The Mutual Influence of Predominant Microbes in Sourdough Fermentation: Focusing on Flavor Formation and Gene Transcription
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Cultural Media
2.2. Wheat Flour and Sourdough Manufacturing
2.3. Determination of the pH and Microbial Loads
2.4. RNA Extraction and Sequencing Library Preparation
2.5. Library Sequencing and Data Treatment
2.6. Determination of Volatile Compounds in the Sourdough
2.7. Standards
2.8. Statistical Analysis
3. Results and Discussion
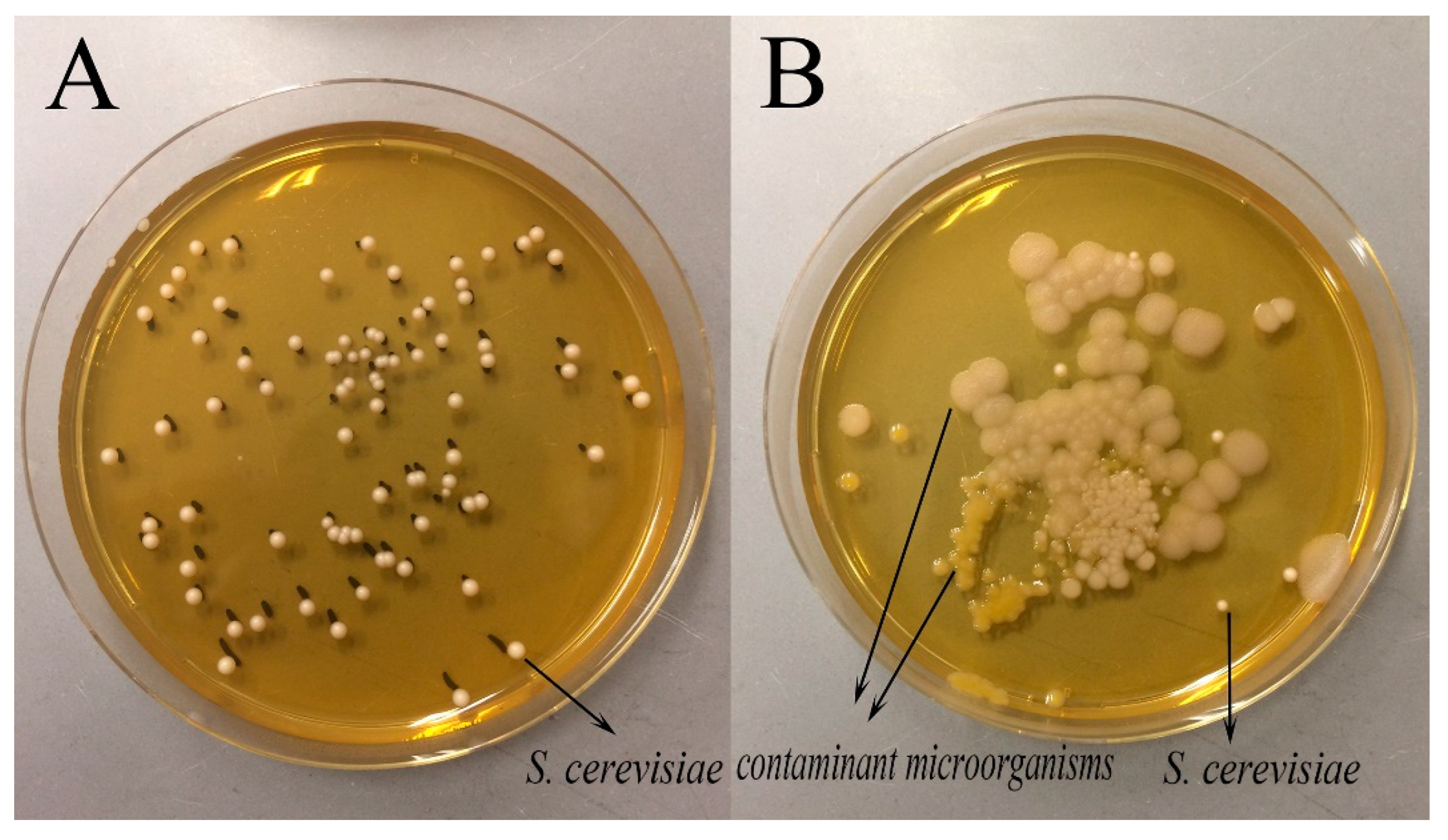
3.1. Acidity and Microbial Loads of the Fermented Dough
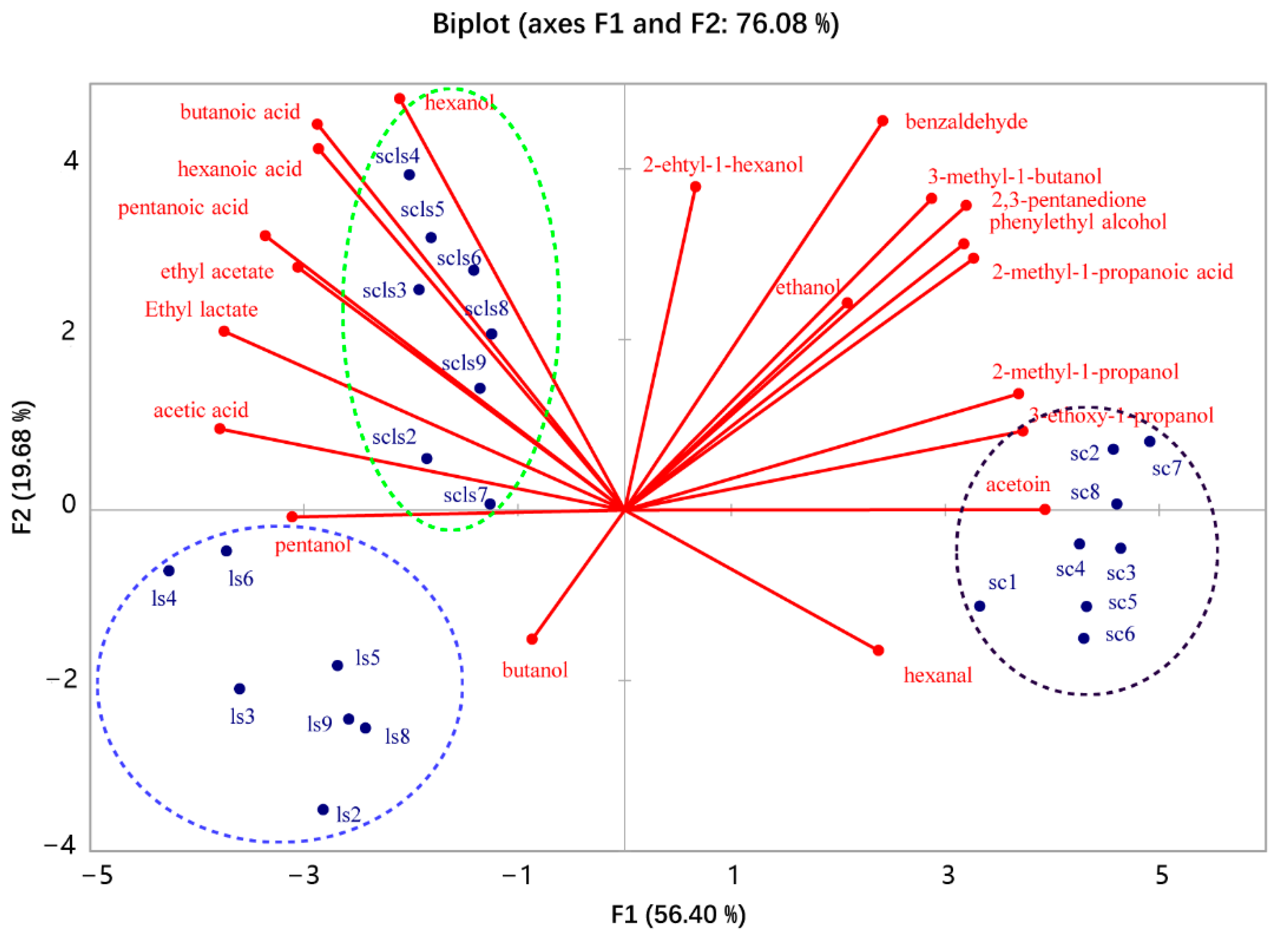
3.2. Volatile Compounds in the Sourdoughs
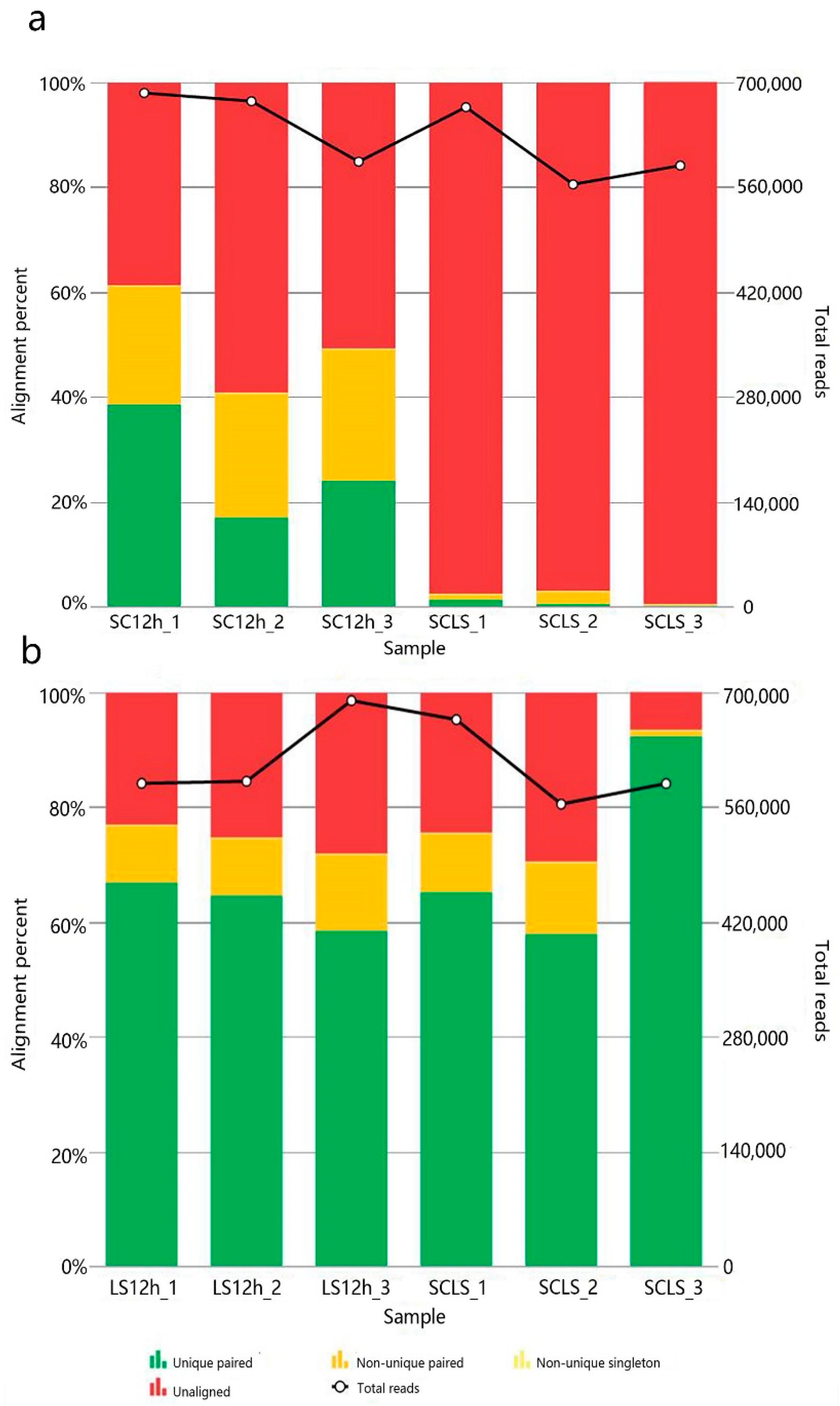
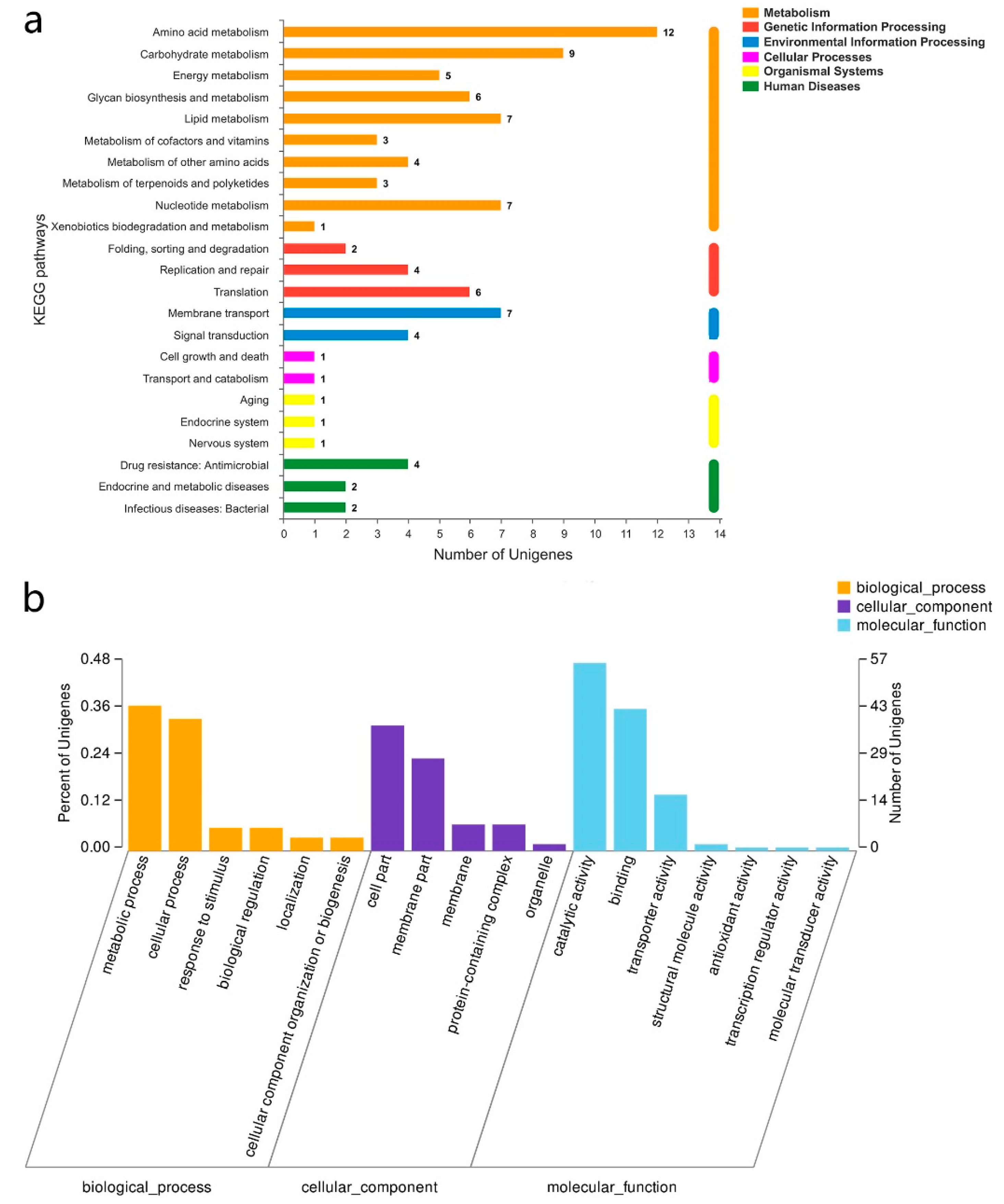
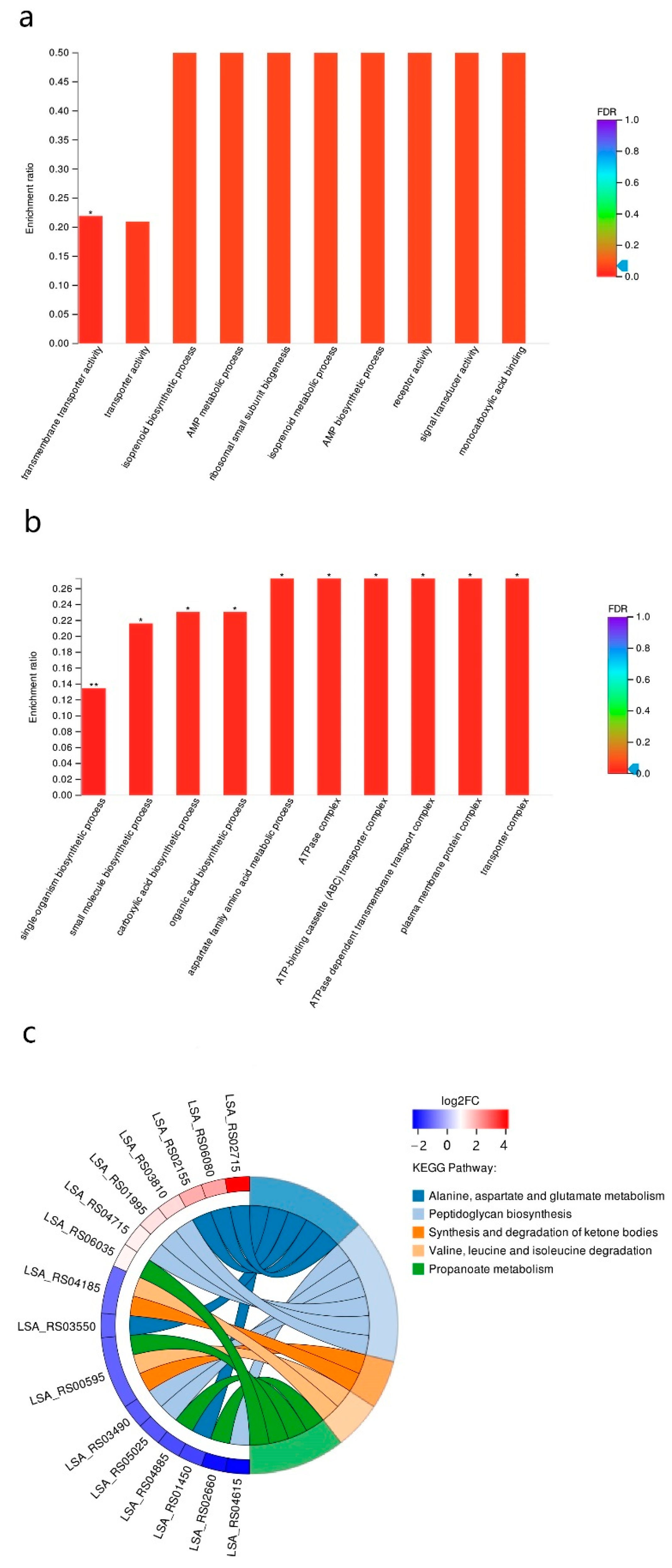
3.3. RNA-Seq Analysis of the Sourdoughs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, H.; Xi, J.; Jin, Y.; Chen, Y.; Guo, L.; Jin, Z.; Xu, X. Improving bread aroma using low-temperature sourdough fermentation. Food Biosci. 2020, 37, 100704. [Google Scholar] [CrossRef]
- De Vuyst, L.; Harth, H.; Van Kerrebroeck, S.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Minervini, F.; Pontonio, E.; Di Cagno, R.; De Angelis, M. Drivers for the establishment and composition of the sourdough lactic acid bacteria biota. Int. J. Food Microbiol. 2016, 239, 3–18. [Google Scholar] [CrossRef]
- Van Kerrebroeck, S.; Maes, D.; De Vuyst, L. Sourdoughs as a function of their species diversity and process conditions, a meta-analysis. Trends Food Sci. Technol. 2017, 68, 152–159. [Google Scholar] [CrossRef]
