Prev. 2014, 21, 235–242. [Google Scholar] [CrossRef]
- Su, H.Y.; Yang, C.; Liang, D.; Liu, H.F. Research Advances in the Mechanisms of Hyperuricemia-Induced Renal Injury. BioMed Res. Int. 2020, 2020, 5817348. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Y.; Zhong, H.; Chen, F.; Regenstein, J.; Hu, X.S.; Cai, L.Y.; Feng, F.Q. The gut microbiota as a target to control hyperuricemia pathogenesis: Potential mechanisms and therapeutic strategies. Crit. Rev. Food Sci. Nutr. 2021, 62, 3979–3989. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, J.; Zhang, Y.; Shao, T.J.; Li, H.C.; Wang, M.J.; Zhang, L.; Bian, H.; Wen, C.P.; Xie, Z.J.; et al. Effect of a Traditional Chinese Medicine Formula (CoTOL) on Serum Uric Acid and Intestinal Flora in Obese Hyperuricemic Mice Inoculated with Intestinal Bacteria. Evid. Based Complementary Altern. Med. 2020, 2020, 8831937. [Google Scholar] [CrossRef]
- Dalbeth, N.; Merriman, T.R.; Stamp, L.K. Gout. Lancet 2016, 388, 2039–2052. [Google Scholar] [CrossRef]
- Kohagura, K.; Tana, T.; Higa, A.; Yamazato, M.; Ishida, A.; Nagahama, K.; Sakima, A.; Iseki, K.; Ohya, Y. Effects of xanthine oxidase inhibitors on renal function and blood pressure in hypertensive patients with hyperuricemia. Hypertens. Res. 2016, 39, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Bailén, R.; González Senac, N.M.; López, M.M.; Luisa Llena, M.; Migoya, M.; Teresa Rodriguez, M.; de Miguel, E.; Torres, R.J.; Puig, J.G. Efficacy and Safety of a Urate Lowering Regimen in Primary Gout. Nucleos. Nucleot. Nucl. 2014, 33, 174–180. [Google Scholar] [CrossRef]
- Abd El-Maksoud, A.A.; Korany, R.M.S.; Abd El-Ghany, I.H.; El-Beltagi, S.H.; de Gouveia, G.M.A.F. Dietary solutions to dyslipidemia: Milk protein–polysaccharide conjugates as liver biochemical enhancers. J. Food Biochem. 2020, 44, e13142. [Google Scholar] [CrossRef]
- Qi, X.F.; Chen, H.R.; Guan, K.F.; Wang, R.C.; Ma, Y. Anti-hyperuricemic and nephroprotective effects of whey protein hydrolysate in potassium oxonate induced hyperuricemic rats. J. Sci. Food Agric. 2021, 101, 4916–4924. [Google Scholar] [CrossRef]
- Li, Y.J.; Kang, X.Y.; Li, Q.Y.; Shi, C.C.; Lian, Y.Y.; Yuan, E.D.; Zhou, M.; Ren, J.Y. Anti-hyperuricemic peptides derived from bonito hydrolysates based on in vivo hyperuricemic model and in vitro xanthine oxidase inhibitory activity. Peptides 2018, 107, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Q.; He, H.; Hou, T. Purification, identification, and computational analysis of xanthine oxidase inhibitory peptides from kidney bean. J. Food Sci. 2021, 86, 1081–1088. [Google Scholar] [CrossRef]
- He, W.W.; Su, G.W.; Sun-Waterhouse, D.; Waterhouse, G.I.N.; Zhao, M.M.; Liu, Y. In vivo anti-hyperuricemic and xanthine oxidase inhibitory properties of tuna protein hydrolysates and its isolated fractions. Food Chem. 2019, 272, 453–461. [Google Scholar] [CrossRef]
