Metataxonomic Mapping of the Microbial Diversity of Irish and Eastern Mediterranean Cheeses
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Metagenomic DNA Extraction
2.3. Quantification of Total DNA
2.4. Barcoded Illumina MiSeq Amplicon Sequencing of Bacterial 16s rRNA Gene and of Fungal ITS Region
2.5. Microbiome and Statistical Analysis
3. Results
3.1. Abundance and Alpha Diversity of the Cheese Microbiota
3.2. Microbial Beta Biodiversity among the Different Regions
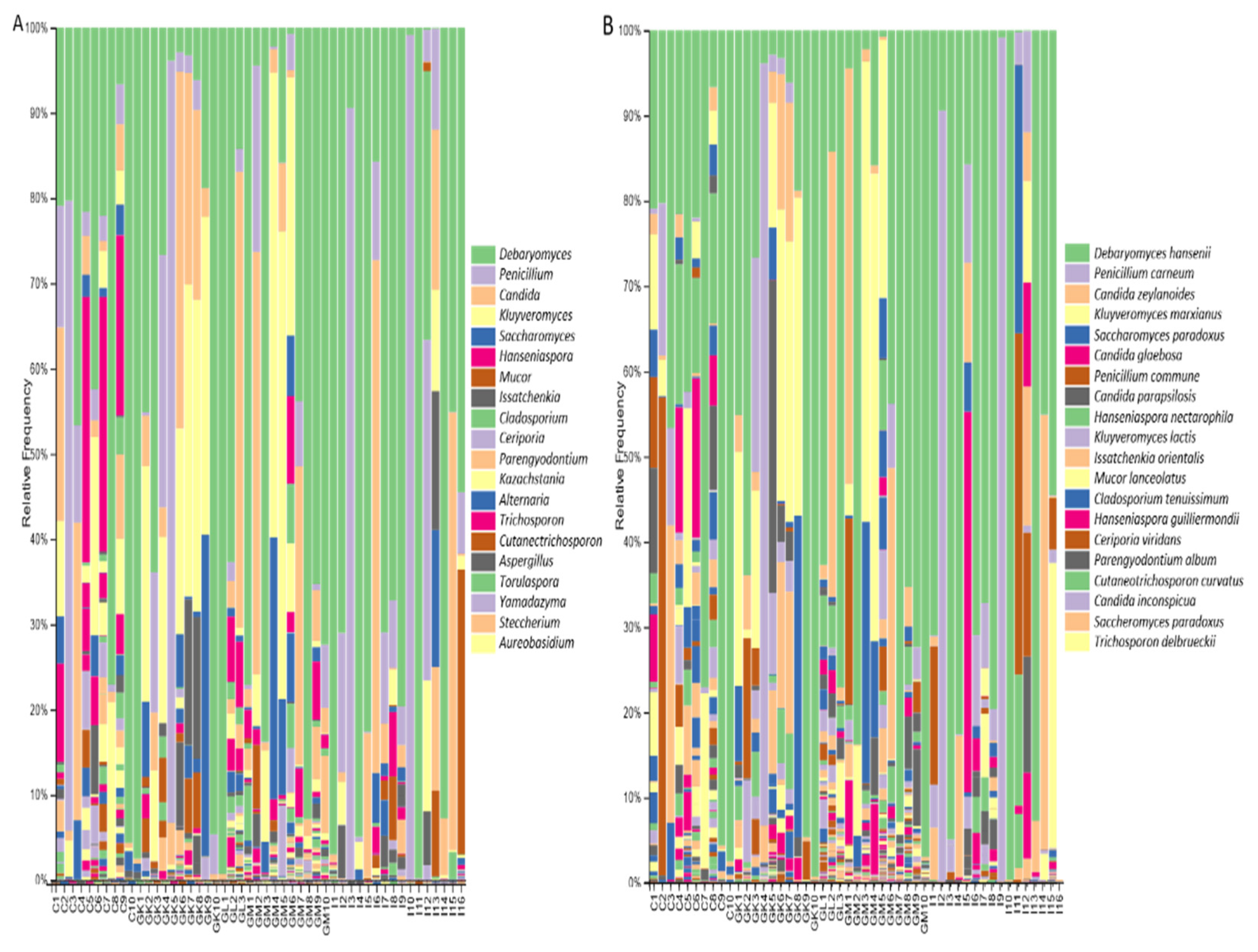
3.3. Taxonomic Composition of Microbial Communities
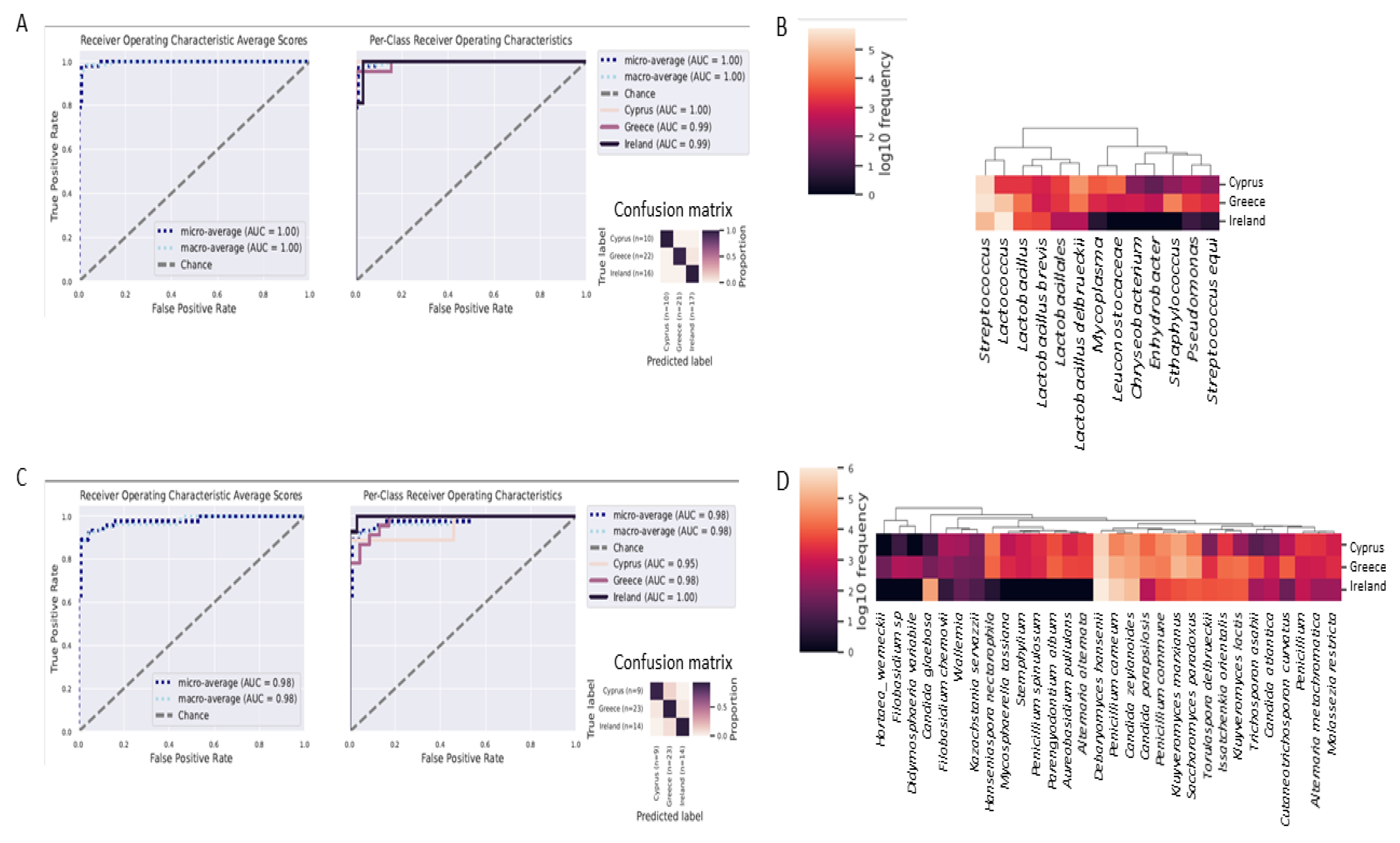
3.4. Identification of Microbial Biomarkers
3.5. Differentiation of Cheeses from Each Country with Great Accuracy, Using Random Forests Classifier
3.6. Microbial Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamilari, E.; Tomazou, M.; Antoniades, A.; Tsaltas, D. High Throughput Sequencing Technologies as a New Toolbox for Deep Analysis, Characterization and Potentially Authentication of Protection Designation of Origin Cheeses? Int. J. Food Sci. 2019, 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yeluri Jonnala, B.R.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the Cheese Microbiome and Its Relevance to Industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef] [PubMed]
- Parente, E.; Zotta, T.; Ricciardi, A. Microbial association networks in cheese: A meta-analysis. BioRxiv 2021, 107, 2411–2502. [Google Scholar] [CrossRef]
- Kamimura, B.A.; de Filippis, F.; Sant’Ana, A.S.; Ercolini, D. Large-scale mapping of microbial diversity in artisanal Brazilian cheeses. Food Microbiol. 2019, 80, 40–49. [Google Scholar] [CrossRef]
- Xue, J.; Yang, Y.; Wang, Z.; Guo, Y.; Shao, Y. Bacterial diversity in Chinese Rushan Cheese from different geographical origins. Front. Microbiol. 2018, 9, 1920–1944. [Google Scholar] [CrossRef]
- Camargo, A.C.; Costa, E.A.; Fusieger, A.; de Freitas, R.; Nero, L.A.; de Carvalho, A.F. Microbial shifts through the ripening of the ‘Entre Serras’ Minas artisanal cheese monitored by high-throughput sequencing. Food Res. Int. 2021, 139, 109803. [Google Scholar] [CrossRef]
- Aldrete-Tapia, A.; Escobar-Ramírez, C.M.; Tamplin, M.L.; Hernández-Iturriaga, M. Characterization of Bacterial Communities in Mexican Artisanal Raw Milk ‘Bola de Ocosingo’ Cheese by High-Throughput Sequencing. Front. Microbiol. 2018, 9, 2598. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef]
- Kelleher, P.; Murphy, J.; Mahony, J.; van Sinderen, D. Next-generation sequencing as an approach to dairy starter selection. Dairy Sci. Technol. 2015, 95, 545–568. [Google Scholar] [CrossRef]
- Lavoie, K.; Touchette, M.; St-Gelais, D.; Labrie, S. Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Sci. Technol. 2012, 92, 455–468. [Google Scholar] [CrossRef]