- Yiming, Z.; Xuanming, S.; Siyi, Z.; Xiaoli, Z. The study of microbial diversity and volatile compounds in Tartary buckwheat sourdoughs. Food Chem. X 2022, 14, 100353. [Google Scholar]
- Gobbetti, M. The sourdough microflora: Interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 1998, 9, 267–274. [Google Scholar] [CrossRef]
- Sugihara, T.; Kline, L.; McCready, L. Nature of the San Francisco sour dough French bread process. II. Microbiological aspects. Bak. Digest. 1970, 44, 48. [Google Scholar]
- Sieuwerts, S.; Bron, P.A.; Smid, E.J. Mutually stimulating interactions between lactic acid bacteria and Saccharomyces cerevisiae in sourdough fermentation. LWT 2018, 90, 201–206. [Google Scholar] [CrossRef]
- Smid, E.J.; Lacroix, C. Microbe-microbe interactions in mixed culture food fermentations. Curr. Opin. Biotechnol. 2013, 24, 148–154. [Google Scholar] [CrossRef]
- Taylor, A.J.; Hort, J. Modifying Flavour in Food; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Irlinger, F.; In Yung, S.A.Y.; Sarthou, A.-S.; Delbès-Paus, C.; Montel, M.-C.; Coton, E.; Coton, M.; Helinck, S. Ecological and aromatic impact of two Gram-negative bacteria (Psychrobacter celer and Hafnia alvei) inoculated as part of the whole microbial community of an experimental smear soft cheese. Int. J. Food Microbiol. 