- Li, Q.; Kang, X.; Shi, C.; Li, Y.; Majumder, K.; Ning, Z.; Ren, J. Moderation of hyperuricemia in rats via consuming walnut protein hydrolysate diet and identification of new antihyperuricemic peptides. Food Funct. 2018, 9, 107. [Google Scholar] [CrossRef]
- Chirinos, R.; Zuloeta, G.; Pedreschi, R.; Mignolet, E.; Larondelle, Y.; Campos, D. Sacha inchi (Plukenetia volubilis): A seed source of polyunsaturated fatty acids, tocopherols, phytosterols, phenolic compounds and antioxidant capacity. Food Chem. 2013, 141, 1732–1739. [Google Scholar] [CrossRef]
- Torres Sánchez, E.G.; Hernández-Ledesma, B.; Gutiérrez, L.F. Sacha Inchi Oil Press-cake: Physicochemical Characteristics, Food-related Applications and Biological Activity. Food Rev. Int. 2021, 5, 1–12. [Google Scholar] [CrossRef]
- Chirinos, R.; Aquino, M.; Pedreschi, R.; Campos, D. Optimized Methodology for Alkaline and Enzyme-Assisted Extraction of Protein from Sacha Inchi (Plukenetia volubilis) Kernel Cake. J. Food Process. Eng. 2017, 40, e12412. [Google Scholar] [CrossRef]
- Lai, X.; Pan, S.; Zhang, W.; Sun, L.; Li, Q.; Chen, R.; Sun, S. Properties of ACE inhibitory peptide prepared from protein in green tea residue and evaluation of its anti-hypertensive activity. Process. Biochem. 2022, 92, 277–287. [Google Scholar] [CrossRef]
- Chirinos, R.; Pedreschi, R.; Campos, D. Enzyme-assisted hydrolysates from sacha inchi (Plukenetia volubilis) protein with in vitro antioxidant and antihypertensive properties. J. Food Process. Preserv. 2020, 44, e14969. [Google Scholar] [CrossRef]
- Rawdkuen, S.; Rodzi, N.; Pinijsuwan, S. Characterization of sacha inchi protein hydrolysates produced by crude papain and Calotropis proteases. Lwt-Food Sci. Technol. 2018, 98, 18–24. [Google Scholar] [CrossRef]
- Perez-Galvez, R.; Morales-Medina, R.; Espejo-Carpio, F.; Guadix, A.; Guadix, E.M. Modelling of the production of ACE inhibitory hydrolysates of horse mackerel using proteases mixtures. Food Funct. 2016, 7, 3890–3901. [Google Scholar] [CrossRef]
- Benjakul, S.; Oungbho, K.; Visessanguan, W.; Thiansilakul, Y.; Roytrakul, S. Characteristics of gelatin from the skins of bigeye snapper, Priacanthus tayenus and Priacanthus macracanthus. Food Chem. 2009, 116, 445–451. [Google Scholar] [CrossRef]
- Li, G.S.; Zhan, J.Q.; Hu, L.P.; Yuan, C.H.; Takaki, K.; Ying, X.G.; Hu, Y.Q. Identification of a new antioxidant peptide from porcine plasma by in vitro digestion and its cytoprotective effect on H2O2 induced HepG2 model. J. Funct. Foods 2021, 86, 104679. [Google Scholar] [CrossRef]