- Ledenbach, L.H.; Marshall, R.T. Microbiological Spoilage of Dairy Products. In Compendium of the Microbiological Spoilage of Foods and Beverages; Sperber, W.H., Doyle, M.P., Eds.; Springer: New York, NY, USA, 2009; pp. 41–67. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef] [PubMed]
- COUSIN, M.A. Presence and Activity of Psychrotrophic Microorganisms in Milk and Dairy Products: A Review. J. Food Prot. 1982, 45, 172. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.; Eyi, A.; Özdemir, F. Spoilage potentials and antimicrobial resistance of Pseudomonas sisolated from cheeses. J. Dairy Sci. 2011, 94, 5851–5856. [Google Scholar] [CrossRef] [PubMed]
- De Santana, E.H.W.; Luiz, L.L.; Pasquim, P.D.S.; Pinto, L.D.F.B.; Pereira, F.D.A.B.; Gasparini, G.B.F.B.; Lorenzetti, E.; Bruzaroski, S.R.; Eleodoro, J.I. Psychrotrophic microorganisms in raw milk and the cheese quality. Res. Soc. Dev. 2020, 9, 9. [Google Scholar] [CrossRef]
- Skovgaard, N. Fundamental food microbiology. Third edition. Int. J. Food Microbiol. 2004, 97, 1. [Google Scholar] [CrossRef]
- Trmčić, A.; Chauhan, K.; Kent, D.J.; Ralyea, R.D.; Martin, N.H.; Boor, K.J.; Wiedmann, M. Coliform detection in cheese is associated with specific cheese characteristics, but no association was found with pathogen detection. J. Dairy Sci. 2016, 99, 6105–6120. [Google Scholar] [CrossRef]
- Laleye, L.C.; Simard, R.E.; Lee, B.H.; Holley, R.A. Control of Heterofermentative Bacteria During Cheddar Cheese Maturation by Homofermentative Lactobacillus Starters. J. Dairy Sci. 1989, 72, 3134–3142. [Google Scholar] [CrossRef]
- Martín, J.F.; Liras, P. Secondary Metabolites in Cheese Fungi. In Fungal Metabolites; Merillon, J.M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 293–315. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Kamilaris, A.; Tsaltas, D. Characterizing Halloumi cheese’s bacterial communities through metagenomic analysis. LWT 2020, 126, 109298. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef]
- Kamilari, E.; Mina, M.; Karallis, C.; Tsaltas, D. Metataxonomic Analysis of Grape Microbiota During Wine Fermentation Reveals the Distinction of Cyprus Regional terroirs. Front. Microbiol. 2021, 12, 726483. [Google Scholar] [CrossRef]
- Papademas, P.; Kamilari, E.; Aspri, M.; Anagnostopoulos, D.A.; Mousikos, P.; Kamilaris, A.; Tsaltas, D. Investigation of the Cyprus donkey milk bacterial diversity by 16S rDNA high-throughput sequencing on a Cyprus donkey farm. J. Dairy Sci. 2020, 104, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 12. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 5. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Efthymiou, M.; Tretiak, S.; Tsaltas, D. Snapshot of Cyprus Raw Goat Milk Bacterial Diversity via 16S rDNA High-Throughput Sequencing; Impact of Cold Storage Conditions. Fermentation 2020, 6, 100. [Google Scholar] [CrossRef]