2012, 153, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Ding, Y.; Chen, N.; Wang, L.; Gao, F.; Zhang, L.; Song, R.; Liu, Y.; Li, H.; Wang, H. Diversity and dynamics of microbial communities during spontaneous fermentation of Cabernet Sauvignon (Vitis vinifera L.) from different regions of China and their relationship with the volatile components in the wine. Food Res. Int. 2022, 156, 111372. [Google Scholar] [CrossRef] [PubMed]
- Wenjing, J.; Fei, X.; Lei, G.; Liquan, D.; Yanxia, W.; Jian, Z.; Guiqiang, H. Identification of core microbiota in the fermented grains of a Chinese strong-flavor liquor from Sichuan. LWT 2022, 158, 113140. [Google Scholar]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- Paramithiotis, S.; Gioulatos, S.; Tsakalidou, E.; Kalantzopoulos, G. Interactions between Saccharomyces cerevisiae and lactic acid bacteria in sourdough. Process Biochem. 2006, 41, 2429–2433. [Google Scholar] [CrossRef]
- Pétel, C.; Onno, B.; Prost, C. Sourdough volatile compounds and their contribution to bread: A review. Trends Food Sci. Technol. 2017, 59, 105–123. [Google Scholar] [CrossRef]
- Chen, W. Demystification of fermented foods by omics technologies. Curr. Opin. Food Sci. 2022, 46, 100845. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Chen, J.; Sadiq, F.A.; Zhang, G.; Li, Y.; He, G. Prevalence and diversity of lactic acid bacteria in Chinese traditional sourdough revealed by culture dependent and pyrosequencing approaches. LWT-Food Sci. Technol. 2016, 68, 91–97. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Sadiq, F.A.; Yang, H.; Gu, J.; Yuan, L.; Lee, Y.K.; He, G. Predominant yeasts in Chinese traditional sourdough and their influence on aroma formation in Chinese steamed bread. Food Chem. 2018, 242, 404–411. [Google Scholar] [CrossRef]
