Improving the Specific Activity and Thermostability of Psychrophilic Xylosidase AX543 by Comparative Mutagenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Biochemical Reagents
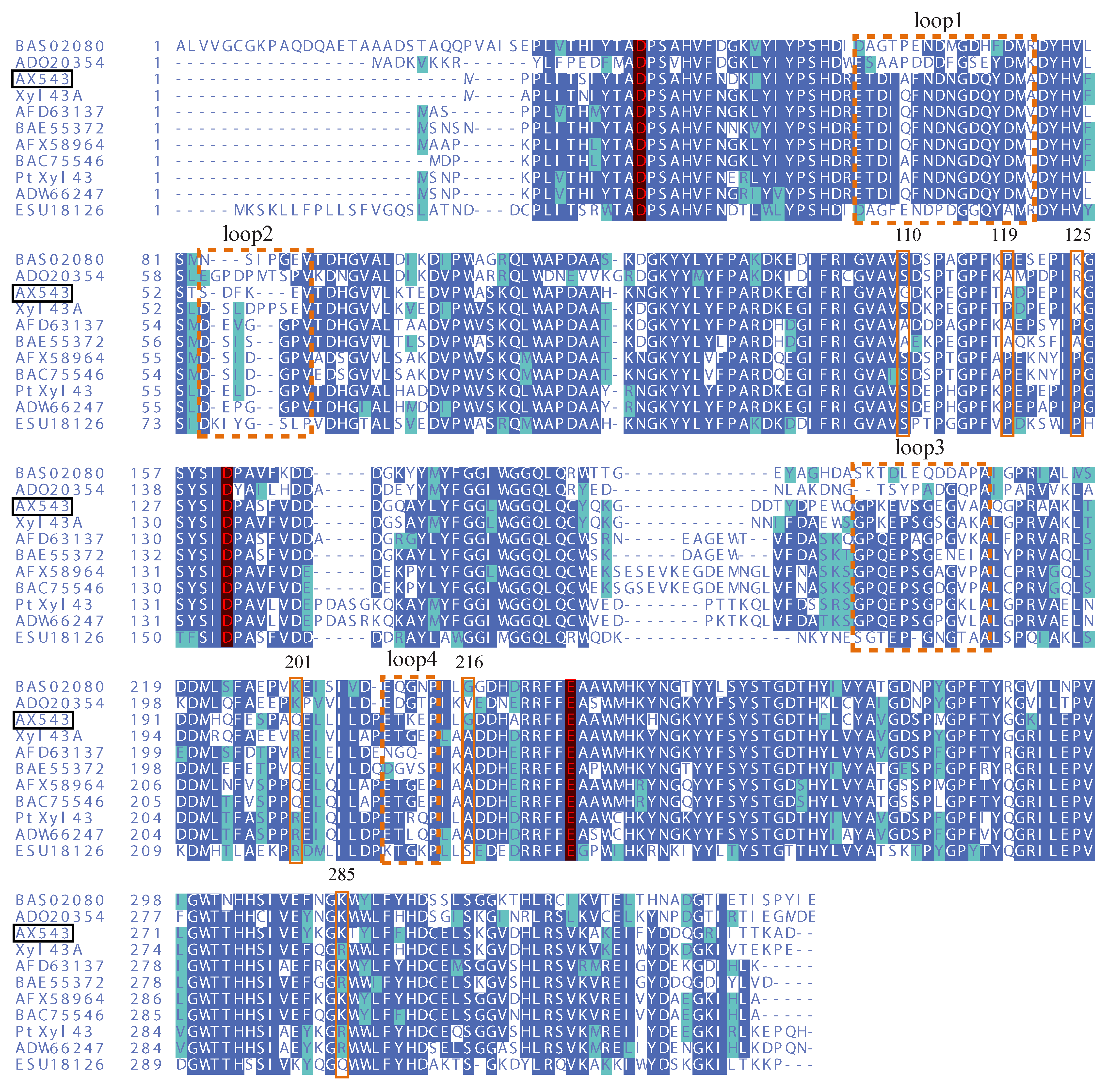
2.2. Protein Sequence and Structure Analysis
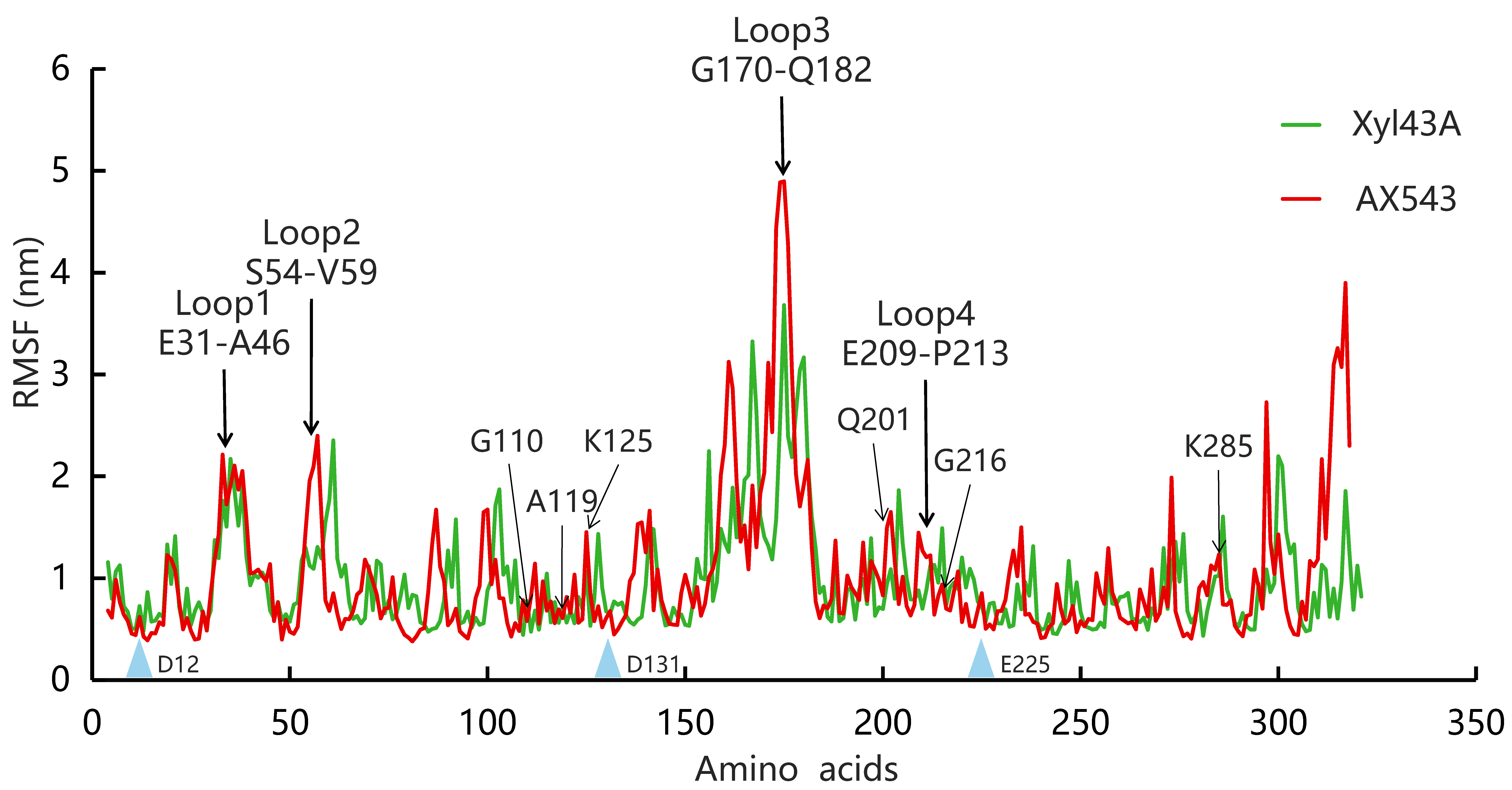
2.3. Molecular Dynamic Simulation
2.4. Site-Directed Mutagenesis
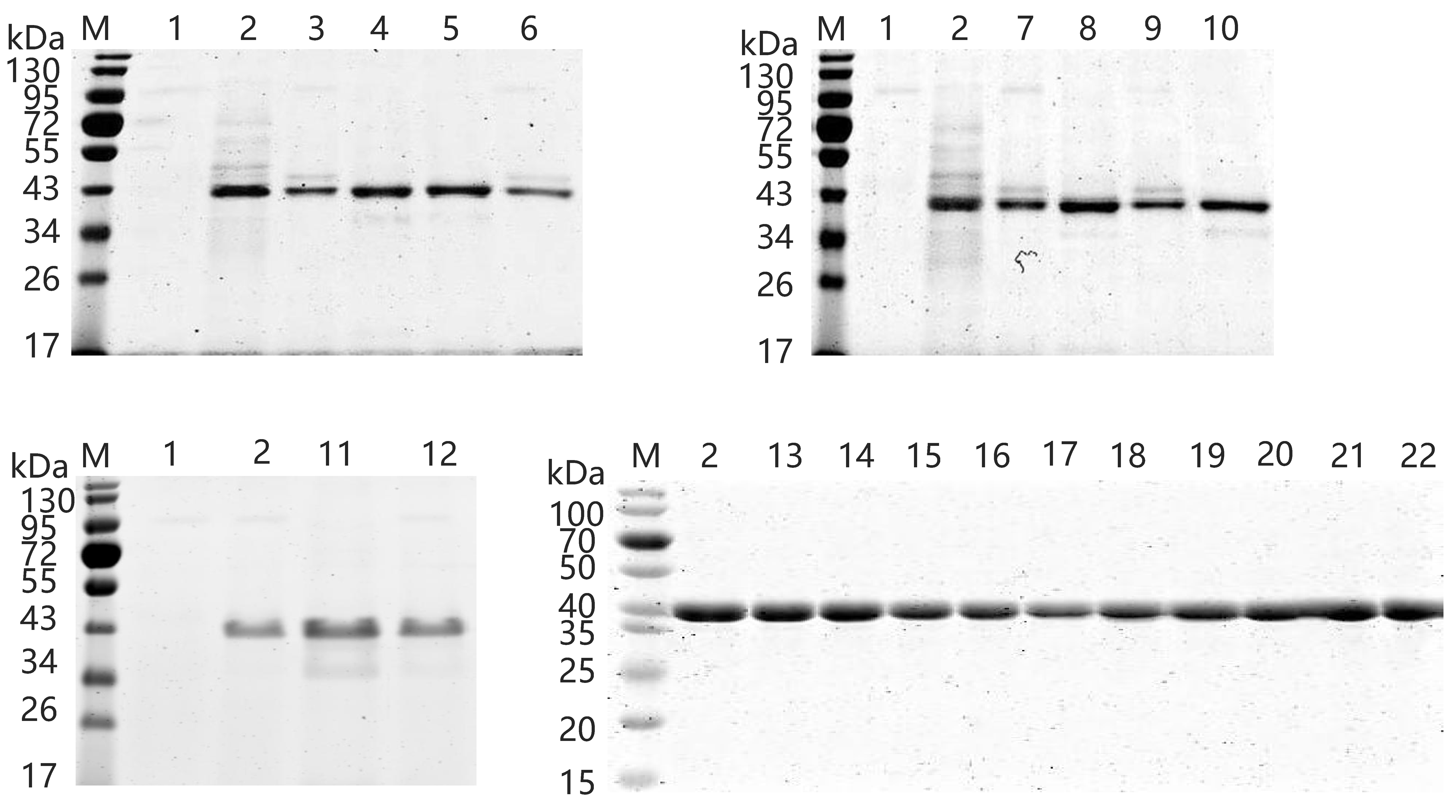
2.5. Expression and Purification of Wild-Type and Variants
2.6. Enzyme Activity Assays and Biochemical Characterization
3. Results and Discussions
3.1. Expression and Biochemical Characterization of Xylosidase AX543 in E. coli
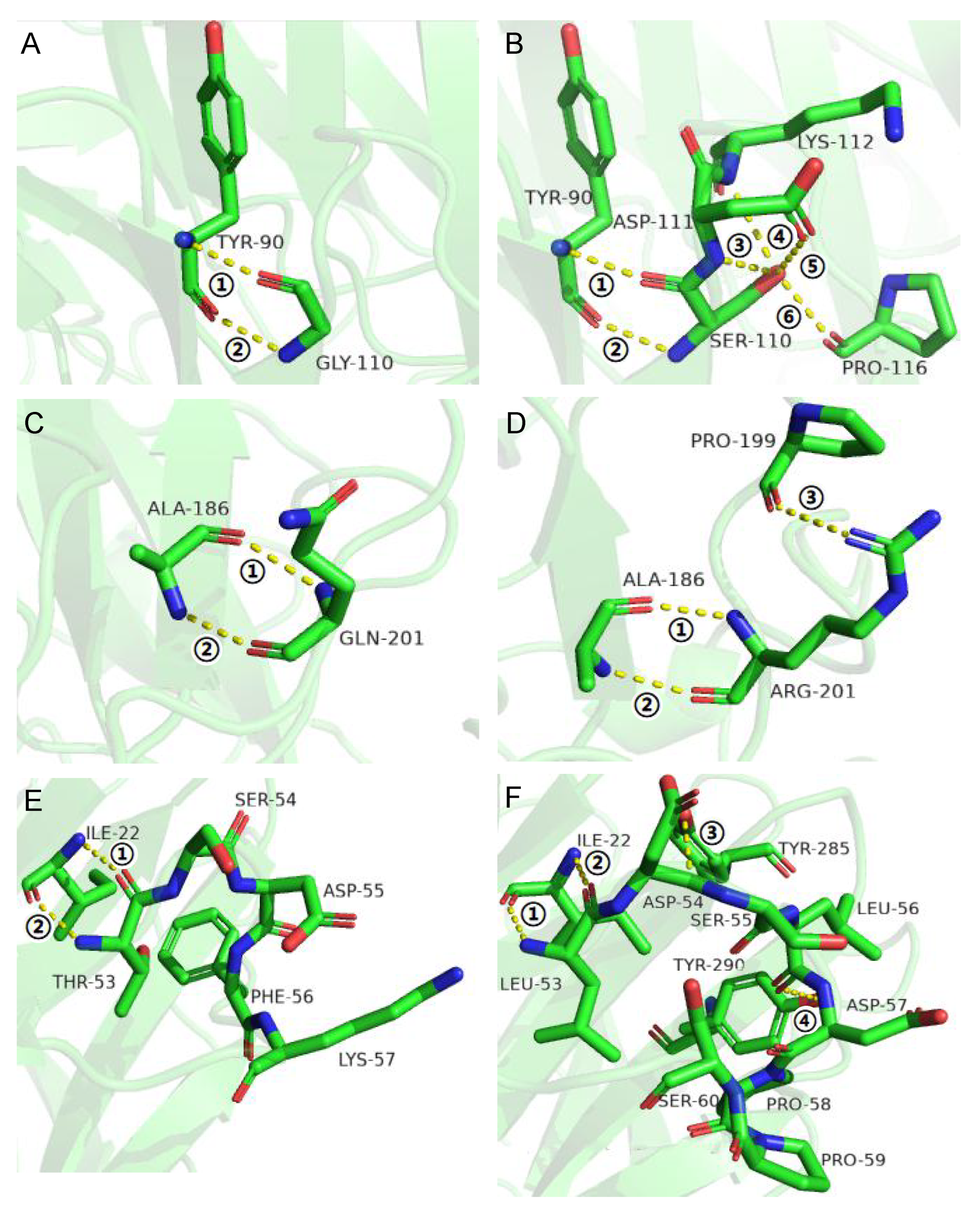
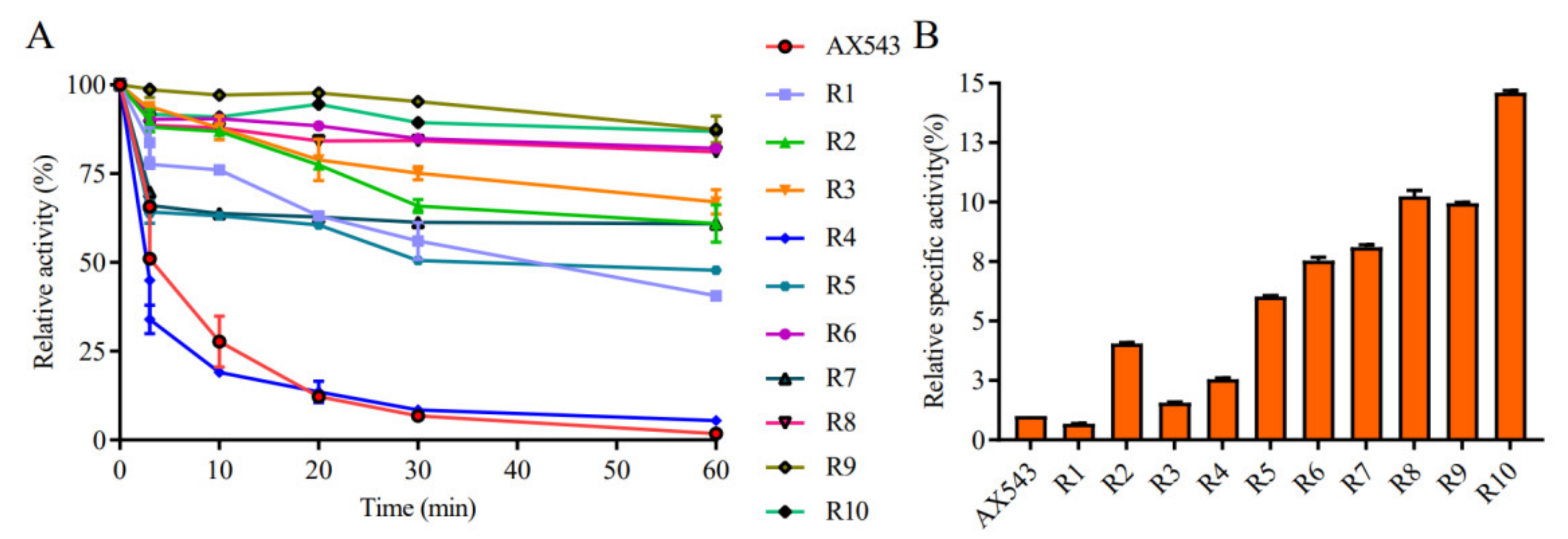
3.2. Improvement of Specific Activity and Thermostability by Site-Directed Mutagenesis
3.3. Effects of Flexible Loop Regions on Cold-Adaptation of AX543
3.4. Homologous Module Substitution between Psychrophilic and Thermophilic Xylosidases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Dilokpimol, A.; Kabel, M.A.; de Vries, R.P. Fungal xylanolytic enzymes: Diversity and applications. Bioresour. Technol. 2022, 344, 126290. [Google Scholar] [CrossRef] [PubMed]
- Rohman, A.; Dijkstra, B.W.; Puspaningsih, N.N.T. β-Xylosidases: Structural diversity, catalytic mechanism, and inhibition by Monosaccharides. Int. J. Mol. Sci. 2019, 20, 5524. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mukhia, S.; Kumar, R. Industrial applications of cold-adapted enzymes: Challenges, innovations and future perspective. 3 Biotech 2021, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- Yusof, N.A.; Hashim, N.H.F.; Bharudin, I. Cold Adaptation strategies and the potential of psychrophilic enzymes from the antarctic yeast, Glaciozyma antarctica PI12. J. Fungi 2021, 7, 528. [Google Scholar] [CrossRef]