- ANDERSON, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2008, 26, 32–46. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi-recent updates and future perspectives. New Phytologist. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Segata, N.; Abubucker, S.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Waldron, L.; Zucker, J.; Thiagarajan, M.; et al. Microbial community function and biomarker discovery in the human microbiome. Genome Biol. 2011, 12, 47. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Dillon, M.R.; Bolyen, E.; Kaehler, B.D.; Huttley, G.A.; Caporaso, J.G. q2-sample-classifier: Machine-learning tools for microbiome classification and regression. J. Open Res. Softw. 2018, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Res 2016, 5, 1519. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016, 10, 1–11. [Google Scholar] [CrossRef]
- Ju, F.; Xia, Y.; Guo, F.; Wang, Z.; Zhang, T. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ. Microbiol. 2014, 16, 2421–2432. [Google Scholar] [CrossRef]
- Grass, J.E.; Gould, L.H.; Mahon, B.E. Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998–2010. Foodborne Pathog. Dis. 2013, 10, 131–136. [Google Scholar] [CrossRef]
- M Zago, M.; Bardelli, T.; Rossetti, L.; Nazzicari, N.; Carminati, D.; Galli, A.; Giraffa, G. Evaluation of bacterial communities of Grana Padano cheese by DNA metabarcoding and DNA fingerprinting analysis. Food Microbiol. 2021, 93, 103613. [Google Scholar] [CrossRef]
- Yang, C.; You, L.; Kwok, L.-Y.; Jin, H.; Peng, J.; Zhao, Z.; Sun, Z. Strain-level multiomics analysis reveals significant variation in cheeses from different regions. LWT 2021, 151, 112043. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Facility-specific ‘house’ microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 2013, 79, 5214–5223. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Tidona, F.; Francolino, S.; Ghiglietti, R.; Locci, F.; Carminati, D.; LaForce, P.; Giraffa, G. Characterization and pre-industrial validation of Streptococcus thermophilus strains to be used as starter cultures for Crescenza, an Italian soft cheese. Food Microbiol. 2020, 92, 103599. [Google Scholar] [CrossRef] [PubMed]
- Ruggirello, M.; Cocolin, L.; Dolci, P. Fate of Lactococcus lactis starter cultures during late ripening in cheese models. Food Microbiol. 2016, 59, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Dolci, P.; Ferrocino, I.; Giordano, M.; Pramotton, R.; Vernetti-Prot, L.; Zenato, S.; Barmaz, A. Impact of Lactococcus lactis as starter culture on microbiota and metabolome profile of an Italian raw milk cheese. Int. Dairy J. 2020, 110, 104804. [Google Scholar] [CrossRef]
- Papademas, P.; Aspri, M.; Mariou, M.; Dowd, S.E.; Kazou, M.; Tsakalidou, E. Conventional and omics approaches shed light on Halitzia cheese, a long-forgotten white-brined cheese from Cyprus. Int. Dairy J. 2019, 98, 72–83. [Google Scholar] [CrossRef]