- Kline, L.; Sugihara, T. Microorganisms of the San Francisco sour dough bread process II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl. Microbiol. 1971, 21, 459–465. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Yang, Y.; Yi, H.; Zhang, L.; He, G. The influence of different lactic acid bacteria on sourdough flavor and a deep insight into sourdough fermentation through RNA sequencing. Food Chem. 2020, 307, 125529. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Abrams, Z.; Johnson, T.; Huang, K.; Payne, P.; Coombes, K. A protocol to evaluate RNA sequencing normalization methods. BMC Bioinform. 2019, 20, 1–7. [Google Scholar] [CrossRef]
- Ruiz, J.A.; Quilez, J.; Mestres, M.; Guasch, J. Solid-phase microextraction method for headspace analysis of volatile compounds in bread crumb. Cereal Chem. 2003, 80, 255–259. [Google Scholar] [CrossRef]
- Di Cagno, R.; Pontonio, E.; Buchin, S.; De Angelis, M.; Lattanzi, A.; Valerio, F.; Gobbetti, M.; Calasso, M. Diversity of the Lactic Acid Bacterium and Yeast Microbiota in the Switch from Firm-to Liquid-Sourdough Fermentation. Appl. Environ. Microbiol. 2014, 80, 3161–3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, R.F.; Pavlovic, M.; Ehrmann, M.A.; Wiezer, A.; Liesegang, H.; Offschanka, S.; Voget, S.; Angelov, A.; Bocker, G.; Liebl, W. Genomic analysis reveals Lactobacillus sanfranciscensis as stable element in traditional sourdoughs. Microb. Cell Fact. 2011, 10, S6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pico, J.; Bernal, J.; Gomez, M. Wheat bread aroma compounds in crumb and crust: A review. Food Res. Int. 2015, 75, 200–215. [Google Scholar] [CrossRef]