- Carapito, R.; Carapito, C.; Jeltsch, J.M.; Phalip, V. Efficient hydrolysis of hemicellulose by a Fusarium graminearum xylanase blend produced at high levels in Escherichia coli. Bioresour. Technol. 2009, 100, 845–850. [Google Scholar] [CrossRef]
- Zhang, R.; Li, N.; Liu, Y.; Han, X.; Tu, T.; Shen, J.; Xu, S.; Wu, Q.; Zhou, J.; Huang, Z. Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 β-xylosidase: Activity and instability at high neutral salt concentrations. Food Chem. 2019, 301, 125266. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zheng, X.; Pilgaard, B.; Holck, J.; Muschiol, J.; Li, S.; Lange, L. Identification and characterization of GH11 xylanase and GH43 xylosidase from the chytridiomycetous fungus, Rhizophlyctis rosea. Appl. Microbiol. Biotechnol. 2019, 103, 777–791. [Google Scholar] [CrossRef]
- Suzuki, S.; Fukuoka, M.; Ookuchi, H.; Sano, M.; Ozeki, K.; Nagayoshi, E.; Takii, Y.; Matsushita, M.; Tada, S.; Kusumoto, K.I.; et al. Characterization of Aspergillus oryzae glycoside hydrolase family 43 beta-xylosidase expressed in Escherichia coli. J. Biosci. Bioeng. 2010, 109, 115–117. [Google Scholar] [CrossRef]
- Ito, T.; Yokoyama, E.; Sato, H.; Ujita, M.; Funaguma, T.; Furukawa, K.; Hara, A. Xylosidases associated with the cell surface of Penicillium herquei IFO 4674. J. Biosci. Bioeng. 2003, 96, 354–359. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, L.; Zheng, D.; Xu, Z.; Li, Y.; Shao, S.; Zhang, Y.; Ge, X.; Lu, F. Biochemical characterization of a novel GH43 family β-xylosidase from Bacillus pumilus. Food Chem. 2019, 295, 653–661. [Google Scholar] [CrossRef]