- Bachmann, H.P.; Bütikofer, U.; Fröhlich-Wyder, M.T.; Isolini, D.; Jakob, E. Cheese: Swiss-Type Cheeses. In Encyclopedia of Dairy Sciences, 2nd ed.; Academic Press: London, UK, 2011; pp. 712–720. [Google Scholar] [CrossRef]
- Walsh, A.M.; Macori, G.; Kilcawley, K.N.; Cotter, P.D. Meta-analysis of cheese microbiomes highlights contributions to multiple aspects of quality. Nat. Food 2020, 1, 500–510. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef]
- Michailidou, S.; Pavlou, E.; Pasentsis, K.; Rhoades, J.; Likotrafiti, E.; Argiriou, A. Microbial profiles of Greek PDO cheeses assessed with amplicon metabarcoding. Food Microbiol. 2021, 99, 103836. [Google Scholar] [CrossRef]
- Spyrelli, E.D.; Stamatiou, A.; Tassou, C.C.; Nychas, G.J.E.; Doulgeraki, A.I. Microbiological and metagenomic analysis to assess the effect of container material on the microbiota of Feta cheese during ripening. Fermentation 2020, 6, 12. [Google Scholar] [CrossRef]
- Ceugniez, A.; Taminiau, B.; Coucheney, F.; Jacques, P.; Delcenserie, V.; Daube, G.; Drider, D. Use of a metagenetic approach to monitor the bacterial microbiota of ‘Tomme d’Orchies’ cheese during the ripening process. Int. J. Food Microbiol. 2017, 247, 65–69. [Google Scholar] [CrossRef]
- Desfossés-Foucault, É.; LaPointe, G.; Roy, D. Dynamics and rRNA transcriptional activity of lactococci and lactobacilli during Cheddar cheese ripening. Int. J. Food Microbiol. 2013, 166, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Ruggirello, M.; Dolci, P.; Cocolin, L. Detection and viability of Lactococcus lactis throughout cheese ripening. PLoS ONE 2014, 9, e114280. [Google Scholar] [CrossRef] [PubMed]
- Dal Bello, B.; Cocolin, L.; Zeppa, G.; Field, D.; Cotter, P.D.; Hill, C. Technological characterization of bacteriocin producing Lactococcus lactis strains employed to control Listeria monocytogenes in Cottage cheese. Int. J. Food Microbiol. 2012, 153, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical applications of nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef]
- Helinck, S.; le Bars, D.; Moreau, D.; Yvon, M. Ability of thermophilic lactic acid bacteria to produce aroma compounds from amino acids. Appl. Environ. Microbiol. 2004, 70, 3855–3861. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Danielsen, M.; Valina, O.; Garrigues, C.; Johansen, E.; Pedersen, M.B. Streptococcus thermophilus core genome: Comparative genome hybridization study of 47 strains. Appl. Environ. Microbiol. 2008, 74, 4703–4710. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Renault, P.; Sorokine, A.; Ehrlich, S.D.; Kulakauskas, S.; Lapidus, A.; Goltsman, E.; Mazur, M.M.; Pusch, G.D.; et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004, 22, 1554–1558. [Google Scholar] [CrossRef]