- Hazelwood, L.A.; Daran, J.M.; van Maris, A.J.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Deng, Y.; Peters, B.M.; Li, L.; Li, B.; Chen, L.; Xu, Z.; Shirtliff, M.E. Transcriptomic analysis on the formation of the viable putative non-culturable state of beer-spoilage Lactobacillus acetotolerans. Sci. Rep. 2016, 6, 36753. [Google Scholar] [CrossRef]
- Wang, H.K.; Ng, Y.K.; Koh, E.; Yao, L.; Chien, A.S.; Lin, H.X.; Lee, Y.K. RNA-Seq reveals transcriptomic interactions of Bacillus subtilis natto and Bifidobacterium animalis subsp. lactis in whole soybean solid-state co-fermentation. Food Microbiol. 2015, 51, 25–32. [Google Scholar] [CrossRef]
- Mouilleron, S.; Badet-Denisot, M.A.; Golinelli-Pimpaneau, B. Glutamine binding opens the ammonia channel and activates glucosamine-6P synthase. J. Biol. Chem. 2006, 281, 4404–4412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price Evans, D.A. N-acetyltransferase. Pharmacol. Ther. 1989, 42, 157–234. [Google Scholar] [CrossRef]
- Rosenberg, E.; DeLong, E.F.; Lory, S.; Stackebrandt, E.; Thompson, F.; Lory, S. The Prokaryotes: Prokaryotic Physiology and Biochemistry; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Yazar, G.; Tavman, S. Functional and Technological Aspects of Sourdough Fermentation with Lactobacillus sanfranciscensis. Food Eng. Rev. 2012, 4, 171–190. [Google Scholar] [CrossRef]