- Liao, W.W.; Chen, H.; Jin, W.G.; Yang, Z.N.; Cao, Y.; Miao, J.Y. Three Newly Isolated Calcium-Chelating Peptides from Tilapia Bone Collagen Hydrolysate Enhance Calcium Absorption Activity in Intestinal Caco-2 Cells. J. Agric. Food Chem. 2020, 68, 2091–2098. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Tan, M.L.; Li, K.K.; Leung, P.C.; Ko, C.H. Green tea polyphenols decreases uric acid level through xanthine oxidase and renal urate transporters in hyperuricemic mice. J. Ethnopharmacol. 2015, 175, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.M.; Zhao, J.H.; Zhang, M.; Chen, Z.F.; Ma, Q.Y.; Liu, H.C.; Nie, C.X.; Zhang, Z.Q.; An, W.; Li, J.X. Lycium ruthenicum Anthocyanins Attenuate High-Fat Diet-Induced Colonic Barrier Dysfunction and Inflammation in Mice by Modulating the Gut Microbiota. Mol. Nutr. Food Res. 2021, 65, 2000745. [Google Scholar] [CrossRef]
- Wang, Y.N.; Zhao, M.; Xin, Y.; Liu, J.J.; Wang, M.; Zhao, C.J. 1H NMR and MS based metabolomics study of the therapeutic effect of Cortex Fraxini on hyperuricemic rats. J. Ethnopharmacol. 2016, 185, 272–281. [Google Scholar] [CrossRef]
- Wang, C.; Pan, Y.; Zhang, Q.Y.; Wang, F.M.; Kong, L.D. Quercetin and allopurinol ameliorate kidney injury in STZ-treated rats with regulation of renal NLRP3 inflammasome activation and lipid accumulation. PLoS ONE 2012, 7, e38285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Fang, L.; Jiang, L.; Wen, P.; Cao, H.D.; He, W.C.; Dai, C.S.; Yang, J.W. Uric acid induces renal inflammation via activating tubular NF-kappaB signaling pathway. PLoS ONE 2012, 7, e39738. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.Y.; Shi, C.C.; Wang, M.; Zhou, M.; Liang, M.; Zhang, T.; Yuan, E.D.; Wang, Z.; Yao, M.J.; Ren, J.Y. Tryptophan residue enhances in vitro walnut protein-derived peptides exerting xanthine oxidase inhibition and antioxidant activities. J. Funct. Foods 2019, 53, 276–285. [Google Scholar] [CrossRef]
- Liu, N.X.; Wang, Y.; Zeng, L.; Yin, S.G.; Hu, Y.; Li, S.S.; Fu, Y.; Zhang, X.P.; Xie, C.; Shu, L.J.; et al. RDP3, A Novel Antigout Peptide Derived from Water Extract of Rice. J. Agric. Food Chem. 2020, 68, 7143–7151. [Google Scholar] [CrossRef]
- Wei, L.Y.; Ji, H.W.; Song, W.K.; Peng, S.; Zhan, S.H.; Qu, Y.S.; Chen, M.; Zhang, D.; Liu, S.C. Hypouricemic, hepatoprotective and nephroprotective roles of oligopeptides derived from Auxis thazard protein in hyperuricemic mice. Food Funct. 2021, 12, 11838–11848. [Google Scholar] [CrossRef]
- Han, J.J.; Wang, X.F.; Tang, S.S.; Lu, C.Y.; Wan, H.T.; Zhou, J.; Li, Y.; Ming, T.H.; Wang, Z.J.; Su, X.R. Protective effects of tuna meat oligopeptides (TMOP) supplementation on hyperuricemia and associated renal inflammation mediated by gut microbiota. FASEB J. 2020, 34, 5061–5076. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.H.; Zhang, C.; Xing, X.H. Xanthine dehydrogenase: An old enzyme with new knowledge and prospects. Bioengineered 2016, 7, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, J.B.; Ealick, S.E. Structural and kinetic insights into the mechanism of 5-hydroxyisourate hydrolase from Klebsiella pneumoniae. Acta. Crystallogr. D 2011, 67, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Shi, M.; Guo, F.; Ma, L.; Fu, P. Pharmacologic inhibiting STAT3 delays the progression of kidney fibrosis in hyperuricemia-induced chronic kidney disease. Life Sci. 2021, 285, 119946. [Google Scholar] [CrossRef] [PubMed]