- Buchin, S.; Duboz, G.; Salmon, J.C. Lactobacillus delbrueckii subslactis as a starter culture significantly affects the dynamics of volatile compound profiles of hard cooked cheeses. Eur. Food Res. Technol. 2017, 243, 1943–1955. [Google Scholar] [CrossRef]
- Rizzello, C.G.; De Angelis, M.; Lactobacillus delbrueckii Group. Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 91–95. [Google Scholar] [CrossRef]
- Charlet, M.; Duboz, G.; Faurie, F.; Le Quéré, J.L.; Berthier, F. Multiple interactions between Streptococcus thermophilus, Lactobacillus helveticus and Lactobacillus delbrueckii strongly affect their growth kinetics during the making of hard cooked cheeses. Int. J. Food Microbiol. 2009, 131, 10–19. [Google Scholar] [CrossRef]
- Mladenovic, K.G.; Grujovic, M.Z.; Kis, M.; Furmeg, S.; Tkalec, V.J.; Stefanovic, O.D.; Kocic-Tanackov, S.D. Enterobacteriaceae in food safety with an emphasis on raw milk and meat. Appl. Microbiol. Biotechnol. 2021, 105, 8615–8627. [Google Scholar] [CrossRef]
- Al-Nabulsi, A.A.; Osaili, T.M.; AbuNaser, R.A.; Olaimat, A.N.; Ayyash, M.; Al-Holy, M.A.; Kadora, K.M.; Holley, R.A. Factors affecting the viability of Staphylococcus aureus and production of enterotoxin during processing and storage of white-brined cheese. J. Dairy Sci. 2020, 103, 6869–6881. [Google Scholar] [CrossRef] [PubMed]
- Uzal, F.A.; Freedman, J.C.; Shrestha, A.; Theoret, J.R.; Garcia, J.; Awad, M.M.; Adams, V.; Moore, R.J.; Rood, J.I.; Mc Clane, B.A. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014, 9, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.M.; Westall, S.; Jespersen, L. Microbial succession of Debaryomyces hansenii strains during the production of Danish surfaced-ripened cheeses. J. Dairy Sci. 2002, 85, 478–486. [Google Scholar] [CrossRef]
- Ogawa, Y.; Hirose, D.; Akiyama, A.; Ichinoe, M. Examination of the Taxonomic Position of Penicillium Strains Used in Blue Cheese Production Based on the Partial Sequence of β-Tubulin. J. Food Hyg. Soc. Jpn. 2014, 55, 157–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garnier, L.; Valence, F.; Mounier, J. Diversity and control of spoilage fungi in dairy products: An update. Microorganisms 2017, 5, 42. [Google Scholar] [CrossRef]
- Jurado, M.; Vicente, C.J. Penicillium commune affects textural properties and water distribution of hard and extra-hard cheeses. J. Dairy Res. 2020, 87, 117–122. [Google Scholar] [CrossRef]
- Tofalo, R.; Fasoli, G.; Schirone, M.; Perpetuini, G.; Pepe, A.; Corsetti, A.; Suzzi, G. The predominance, biodiversity and biotechnological properties of Kluyveromyces marxianus in the production of Pecorino di Farindola cheese. Int. J. Food Microbiol. 2014, 187, 41–49. [Google Scholar] [CrossRef]
- Belloch, C.; Querol, A.; Barrio, E. Yeasts and Molds: Kluyveromyces spp. In Encyclopedia of Dairy Sciences, 2nd ed.; Elsevier: Edinburgh, UK, 2011; Volume 3, pp. 1417–1428. [Google Scholar] [CrossRef]