- Bao, L.; Huang, Q.; Chang, L.; Sun, Q.; Zhou, J.; Lu, H. Cloning and characterization of two β-glucosidase/xylosidase enzymes from yak rumen metagenome. Appl. Biochem. Biotechnol. 2012, 166, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, Z.; Qiu, H.; Wang, C.; Wang, H.; Zhang, T. Cloning and characterization of a novel cold-active bifunctional xylosidase/arabinfuranosidease AX543. Biotechnol. Bull. 2016, 32, 9. (In Chinese) [Google Scholar]
- Kumari, M.; Padhi, S.; Sharma, S.; Phukon, L.C.; Singh, S.P.; Rai, A.K. Biotechnological potential of psychrophilic microorganisms as the source of cold-active enzymes in food processing applications. 3 Biotech 2021, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Shukla, P. Current trends in protein engineering: Updates and progress. Curr. Protein Pept. Sci. 2019, 20, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Satyanarayana, T. Microbial glucoamylases: Characteristics and applications. Crit. Rev. Biotechnol. 2009, 29, 225–255. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and applications of extremozymes from deep-Sea extremophilic microorganisms: A mini review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef] [PubMed]
- Osire, T.; Yang, T.; Xu, M.; Zhang, X.; Li, X.; Niyomukiza, S.; Rao, Z. Lys-Arg mutation improved the thermostability of Bacillus cereus neutral protease through increased residue interactions. World J. Microbiol. Biotechnol. 2019, 35, 173. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Yamanaka, K.; Kazuoka, T.; Kanzawa, N.; Soda, K. Psychrophilic valine dehydrogenase of the antarctic psychrophile, Cytophaga sp. KUC-1: Purification, molecular characterization and expression. Eur. J. Biochem. 2001, 268, 4375–4383. [Google Scholar] [CrossRef] [PubMed]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level. Structure 1998, 6, 1503–1516. [Google Scholar] [CrossRef]
