Bluetooth-Connected Pocket Spectrometer and Chemometrics for Olive Oil Applications
Abstract
:1. Introduction
- -
- advanced consumer applications: that is, instantly probing olive oil quality just shining light on the bottle,
- -
- importers, bottlers, and retailers, who wish to check the quality of the olive oil they bottle and sell, particularly the different batches they deal with,
- -
- small/medium-size producers in low-resource settings, for detecting the olive oil quality during production and storage, in addition to better product marketing. In fact, the measurement of nutraceutical and quality indicators is a precursor of generating a QR code, which points through the internet to the values of the measured parameters stored in a data warehouse. The possibility of creating a QR code for labeling leads to an innovative and modern marketing strategy, based on an interactive and rapid communication of oil quality to end consumers, giving to the product the chance of a digital life. Consumers who do not own a spectrometer but have a smartphone can retrieve product information, made available online by producers, simply scanning the QR code.
2. Materials and Methods
2.1. Olive Oil Collection
- -
- Extra virgin olive oils (EVOO) comprised 80 samples produced in two different years, thus considering the production variability. Some of them were provided by Fattoria Castel Ruggero Pellegrini, and some others were kindly supplied by Analytical Food, the official analytical laboratory hired for making reference analyses.
- -
- Lower quality edible oils (OO) comprised 36 samples; they were 24 samples of non-extra virgin olive oils, 8 samples of mixed pomace and olive oils, and 4 lampante oils (very low-quality virgin oil, not suitable for human consumption). All of them were supplied by Analytical Food or purchased on the market.