- Geronikou, A.; Srimahaeak, T.; Rantsiou, K.; Triantafillidis, G.; Larsen, N.; Jespersen, L. Occurrence of Yeasts in White-Brined Cheeses: Methodologies for Identification, Spoilage Potential and Good Manufacturing Practices. Front. Microbiol. 2020, 11, 582778. [Google Scholar] [CrossRef]






| Sample-id | Country of Origin | Area of Production | Type of Cheese | Milk Source | Raw/Pasteurized Milk | Starter Culture | Ripening | Smoking |
|---|---|---|---|---|---|---|---|---|
| C1 | Cyprus | Nicosia | Kefalotyri | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| C2 | Cyprus | Nicosia | Kefalotyri | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| C3 | Cyprus | Famachusta | Kefalotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| C4 | Cyprus | Nicosia | Kefalotyri | sheep | Pasteurized | lactic acid bacteria | hard | no |
| C5 | Cyprus | Nicosia | Kefalotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| C6 | Cyprus | Nicosia | Kefalotyri | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| C7 | Cyprus | Nicosia | Kefalotyri | sheep | Pasteurized | lactic acid bacteria | semi | no |
| C8 | Cyprus | Nicosia | Kefalotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| C9 | Cyprus | Paphos | Kefalotyri | sheep | Pasteurized | lactic acid bacteria | hard | no |
| C10 | Cyprus | Paphos | Kefalotyri | sheep | Pasteurized | lactic acid bacteria | hard | no |
| GK1 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK2 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK3 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK4 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK5 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK6 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK7 | Greece | Crete | Graviera | sheep | Pasteurized | lactic acid bacteria | hard | no |
| GK8 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK9 | Greece | Crete | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GK10 | Greece | Crete | Graviera | sheep | Pasteurized | lactic acid bacteria | hard | no |
| GL1 | Greece | Limnos | Kaskavalli | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| GL2 | Greece | Limnos | Kalathaki | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| GL3 | Greece | Limnos | Melichloro | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GM1 | Greece | Mytillini | Kefalotiri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GM2 | Greece | Mytillini | Ladotyri | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| GM3 | Greece | Mytillini | Kaseri | sheep and goat | Pasteurized | lactic acid bacteria | semi | no |
| GM4 | Greece | Mytillini | Graviera | sheep | Pasteurized | lactic acid bacteria | hard | no |