| Chemical Group | Volatile Compounds | Calculated RI | Kovats RI of Standards | Kovats RI from Literature * | Ions Selected for Integral | Peak Areas of the Identified Volatile Compounds # (Mean Value) ×106 | ||
|---|---|---|---|---|---|---|---|---|
| LS | SC | LS + SC | ||||||
| Alcohols | Ethanol | 941 | 938 | 931 | 31.1 | 87.52 a | 130.85 b | 116.09 b |
| 2-methyl-1-propanol | 1112 | 1097 | 1110 | 43.1 | - | 12.56 a | 5.03 b | |
| butanol | 1130 | 1146 | 56 | 0.38 a | 0.33 a | 0.32 a | ||
| 3-Methyl-1-butanol | 1217 | 1216 | 1210 | 55.1 | 0.84 c | 35.33 a | 28.65 b | |
| Pentanol | 1259 | 1259 | 1251 | 42.1 | 3.47 a | 2.21 b | 3.18 a | |
| Hexanol | 1365 | 1364 | 1352 | 56.1 | 16.1 a | 15.29 a | 18.94 b | |
| 3-ethoxy-1-propanol | 1348 | 1359 | 59 | - | 0.45 b | 0.1 a | ||
| 2-ethyl-1-hexanol | 1459 | 1489 | 57 | 1.23 a | 1.38 a | 1.44 a | ||
| Phenethyl alcohol | 1927 | 1921 | 1935 | 91 | 1.92 a | 17.66 b | 10.97 c | |
| Aldehydes | Hexanal | 1084 | 1082 | 1080 | 44 | 2.11 a | 2.08 a | 1.24 a |
| benzaldehyde | 1524 | 1524 | 1522 | 77 | 0.3 a | 0.67 b | 0.64 b | |
| Ketones | 2,3-pentanedione | 1065 | 1073 | 43 | - | 4.78 a | 3.46 b | |
| acetoin | 1255 | 1264 | 45.1 | - | 2.07 a | 0.33 b | ||
| Acids | Acetic acid | 1453 | 1447 | 1447 | 43 | 55.15 a | 2.9 b | 58.26 a |
| 2-methyl-1-propanoic acid | 1574 | 1562 | 43.1 | 0.17 a | 0.93 b | 0.59 c | ||
| Butanoic acid | 1634 | 1623 | 60 | 0.59 a | 0.19 b | 1.08 c | ||
| Pentanoic acid | 1744 | 1742 | 1732 | 60 | 1.01 a | 0.31 b | 1.21 a | |
| Hexanoic acid | 1852 | 1851 | 1842 | 60 | 4.42 a | 2.14 b | 5.43 a | |