2.2. Spectroscopic Measurements
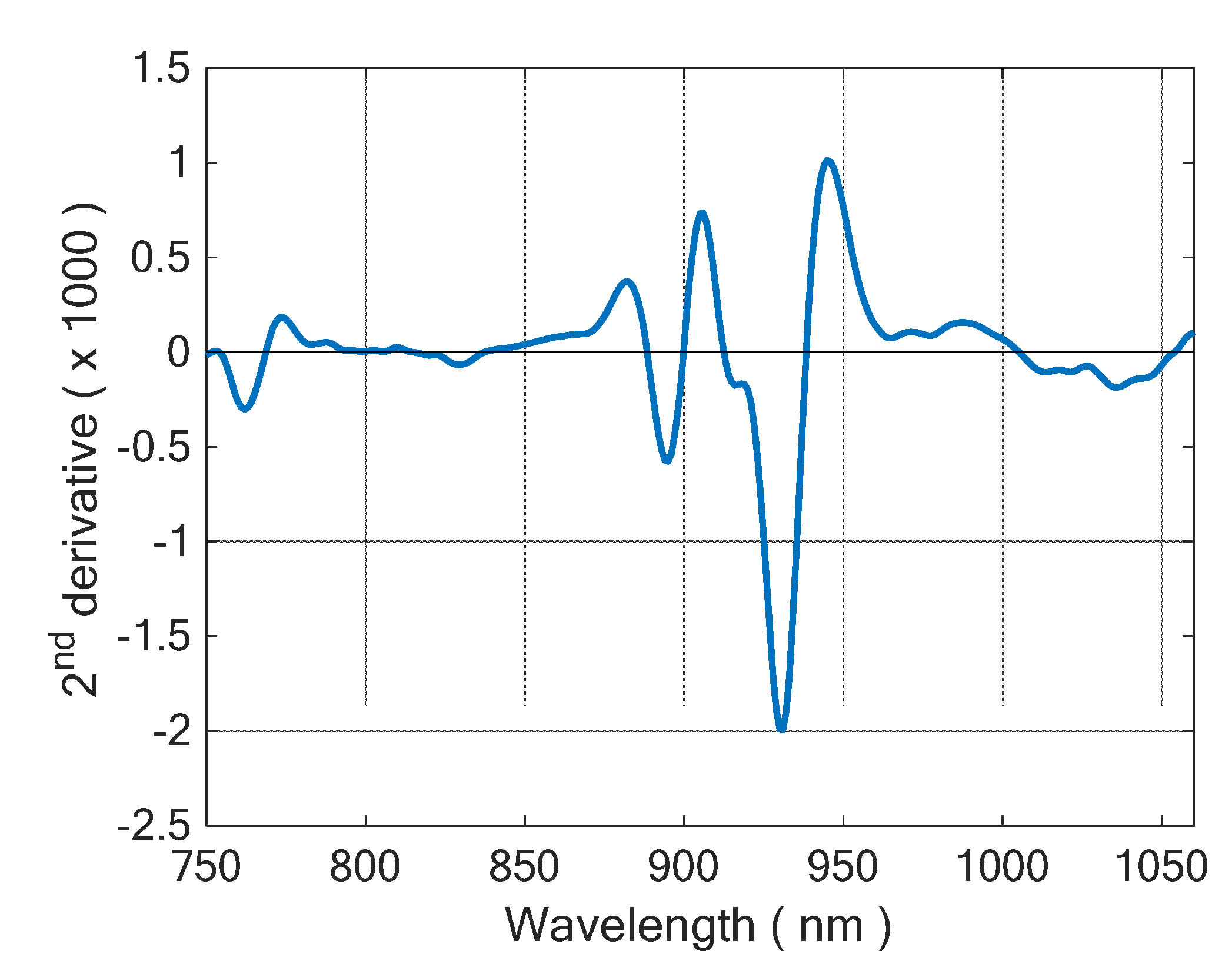
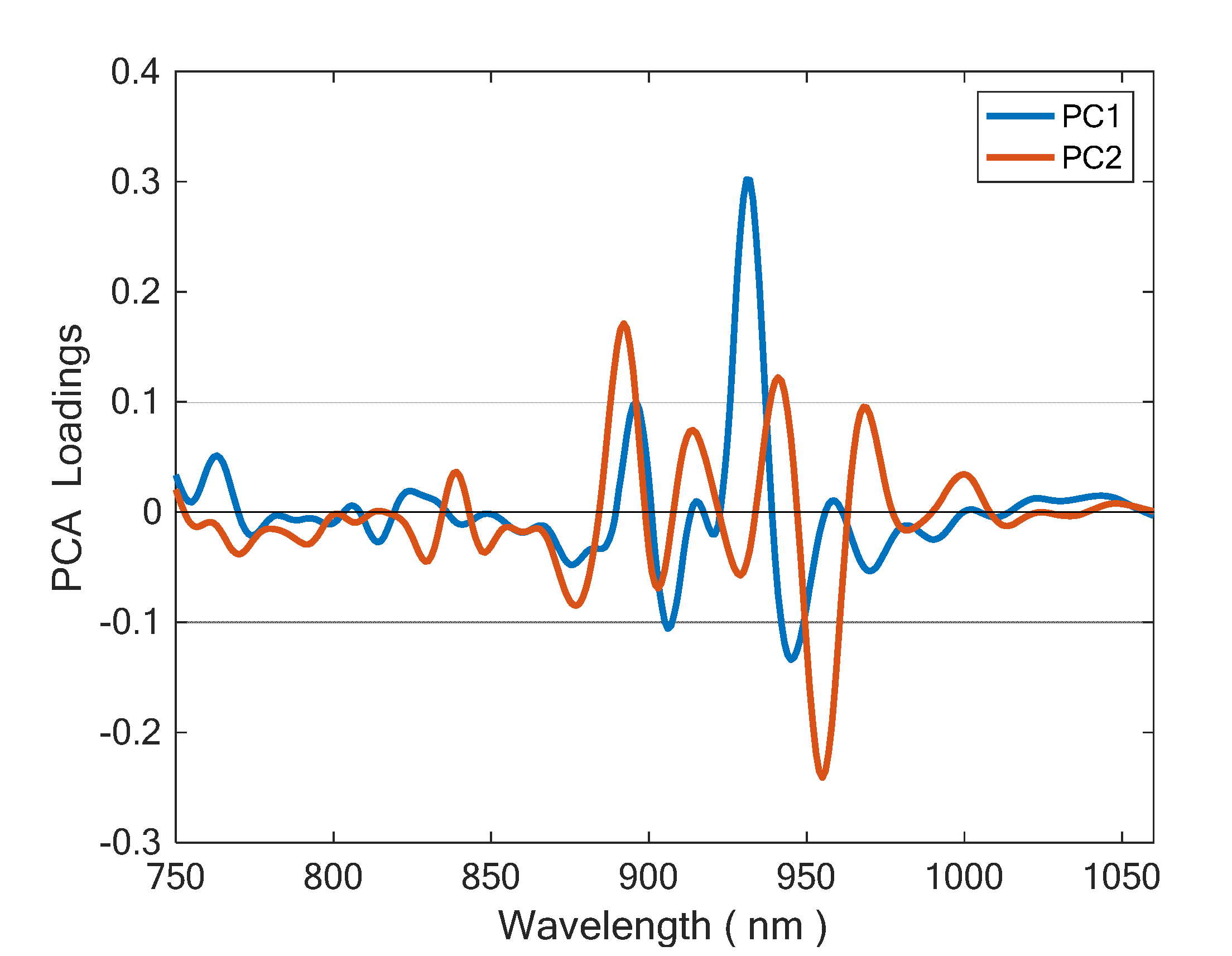
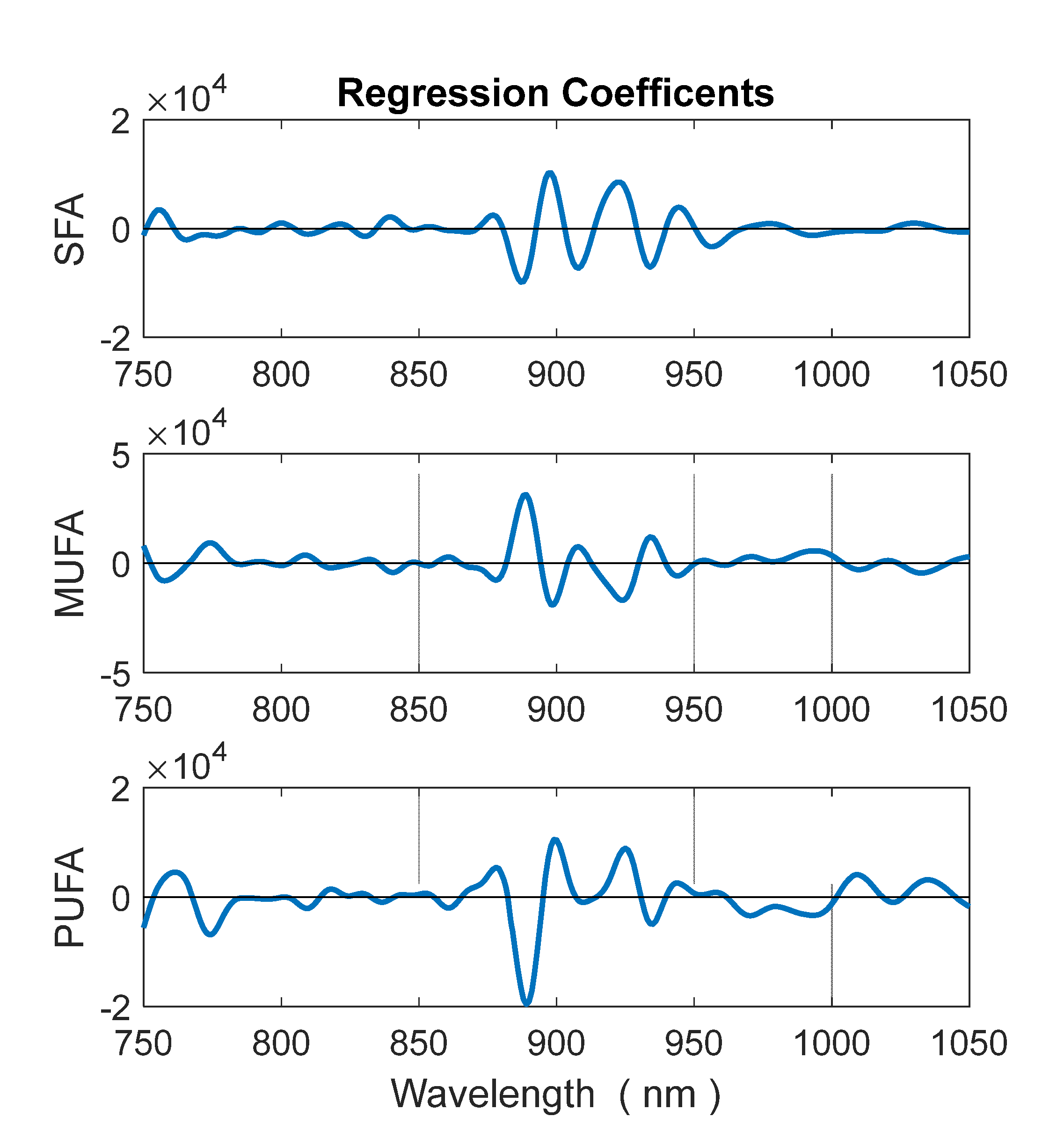
2.3. Data Analysis
- M: number of spectral channels (discrete wavelengths),
- r0 intercept of the linear model,
- rm regression coefficient corresponding to mth wavelength,
- xn m value of the spectrum of nth sample at mth wavelength.
- predicted value of target variable for nth sample.
3. Results and Discussion
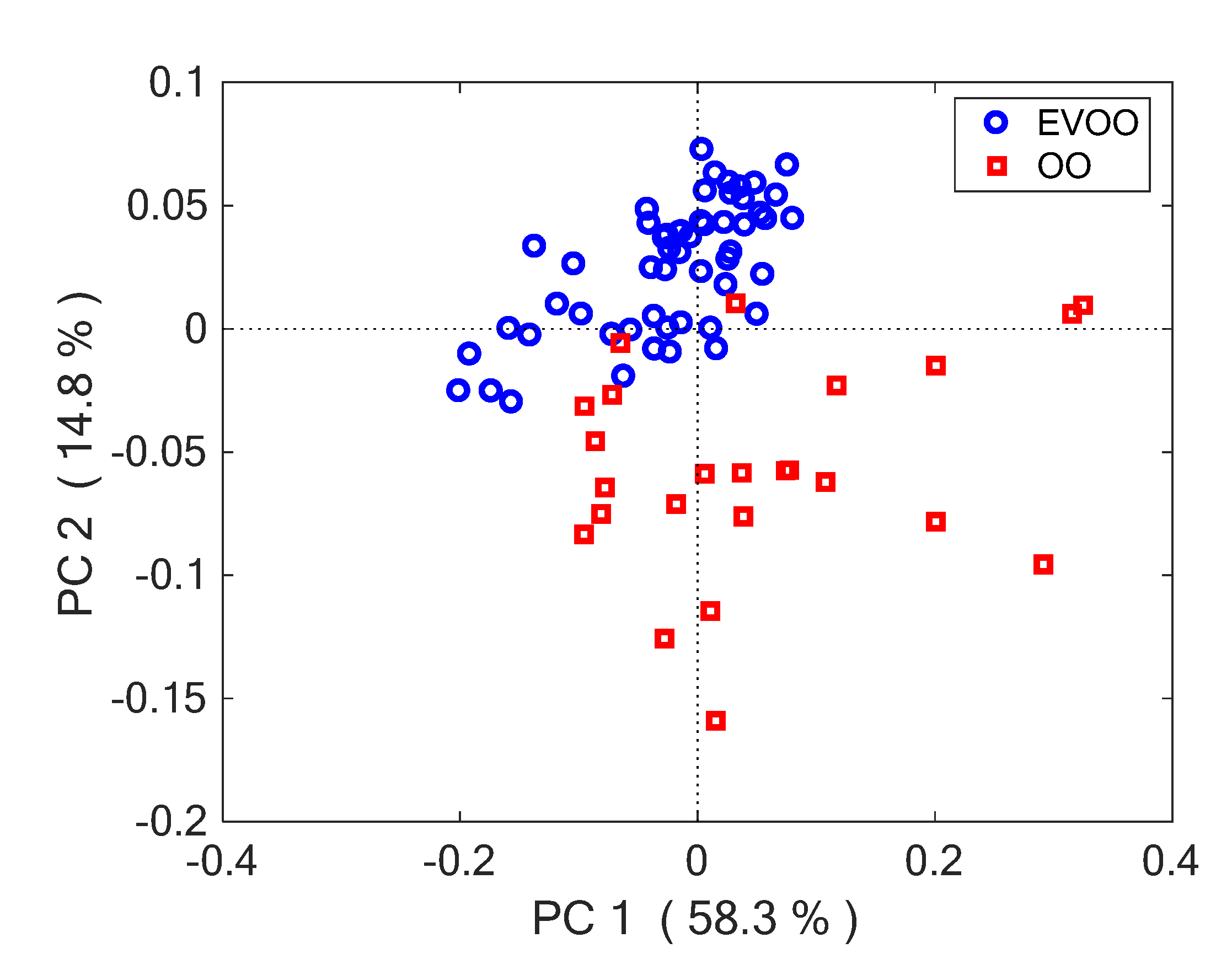
3.1. Classification of Olive Oil Type
- Accuracy: overall percentage of samples correctly classified