- Jaafar, N.R.; Mahadi, N.M.; Mackeen, M.M.; Illias, R.M.; Murad, A.M.A.; Abu Bakar, F.D. Structural and functional characterisation of a cold-active yet heat-tolerant dehydroquinase from Glaciozyma antarctica PI12. J. Biotechnol. 2021, 329, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shi, P.; Huang, H.; Luo, H.; Wang, Y.; Zhang, W.; Yao, B. Two xylose-tolerant GH43 bifunctional β-xylosidase/a-arabinosidases and one GH11 xylanase from Humicola insolens and their synergy in the degradation of xylan. Food Chem. 2014, 148, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.; Jia, H.; Yan, Q.; Zhou, P.; Jiang, Z. High-level expression of extracellular secretion of a β-xylosidase gene from Paecilomyces thermophila in Escherichia coli. Bioresour. Technol. 2011, 102, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Zheng, J.; Wang, X.; Wang, X.; Huang, H.; Yang, H.; Tu, T.; Wang, Y.; Bai, Y.; Yao, B.; et al. Improvement of thermostability and catalytic efficiency of glucoamylase from Talaromyces leycettanus JCM12802 via site-directed mutagenesis to enhance industrial saccharification applications. Biotechnol. Biofuels 2021, 14, 202. [Google Scholar] [CrossRef]
- Feller, G. Psychrophilic enzymes: From folding to function and biotechnology. Scientifica 2013, 2013, 512840. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.L.; Hou, Y.J.; Fan, H.X.; Qu, J.; Qi, C.; Liu, Y.; Li, D.F.; Liu, Z.P. Molecular structural basis for the cold adaptedness of the psychrophilic β-glucosidase BglU in Micrococcus antarcticus. Appl. Environ. Microbiol. 2016, 82, 2021–2030. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Liu, W.; Chen, C.C.; Ko, T.P.; He, M.; Xu, Z.; Liu, M.; Luo, H.; Guo, R.T.; et al. Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-β-1,4-xylanase. J. Struct. Biol. 2016, 193, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Paës, G.; Cortés, J.; Siméon, T.; O’Donohue, M.J.; Tran, V. Thumb-loops up for catalysis: A structure/function investigation of a functional loop movement in a GH11 xylanase. Comput. Struct. Biotechnol. J. 2012, 1, e201207001. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shi, P.; Huang, H.; Li, Z.; Yuan, T.; Yang, P.; Luo, H.; Bai, Y.; Yao, B. A novel thermoacidophilic and thermostable endo-β-1,4-glucanase from Phialophora sp. G5: Its thermostability influenced by a distinct β-sheet and the carbohydrate-binding module. Appl. Microbiol. Biotechnol 2012, 95, 947–955. [Google Scholar] [CrossRef]
- Vieira, D.S.; Ward, R.J. Conformation analysis of a surface loop that controls active site access in the GH11 xylanase a from Bacillus subtilis. J. Mol. Model. 2012, 18, 1473–1479. [Google Scholar] [CrossRef]
- Yang, H.; Shi, P.; Liu, Y.; Xia, W.; Wang, X.; Cao, H.; Ma, R.; Luo, H.; Bai, Y.; Yao, B. Loop 3 of Fungal endoglucanases of glycoside hydrolase family 12 modulates catalytic efficiency. Appl. Environ. Microbiol. 2017, 83, e03123-16. [Google Scholar] [CrossRef]
- Tu, T.; Meng, K.; Luo, H.; Turunen, O.; Zhang, L.; Cheng, Y.; Su, X.; Ma, R.; Shi, P.; Wang, Y.; et al. New insights into the role of T3 loop in determining catalytic efficiency of GH28 endo-polygalacturonases. PLoS ONE 2015, 10, e0135413. [Google Scholar] [CrossRef] [PubMed]
- Purmonen, M.; Valjakka, J.; Takkinen, K.; Laitinen, T.; Rouvinen, J. Molecular dynamics studies on the thermostability of family 11 xylanases. Protein Eng. Des. Sel. 2007, 20, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Bommarius, A.S.; Paye, M.F. Stabilizing biocatalysts. Chem. Soc. Rev. 2013, 42, 6534–6565. [Google Scholar] [CrossRef] [PubMed]