| Esters | Ethyl acetate | 903 | 892 | 884 | 61 | 9.66 a | 5.38 b | 14.13 c |
| Ethyl lactate | 1351 | 1343 | 45.1 | 25.3 a | - | 28.73 a | ||
| Gene ID | Gene Description | Change Fold | p Adjust | |
|---|---|---|---|---|
| Upregulated genes | ||||
| LSA_RS01375 | N-acetyltransferase | 28.40 | 4.03 × 10−11 | |
| LSA_RS01370 | ammonium transporter | 22.07 | 4.52 × 10−54 | |
| LSA_RS02715 | glutamine—fructose-6-phosphate transaminase (isomerizing) | 21.02 | 8.99 × 10−58 | |
| LSA_RS00920 | amino acid ABC transporter permease | 20.00 | 3.82 × 10−4 | |
| LSA_RS00925 | amino acid ABC transporter ATP-binding protein | 17.54 | 8.63 × 10−4 | |
| LSA_RS00930 | amino acid ABC transporter substrate-binding protein | 13.22 | 1.48 × 10−3 | |
| LSA_RS06730 | amino acid ABC transporter ATP-binding protein | 6.88 | 2.37 × 10−35 | |
| LSA_RS00235 | diaminopimelate decarboxylase | 6.05 | 3.75 × 10−5 | |
| LSA_RS06725 | glutamine ABC transporter substrate-binding protein | 5.74 | 8.69 × 10−17 | |
| LSA_RS00340 | amino acid permease | 5.48 | 2.34 × 10−13 | |
| LSA_RS03825 | GatB/YqeY domain-containing protein | 5.39 | 1.72 × 10−11 | |
| LSA_RS06720 | amino acid ABC transporter permease | 5.02 | 2.55 × 10−13 | |
| Downregulated genes | ||||
| LSA_RS00535 | hypothetical protein | 11.11 | 0.03 | |
| LSA_RS00005 | chromosomal replication initiator protein DnaA | 6.90 | 6.35 × 10−18 | |
| LSA_RS01215 | alpha/beta hydrolase | 6.58 | 6.20 × 10−11 | |
| LSA_RS05425 | DUF1304 domain-containing protein | 5.78 | 8.57 × 10−10 | |