- Sensitivity: true positive (EVOO) classification rate (ability to recognize an EVOO).
- Specificity: true negative (OO) classification rate (ability to reject an OO).
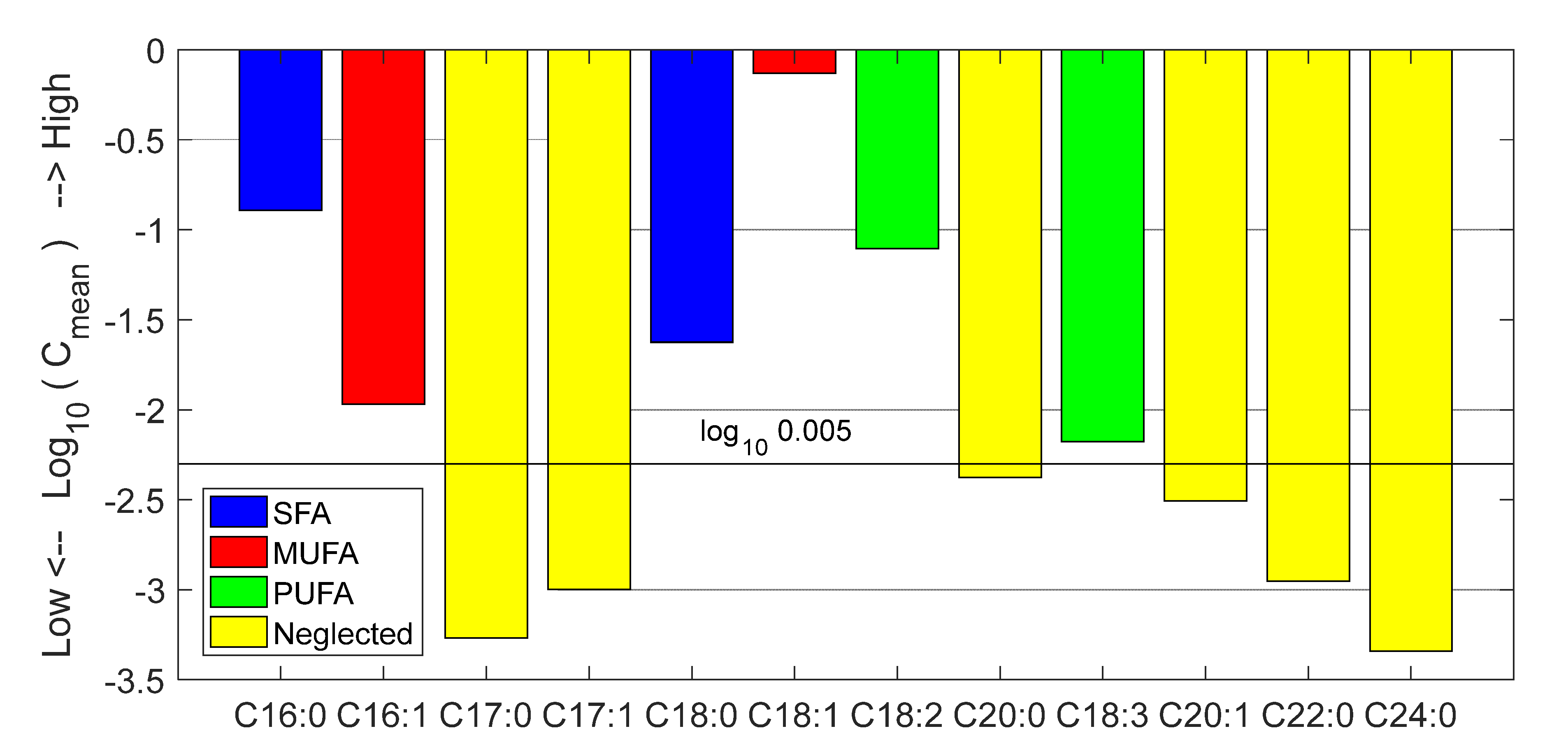
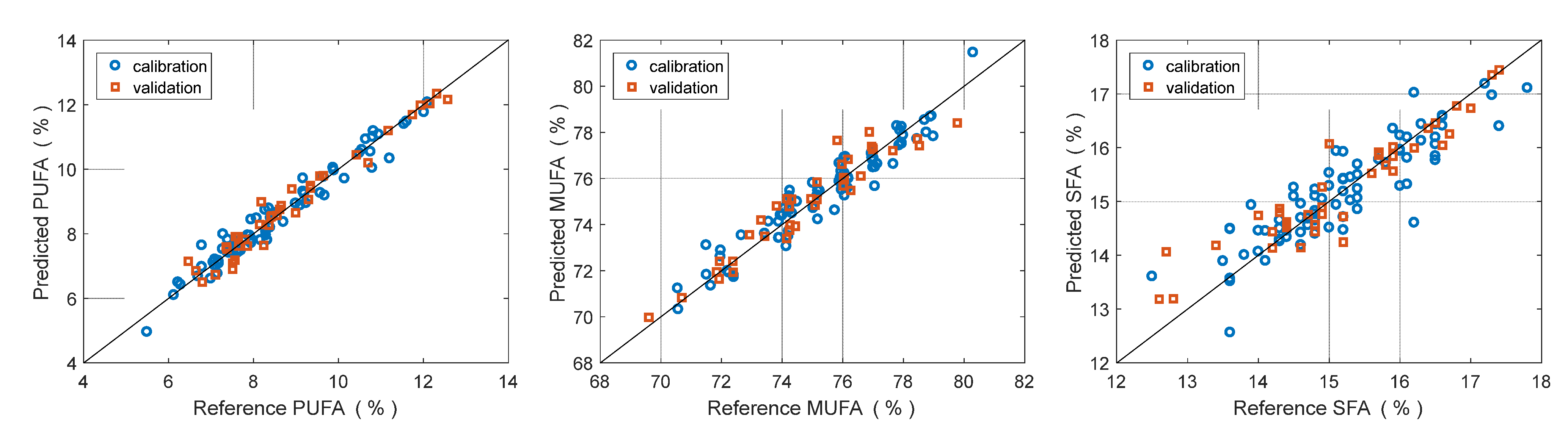
3.2. Prediction of Fatty Acid Content
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- BankMyCell. Available online: https://www.bankmycell.com/blog/how-many-phones-are-in-the-world#:~:text=How%20Many%20People%20Have%20Mobile%20Devices%20In%20The%20World%3F&text=According%20to%20GSMA%20real%2Dtime,population%20has%20a%20mobile%20device (accessed on 27 June 2022).