- Georgoulis, A.; Louka, M.; Mylonas, S.; Stavros, P.; Nounesis, G.; Vorgias, C.E. Consensus protein engineering on the thermostable histone-like bacterial protein HUs significantly improves stability and DNA binding affinity. Extremophiles 2020, 24, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Wijma, H.J.; Floor, R.J.; Janssen, D.B. Structure- and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr. Opin. Struct. Biol. 2013, 23, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Jun, H.; Bing, Y.; Keying, Z.; Xuemei, D.; Daiwen, C. Thermostable carbohydrate binding module increases the thermostability and substrate-binding capacity of Trichoderma reesei xylanase 2. N. Biotechnol. 2009, 26, 53–59. [Google Scholar] [CrossRef]
- Wang, Q.; Xia, T. Importance of C-terminal region for thermostability of GH11 xylanase from Streptomyces lividans. Appl. Biochem. Biotechnol. 2008, 144, 273–282. [Google Scholar] [CrossRef]
- Ackerman, S.H.; Gatti, D.L. The contribution of coevolving residues to the stability of KDO8P synthase. PLoS ONE 2011, 6, e17459. [Google Scholar] [CrossRef]





| Microorganism or Environment | Protein Name | GenBank Accession No. | Family | Optimal pH | Optimal Temperature | Reference |
|---|---|---|---|---|---|---|
| Acremonium sp. WCQ6A | AX543 | ON730957 | GH43 | 6 | 20 | This study |
| Fusarium graminearum | Xylo | ESU18096 | GH43 | 6 | 20 | [5] |
| Bacillus | rHJ14GH43 | KY391885 | GH43 | 7 | 25 | [6] |
| Rhizophlyctis rosea | RrXyl43A | AYV64572 | GH43 | 7 | 25 | [7] |
| Aspergillus oryzae | XylB | BAE55732 | GH43 | 7 | 30 | [8] |
| Penicillium herquei | S2 | AB093564 | GH43 | 6.5 | 30 | [9] |