- Shan, B.; Chen, T.; Huang, B.X.; Liu, Y.; Chen, J. Untargeted metabolomics reveal the therapeutic effects of Ermiao wan categorized formulas on rats with hyperuricemia. J. Ethnopharmacol. 2021, 281, 114545. [Google Scholar] [CrossRef]
- Ru, L.B.; Scott, P.; Sydlaske, A.; Rose, D.M.; Terkeltaub, R. Innate immunity conferred by Toll-like receptors 2 and 4 and myeloid differentiation factor 88 expression is pivotal to monosodium urate monohydrate crystal-induced inflammation. Arthritis Rheumatol. 2005, 52, 2936–2946. [Google Scholar] [CrossRef]
- Bi, F.F.; Chen, F.; Li, Y.N.; Wei, A.; Cao, W.S. Klotho preservation by Rhein promotes toll-like receptor 4 proteolysis and attenuates lipopolysaccharide-induced acute kidney injury. J. Mol. Med. 2018, 96, 915–927. [Google Scholar] [CrossRef]
- Mehmood, A.; Zhao, L.; Wang, C.T.; Hossen, I.; Raka, R.N.; Zhang, H.M. Stevia residue extract increases intestinal uric acid excretion via interactions with intestinal urate transporters in hyperuricemic mice. Food Funct. 2019, 10, 7900–7912. [Google Scholar] [CrossRef]
- Xie, X.S.; Zhang, L.; Yuan, S.; Li, H.L.; Zheng, C.J.; Xie, S.S.; Sun, Y.B.; Zhang, C.H.; Wang, R.K.; Jin, Y. Val-Val-Tyr-Pro protects against non-alcoholic steatohepatitis in mice by modulating the gut microbiota and gut-liver axis activation. J. Cell. Mol. Med. 2021, 25, 1439–1455. [Google Scholar] [CrossRef]
- Lu, C.Y.; Tang, S.S.; Han, J.J.; Fan, S.Q.; Huang, Y.M.; Zhang, Z.; Zhou, J.; Ming, T.H.; Li, Y.; Su, X.R. Apostichopus japonicus Oligopeptide Induced Heterogeneity in the Gastrointestinal Tract Microbiota and Alleviated Hyperuricemia in a Microbiota-Dependent Manner. Mol. Nutr. Food Res. 2021, 65, 2100147. [Google Scholar] [CrossRef]
- Yu, Y.R.; Liu, Q.P.; Li, H.C.; Wen, C.P.; He, Z.X. Alterations of the Gut Microbiome Associated With the Treatment of Hyperuricaemia in Male Rats. Front. Microbiol. 2018, 9, 2233. [Google Scholar] [CrossRef] [Green Version]
- Solano-Aguilar, G.I.; Jang, S.; Lakshman, S.; Gupta, R.; Beshah, E.; Sikaroodi, M.; Vinyard, B.; Molokin, A.; Gillevet, P.M.; Urban, J.F. The Effect of Dietary Mushroom Agaricus bisporus on Intestinal Microbiota Composition and Host Immunological Function. Nutrients 2018, 10, 1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| SISH | ||
|---|---|---|
| Molecular weight distribution (%) | ||
| <1 kDa | 90.7 | |
| 1~2 kDa | 6.0 | |
| 2~5 kDa | 1.9 | |
| >5 kDa | 1.4 | |
| Amino acid composition (%) | ||
| Glutamate | 16.7 | |