- Li, F.; Bao, Y.; Wang, D.; Wang, W.; Niu, L. Smartphones for sensing. Sci. Bull. 2016, 61, 190–201. [Google Scholar] [CrossRef]
- Hernandez-Neuta, I.; Neumann, F.; Brightmeyer, J.; Ba, T.T.; Madaboosi, N.; Wei, Q.; Ozcan, A.; Nilsson, M. Smartphone-based clinical diagnostics: Towards democratization of evidence-based health care. J. Intern. Med. 2019, 285, 19–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, S.; Deen, M.J. Smartphone sensors for health monitoring and diagnosis. Sensors 2019, 19, 2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roda, A.; Michelini, E.; Zangheri, M.; Di Fusco, M.; Calabria, D.; Simoni, P. Smartphone-based biosensors: A critical review and perspectives. Trac-Trends Anal. Chem. 2016, 79, 317–325. [Google Scholar] [CrossRef]
- Zhao, W.; Tian, S.; Huang, L.; Liu, K.; Dong, L.; Guo, J. A smartphone-based biomedical sensory system. Analyst 2020, 145, 2873–2891. [Google Scholar] [CrossRef] [PubMed]
- Rateni, G.; Dario, P.; Cavallo, F. Smartphone-based food diagnostic technologies: A review. Sensors 2017, 17, 1453. [Google Scholar] [CrossRef]
- Nelis, J.L.D.; Tsagkaris, A.S.; Dillon, M.J.; Hajslova, J.; Elliott, C.T. Smartphone-based optical assays in the food safety field. Trac-Trends Anal. Chem. 2020, 129, 115934. [Google Scholar] [CrossRef]
- Ross, G.M.S.; Bremer, M.G.E.G.; Nielen, M.W.F. Consumer-friendly food allergen detection: Moving towards smartphone-based immunoassays. Anal. Bioanal. Chem. 2018, 410, 5353–5371. [Google Scholar] [CrossRef] [Green Version]
- Tellspec. Available online: http://tellspec.com/howitworks/ (accessed on 27 June 2022).
- Consumer Physics. Available online: https://www.consumerphysics.com/business/ (accessed on 27 June 2022).
- YouTube. Available online: https://www.youtube.com/watch?v=3 (accessed on 27 June 2022).
- YouTube. Available online: https://www.youtube.com/watch?v=y5xsp-KtMkQ/ (accessed on 27 June 2022).
- Das, A.J.; Wahi, A.; Kothari, I.; Raskar, R. Ultra-portable, wireless smartphone spectrometer for rapid, non-destructive testing of fruit ripeness. Sci Rep. 2016, 6, 32504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bickler, M.P.; Rhodes, L.J. Accuracy of detection of carboxyhemoglobin and methemoglobin in human and bovine blood with an inexpensive, pocket-size infrared scanner. PLoS ONE 2018, 13, e0193891. [Google Scholar] [CrossRef] [Green Version]
- Golse, N.; Cosse, C.; Allard, M.A.; Samuel, D.; Sebagh, M.; Vibert, E. Evaluation of a micro-spectrometer for the real-time assessment of liver graft with mild-to-moderate macrosteatosis: A proof of concept study. J. Hepatol. 2018, 70, 423–430. [Google Scholar] [CrossRef]
- Barri, K.; Jahangiri, B.; Davami, O.; Buttlar, W.G.; Alavi, A.H. Smartphone-based molecular sensing for advanced characterization of asphalt concrete materials. Measurement 2020, 151, 107212. [Google Scholar]
- Wiedemair, V.; Huck, C.W. Evaluation of the performance of three hand-held near-infrared spectrometer through investigation of total antioxidant capacity in gluten-free grains. Talanta 2018, 189, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Wiedemair, V.; Langore, D.; Garsleitner, R.; Dillinger, K.; Huck, C. Investigations into the performance of a novel pocket-sized near-infrared spectrometer for cheese analysis. Molecules 2019, 24, 428. [Google Scholar] [CrossRef] [Green Version]
- Mulvey, B.W.; Kennedy, M.P. Fat sensing using low-cost near-infrared spectroscopy. In Proceedings of the 30th Irish Signals and Systems Conference (ISSC), Maynoth, Ireland, 17–19 May 2019. [Google Scholar] [CrossRef]
- Riu, J.; Gorla, G.; Chakif, D.; Boqué, R.; Giussani, B. Rapid analysis of milk using low-cost pocket-size NIR spectrometers and multivariate analysis. Foods 2020, 9, 1090. [Google Scholar] [CrossRef]
- Chen, W.; Li, H.; Zhang, F.; Xiao, W.; Zhang, R.; Chen, Z.; Du, Y. Handheld short-wavelength NIR spectroscopy for rapid determination of sugars and carbohydrate in fresh juice with sampling error profile analysis. Infrared Phys. Technol. 2021, 115, 103732. [Google Scholar]
- Armenta, S.; Moros, J.; Garrigues, S.; de la Guardia, M. The use of near-infrared spectrometry in the olive oil industry. Crit. Rev. Food Sci. Nutr. 2010, 50, 567–582. [Google Scholar] [CrossRef]
- Downey, G.; McIntyre, P.; Davies, A.N. Detecting and quantifying sunflower oil adulteration in extra virgin olive oils from the Eastern Mediterranean by visible and near-infrared spectroscopy. J. Agric. Food Chem. 2002, 50, 5520–5525. [Google Scholar] [CrossRef]