| Fusarium graminearum | XyloA | ESU18126 | GH43 | 6 | 30 | [5] |
| Bacillus pumilus | XYL | AAC97375 | GH43 | 7 | 30 | [10] |
| Rumen metagenome | RuBG3A | ADM89626 | GH3 | 7.5 | 35 | [11] |
| Enzyme | AX543 | Xyl43A | PtXyl43 |
|---|---|---|---|
| Microbial source | Acremonium sp. WCQ6A | Humicola insolens | Paecilomyces thermophila |
| Optimal pH | 6 | 6.5 | 7 |
| Optimal temperature (°C) | 25 | 50 | 55 |
| Relative activity at 10 °C | 20% | ND | ND |
| Relative activity at 0 °C | 5% | ND | ND |
| Accession No. | ON730957 | KC962400 | GU937001 |
| Amino acid identity (%) with AX543 | 100 | 80 | 67 |
| Glycine (%) | 8.95 | 8.87 | 7.99 |
| Proline (%) | 6.48 | 7.03 | 8.58 |
| Arginine (%) | 2.47 | 3.36 | 4.73 |
| Arginine/Lysine ratio | 0.36 | 0.55 | 1.07 |
| Numbers of hydrogen bonds | 221 | 229 | 215 |
| Numbers of hydrogen bonds with arginine | 8 | 11 | 12 |
| Enzyme | Km (mM) | Vmax (μmol/min·mg) | Kcat (1/s) | Kcat/Km (1/s·mM) |
|---|---|---|---|---|
| AX543 | 0.85 ± 0.057 | 5.63 ± 0.566 | 3.94 ± 0.396 | 4.64 ± 0.481 |
| L2 | 1.49 ± 0.063 | 17.28 ± 0.301 | 12.10 ± 0.211 | 8.12 ± 0.256 |
| Q201R | 1.46 ± 0.005 | 20.20 ± 0.057 | 14.14 ± 0.04 | 9.49 ± 0.048 |
| G110S | 1.24 ± 0.042 | 12.80 ± 0.280 | 8.96 ± 0.196 | 7.23 ± 0.238 |
| Enzyme | Km (mM) | Vmax (μmol/min·mg) | Kcat (1/s) | Kcat/Km (1/s·mM) |
|---|---|---|---|---|
| AX543 | 0.85 ± 0.057 | 5.63 ± 0.566 | 3.94 ± 0.396 | 4.64 ± 0.481 |
| R1 | 1.25 ± 0.048 | 3.57 ± 0.069 | 2.50 ± 0.049 | 2.00 ± 0.059 |
| R2 | 1.17 ± 0.033 | 14.07 ± 0.196 | 9.85 ± 0.137 | 8.42 ± 0.166 |
| R3 | 1.00 ± 0.025 | 10.44 ± 0.121 | 7.31 ± 0.085 | 7.31 ± 0.103 |
| R4 | 1.84 ± 0.106 | 10.93 ± 0.366 | 7.65 ± 0.256 | 4.16 ± 0.311 |
| R5 | 1.21 ± 0.056 | 23.51 ± 0.552 | 16.46 ± 0.386 | 13.59 ± 0.469 |
| R6 | 1.19 ± 0.034 | 23.20 ± 0.336 | 16.24 ± 0.235 | 13.64 ± 0.286 |