| GM5 | Greece | Mytillini | Ladotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GM6 | Greece | Mytillini | Ladotyri | sheep | Pasteurized | lactic acid bacteria | hard | no |
| GM7 | Greece | Mytillini | Ladotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GM8 | Greece | Mytillini | Ladotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GM9 | Greece | Mytillini | Graviera | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| GM10 | Greece | Mytillini | Ladotyri | sheep and goat | Pasteurized | lactic acid bacteria | hard | no |
| I1 | Ireland | Clare | Cheddar | goat | Raw | no | semi | no |
| I2 | Ireland | Tipperary | Blue cheese | sheep | Pasteurized | lactic acid bacteria and Penicillum roquefortii | semi | no |
| I3 | Ireland | Cork | Artisan | cow | Pasteurized | Starter culture | semi | no |
| I4 | Ireland | Cavan | Artisan | goat, sheep and cow | Raw | Starter culture | hard | no |
| I5 | Ireland | Cork | Artisan | goat | Pasteurized | lactic acid bacteria | hard | yes |
| I6 | Ireland | Cork | Artisan | cow | Pasteurized | Starter culture | semi | yes |
| I7 | Ireland | Cork | Artisan | cow | Pasteurized | Starter culture | hard | no |
| I8 | Ireland | Cork | Fine Swiss | cow | Raw | Starter cultures | hard | no |
| I9 | Ireland | Tipperary | Blue cheese | cow | Pasteurized | starter cultures and Penicillum roquefortii | semi | no |
| I10 | Ireland | Cork | Artisan | cow | Pasteurized | Starter cultures | semi | no |
| I11 | Ireland | Waterford | Cheddar | cow | Pasteurized | Starter cultures | hard | no |
| I12 | Ireland | Waterford | Cheddar | cow | Pasteurized | Starter cultures | hard | no |
| I13 | Ireland | Waterford | Cheddar | cow | Raw | Starter cultures | hard | no |
| I14 | Ireland | Waterford | Cheddar | cow | Raw | Starter cultures | hard | no |
| I15 | Ireland | Offaly | Cheddar | cow | Pasteurized | Starter cultures | hard | no |
| I16 | Ireland | Tipperary | Artisan | sheep | Pasteurized | Starter cultures | hard | no |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamilari, E.; Tsaltas, D.; Stanton, C.; Ross, R.P. Metataxonomic Mapping of the Microbial Diversity of Irish and Eastern Mediterranean Cheeses. Foods 2022, 11, 2483. https://doi.org/10.3390/foods11162483
Kamilari E, Tsaltas D, Stanton C, Ross RP. Metataxonomic Mapping of the Microbial Diversity of Irish and Eastern Mediterranean Cheeses. Foods. 2022; 11(16):2483. https://doi.org/10.3390/foods11162483
Chicago/Turabian StyleKamilari, Eleni, Dimitrios Tsaltas, Catherine Stanton, and R. Paul Ross. 2022. "Metataxonomic Mapping of the Microbial Diversity of Irish and Eastern Mediterranean Cheeses" Foods 11, no. 16: 2483. https://doi.org/10.3390/foods11162483
APA StyleKamilari, E., Tsaltas, D., Stanton, C., & Ross, R. P. (2022). Metataxonomic Mapping of the Microbial Diversity of Irish and Eastern Mediterranean Cheeses. Foods, 11(16), 2483. https://doi.org/10.3390/foods11162483