- Christy, A.A.; Kasemsumran, S.; Du, Y.; Ozaki, Y. The detection and quantification of adulteration in olive oil by near-infrared spectroscopy and chemometrics. Anal. Sci. 2004, 20, 935–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Öztürk, A.; Yalçin, A.; Özdemir, D. Determination of olive oil adulteration with vegetable oils by near infrared spectroscopy coupled with multivariate calibration. J. Near Infrared Spectrosc. 2010, 18, 191–201. [Google Scholar]
- Mignani, A.G.; Ciaccheri, L.; Ottevaere, H.; Thienpont, H.; Conte, L.; Marega, M.; Cichelli, A.; Attilio, C.; Cimato, A. Visible and near-infrared absorption spectroscopy by an integrating sphere and optical fibers for quantifying and discriminating the adulteration of extra virgin olive oil from Tuscany. Anal. Bioanal. Chem. 2011, 399, 315–1324. [Google Scholar] [CrossRef]
- Mendes, T.O.; da Rocha, R.A.; Porto, B.L.S.; de Oliveira, A.A.L.; dos Anjos, V.D.C.; Bell, M.J.V. Quantification of extra-virgin olive oil adulteration with soybean oil: A comparative study of NIR, MIR and Raman spectroscopy associated with chemometric approaches. Food Anal. Meth. 2015, 8, 2339–2346. [Google Scholar] [CrossRef]
- Li, Y.; Xiong, Y.; Min, S. Data fusion strategy in quantitative analysis of spectroscopy relevant to olive oil adulteration. Vib. Spectrosc. 2019, 101, 20–27. [Google Scholar] [CrossRef]
- Cayuela-Sánchez, J.A.; Palarea-Albaladejo, J.; García, M.; Pérez-Camino, M. Olive oil nutritional labelling by using Vis/NIR spectroscopy and compositional statistical methods. Innov. Food Sci. Emerg. Technol. (IFSET) 2019, 51, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Armenta, S.; Garrigues, S.; de la Guardia, M. Determination of edible oil parameters by near infrared spectrometry. Anal. Chim. Acta 2007, 596, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Casale, M.; Oliveri, P.; Casolino, C.; Sinelli, N.; Zunin, P.; Armanino, C.; Forina, M.; Lanteri, S. Characterisation of PDO olive oil Chianti Classico by non-selective (UV-visible, NIR and MIR spectroscopy) and selective (fatty acid composition) analytical techniques. Anal. Chim. Acta 2012, 712, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Abu-Khalaf, N.; Hmidat, M. Visible/Near Infrared (VIS/NIR) spectroscopy as an optical sensor for evaluating olive oil quality. Comput. Electron. Agric. 2020, 173, 105445. [Google Scholar] [CrossRef]
- Sinelli, N.; Cerretani, L.; Di Egidio, V.; Bendini, A.; Casiraghi, E. Application of near (NIR) infrared and mid (MIR) infrared spectroscopy as a rapid tool to classify extra virgin olive oil on the basis of fruity attribute intensity. Food Res. Int. 2010, 43, 369–375. [Google Scholar] [CrossRef]
- Cayuela, J.A.; Moreda, W.; García, J.M. Rapid determination of olive oil oxidative stability and its major quality parameters using Vis/NIR transmittance spectroscopy. J. Agric. Food Chem. 2013, 61, 8056–8062. [Google Scholar] [CrossRef]
- Jiménez, M.A. Monitoring carotenoid and chlorophyll pigments in virgin olive oil by visible-near infrared transmittance spectroscopy. On-line application. J. Near Infrared Spectrosc. 2003, 11, 219–226. [Google Scholar]
- Correa, E.C.; Roger, J.M.; Lleó, L.; Hernández-Sánchez, N.; Barreiro, P.; Diezma, B. Optimal management of oil content variability in olive mill batches by NIR spectroscopy. Sci. Rep. 2019, 9, 13974. [Google Scholar] [CrossRef] [Green Version]
- Estruch, R.; Salas-Salvadó, J. Towards an even healthier Mediterranean diet. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 1163–1166. [Google Scholar] [CrossRef]
- Schwingshackl, L.; Morze, J.; Hoffmann, G. Mediterranean diet and health status: Active ingredients and pharmacological mechanisms. Br. J. Pharmacol. 2020, 177, 1241–1257. [Google Scholar] [CrossRef] [Green Version]
- Fuentes de Mendoza, M.; De Miguel Gordillo, C.; Expóxito, J.M.; Sánchez, C.J.; Martínez, C.M.; Martín, V.D.; Baltasar, M.N.F. Chemical composition of virgin olive oils according to the ripening in olives. Food Chem. 2013, 141, 2575–2581. [Google Scholar] [CrossRef] [PubMed]
- Mazzocchi, A.; Leone, L.; Agostoni, C.; Pali-Schöll, I. The secrets of the Mediterranean diet. Does only olive oil matter? Nutrients 2019, 11, 2941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, H.A.; Marnett, L.J. Introduction to lipid biochemistry, metabolism, and signaling. Chem. Rev. 2011, 111, 5817–5820. [Google Scholar] [CrossRef]