| Aspartic acid | 12.6 | |
| Arginine | 10.8 | |
| Glycine | 6.6 | |
| Serine | 5.9 | |
| Lysine | 4.9 | |
| Valine | 5.8 | |
| Threonine | 5.0 | |
| Proline | 4.1 | |
| Leucine | 7.1 | |
| Alanine | 3.9 | |
| Tyrosine | 4.4 | |
| Isoleucine | 4.7 | |
| Phenylalanine | 2.2 | |
| Histidine | 2.2 | |
| Cysteine | 2.4 | |
| Methionine | 1.0 | |
| Essential amino acid content | 30.7 | |
| Hydrophobic amino acid | 24.9 |
| Peptide Sequence | Mass-to-Charge Ratio | Retention Time | Relative Intensity (%) | Peptide Score |
|---|---|---|---|---|
| TGGWSPLK | 423.2326 | 19.9 | 1.19 | 98 |
| WKPW | 308.6679 | 27.18 | 1.55 | 98 |
| FLTMEPR | 447.2341 | 19.97 | 5.29 | 97 |
| VVLDVK | 672.4325 | 16.53 | 4.1 | 97 |
| KVVL | 458.3361 | 21.18 | 7.86 | 97 |
| MVVKK | 302.6903 | 21.15 | 8.66 | 97 |
| LTGLNKL | 379.7451 | 20.72 | 3.76 | 97 |
| RLLVWELER | 607.3524 | 18.13 | 8.13 | 97 |
| KLSLEWWLK | 601.8453 | 17.98 | 2.9 | 96 |
| FVKLL | 310.2144 | 25.61 | 5.04 | 96 |
| LGDLGTKL | 408.7478 | 22.05 | 1.99 | 96 |
| LTGLDKL | 380.2367 | 22.14 | 3.34 | 96 |
| LFAEMDK | 427.2123 | 18.29 | 4.69 | 96 |
| EADGTLR | 381.1941 | 14.02 | 1.11 | 96 |
| VVLFK | 303.2065 | 19.5 | 2.1 | 95 |
| T(+42.01)LLNPR | 378.2258 | 17.03 | 1.04 | 95 |
| AYLTGLK | 383.2322 | 18.84 | 1.01 | 95 |
| WLPDVK | 379.2173 | 21.21 | 7.68 | 95 |
| VLWLPR | 392.2499 | 30.21 | 1.25 | 95 |
| RWQVWEDR | 587.7963 | 21.99 | 2.107 | 95 |
| TVLLPR | 349.7339 | 18.2 | 8.63 | 95 |
| LVRFPK | 380.2433 | 20.96 | 1.6 | 95 |
| TLLFGDK | 397.2278 | 22.5 | 1.56 | 95 |
| WSELVK | 381.2155 | 18.68 | 1.66 | 95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Wu, S.; Li, P.; Xiao, N.; Wen, J.; Lin, J.; Lu, S.; Cai, X.; Xu, Y.; Du, B. Sacha Inchi Oil Press-Cake Protein Hydrolysates Exhibit Anti-Hyperuricemic Activity via Attenuating Renal Damage and Regulating Gut Microbiota. Foods 2022, 11, 2534. https://doi.org/10.3390/foods11162534
Wang K, Wu S, Li P, Xiao N, Wen J, Lin J, Lu S, Cai X, Xu Y, Du B. Sacha Inchi Oil Press-Cake Protein Hydrolysates Exhibit Anti-Hyperuricemic Activity via Attenuating Renal Damage and Regulating Gut Microbiota. Foods. 2022; 11(16):2534. https://doi.org/10.3390/foods11162534
Chicago/Turabian StyleWang, Kun, Shanshan Wu, Pan Li, Nan Xiao, Jiamin Wen, Jinming Lin, Siming Lu, Xin Cai, Yanan Xu, and Bing Du. 2022. "Sacha Inchi Oil Press-Cake Protein Hydrolysates Exhibit Anti-Hyperuricemic Activity via Attenuating Renal Damage and Regulating Gut Microbiota" Foods 11, no. 16: 2534. https://doi.org/10.3390/foods11162534
APA StyleWang, K., Wu, S., Li, P., Xiao, N., Wen, J., Lin, J., Lu, S., Cai, X., Xu, Y., & Du, B. (2022). Sacha Inchi Oil Press-Cake Protein Hydrolysates Exhibit Anti-Hyperuricemic Activity via Attenuating Renal Damage and Regulating Gut Microbiota. Foods, 11(16), 2534. https://doi.org/10.3390/foods11162534