| R7 | 1.63 ± 0.060 | 37.74 ± 0.772 | 26.42 ± 0.540 | 16.22 ± 0.656 |
| R8 | 1.30 ± 0.192 | 64.63 ± 4.915 | 45.24 ± 3.441 | 34.72 ± 4.178 |
| R9 | 1.32 ± 0.059 | 61.83 ± 1.463 | 43.28 ± 1.005 | 32.84 ± 1.221 |
| R10 | 1.87 ± 0.044 | 103.20 ± 1.412 | 72.24 ± 0.988 | 38.64 ± 1.200 |
| Enzyme | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 |
|---|---|---|---|---|---|---|---|---|---|---|
| AX543 | 0.023 | 0.004 | 0.071 | 0.022 | 0.025 | 0.130 | 0.080 | 0.013 | 0.085 | 0.111 |
| Xyl43A | 0.063 | 0.070 | 0.015 | 0.063 | 0.057 | 0.085 | 0.019 | 0.062 | 0.022 | 0.055 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, K.; Liu, Z.; Zhang, Z.; Jin, S.; Yu, Z.; Liu, T.; Zhang, T.; Zhao, J.; Li, Z. Improving the Specific Activity and Thermostability of Psychrophilic Xylosidase AX543 by Comparative Mutagenesis. Foods 2022, 11, 2463. https://doi.org/10.3390/foods11162463
Pan K, Liu Z, Zhang Z, Jin S, Yu Z, Liu T, Zhang T, Zhao J, Li Z. Improving the Specific Activity and Thermostability of Psychrophilic Xylosidase AX543 by Comparative Mutagenesis. Foods. 2022; 11(16):2463. https://doi.org/10.3390/foods11162463
Chicago/Turabian StylePan, Kungang, Zhongqi Liu, Zhengjie Zhang, Shanzheng Jin, Zhao Yu, Tianhui Liu, Tongcun Zhang, Junqi Zhao, and Zhongyuan Li. 2022. "Improving the Specific Activity and Thermostability of Psychrophilic Xylosidase AX543 by Comparative Mutagenesis" Foods 11, no. 16: 2463. https://doi.org/10.3390/foods11162463
APA StylePan, K., Liu, Z., Zhang, Z., Jin, S., Yu, Z., Liu, T., Zhang, T., Zhao, J., & Li, Z. (2022). Improving the Specific Activity and Thermostability of Psychrophilic Xylosidase AX543 by Comparative Mutagenesis. Foods, 11(16), 2463. https://doi.org/10.3390/foods11162463