| LSA_RS04335 | aminoglycoside phosphotransferase | 5.52 | 8.13 × 10−27 | |
| LSA_RS04615 | D-alanine—D-alanine ligase A | 5.49 | 1.51 × 10−23 | |
| LSA_RS06125 | ABC transporter permease | 5.376344 | 1.98 × 10−3 | |
| LSA_RS04340 | amino acid permease | 5.128205 | 8.13 × 10−27 | |
| LSA_RS00545 | ABC transporter ATP-binding protein | 5.05 | 6.84 × 10−3 | |
| LSA_RS01530 | nitronate monooxygenase | 5.00 | 1.68 × 10−15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Shi, Y.; Li, Y.; Yi, H.; Gong, P.; Lin, K.; Zhang, Z.; Zhang, L. The Mutual Influence of Predominant Microbes in Sourdough Fermentation: Focusing on Flavor Formation and Gene Transcription. Foods 2022, 11, 2373. https://doi.org/10.3390/foods11152373
Liu T, Shi Y, Li Y, Yi H, Gong P, Lin K, Zhang Z, Zhang L. The Mutual Influence of Predominant Microbes in Sourdough Fermentation: Focusing on Flavor Formation and Gene Transcription. Foods. 2022; 11(15):2373. https://doi.org/10.3390/foods11152373
Chicago/Turabian StyleLiu, Tongjie, Yixin Shi, Yang Li, Huaxi Yi, Pimin Gong, Kai Lin, Zhe Zhang, and Lanwei Zhang. 2022. "The Mutual Influence of Predominant Microbes in Sourdough Fermentation: Focusing on Flavor Formation and Gene Transcription" Foods 11, no. 15: 2373. https://doi.org/10.3390/foods11152373
APA StyleLiu, T., Shi, Y., Li, Y., Yi, H., Gong, P., Lin, K., Zhang, Z., & Zhang, L. (2022). The Mutual Influence of Predominant Microbes in Sourdough Fermentation: Focusing on Flavor Formation and Gene Transcription. Foods, 11(15), 2373. https://doi.org/10.3390/foods11152373