- Radzikowska, U.; Rinaldi, A.O.; Sözener, Z.C.; Karaguzel, D.; Wojcik, D.; Cypryk, K.; Akdis, M.; Akdis, C.A.; Sokolowska, M. The influence of dietary fatty acids on immune responses. Nutrients 2019, 11, 2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, S.; Bermudez, B.; Montserrat-de la Paz, S.; Pacheco, Y.M.; Ortega-Gomez, A.; Varela, L.M.; Lemus-Conejo, A.; Millan-Linares, M.C.; Rosillo, M.A.; Abia, R.; et al. Oleic acid-the main component of olive oil on postprandial metabolic processes. In Olives and Olive Oil in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; Chapter 53; pp. 639–649. [Google Scholar]
- Palomer, X.; Pizarro-Delgado, J.; Barroso, E.; Vázquez-Carrera, M. Palmitic and Oleic Acid: The Yin and Yang of fatty acids in type 2 diabetes mellitus. Trends Endocrinol. Metab. 2018, 29, 178–190. [Google Scholar] [CrossRef]
- Balić, A.; Vlašić, D.; Žužul, K.; Marinović, B.; Mokos, Z.B. Omega-3 versus omega-6 polyunsaturated fatty acids in the prevention and treatment of inflammatory skin diseases. Int. J. Mol. Sci. 2020, 21, 741. [Google Scholar] [CrossRef] [Green Version]
- Comàn, R.G.; Jackson, R.E.; Reis, J.; Romàn, A.N.; Toledo, J.B.; Toledo, E. Extra-virgin olive oil for potential prevention of Alzheimer disease. Rev. Neurol. 2019, 175, 705–723. [Google Scholar] [CrossRef]
- Araya-Quintanilla, F.; Gutiérrez-Espinoza, H.; Sánchez-Montoya, U.; Muñoz-Yañez, M.J.; Baeza-Vergara, A.; Petersen-Yanjarí, M.; Fernández-Lecaros, L. Effectiveness of omega-3 fatty acid supplementation in patients with Alzheimer disease: A systematic review and meta-analysis. Neurologia 2020, 35, 105–114. [Google Scholar] [CrossRef]
- Feuchtner, G.; Langer, C.; Barbieri, F.; Beyer, C.; Dichtl, W.; Friedrich, G.; Schgoer, W.; Widmann, G.; Plan, F. The effect of omega-3 fatty acids on coronary atherosclerosis quantified by coronary computed tomography angiography. Clin. Nutr. 2021, 40, 1123–1129. [Google Scholar] [CrossRef]
- INTERNATIONAL OLIVE COUNCIL. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2019/11/COI-T.20-Doc.-No-33-Rev.-1-2017.pdf (accessed on 27 June 2022).
- Jackson, J.E. A User’s Guide to Principal Components; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2003. [Google Scholar]
- Naes, T.; Isaksson, T.; Fearn, T.; Davies, T. A User-Friendly Guide to Multivariate Calibration and Classification; NIR Publications: Stockholm, Sweden, 2002. [Google Scholar]
- Massart, D.L.; Vandeginste, B.G.M.; Buydens, L.M.C.; De Jong, S.; Lewi, P.J.; Smeyers-Verbeke, J. Handbook of Chemometrics and Qualimetrics; cap. 33; Elsevier Science BV: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chem. Int. Lab. Sys. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Aspen Technology Inc. Available online: https://www.aspentech.com/en/products/msc/aspen-unscrambler (accessed on 27 June 2022).
- Delacre, M.; Leys, C.; Mora, Y.L.; Lakens, D. Taking Parametric Assumptions Seriously: Arguments for the Use of Welch’s F-test instead of the Classical F-test in One-Way ANOVA. Int. Rev. Soc. Psychol. 2019, 32, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Workman, J.; Weyer, L. Practical Guide to Interpretative Near-Infrared Spectroscopy; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon, UK, 2008; pp. 23–37. [Google Scholar]


| Calibration Set | Validation Set | |||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| SFA | 15.2% | 1.1% | 15.1% | 1.1% |
| MUFA | 75.1% | 2.3% | 75.7% | 2.1% |
| PUFA | 8.7% | 1.6% | 8.2% | 1.5% |
| Training | Validation | ||||
|---|---|---|---|---|---|
| Accuracy 94% Sensitivity 94% Specificity 92% | Accuracy 92% Sensitivity 88% Specificity 100% | ||||
| Predicted class | Actual class | Predicted class | Actual class | ||
| EVOO | Non-EVOO | EVOO | Non-EVOO | ||
| EVOO | 51 | 2 | EVOO | 23 | 0 |
| Non-EVOO | 3 | 22 | Non-EVOO | 3 | 12 |
| SFA | MUFA | PUFA | ||
|---|---|---|---|---|
| Calibration | LV | 6 | 6 | 6 |
| RMSECV | 0.51% | 0.74% | 0.37% | |
| R2 (cal.) | 0.780 | 0.895 | 0.950 | |
| Validation | RMSEP | 0.55% | 0.74% | 0.39% |
| SEP | 0.55% | 0.75% | 0.39% | |
| Bias | 0.06% | −0.11% | 0.03% | |
| R2 (val.) | 0.764 | 0.872 | 0.921 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciaccheri, L.; Adinolfi, B.; Mencaglia, A.A.; Mignani, A.G. Bluetooth-Connected Pocket Spectrometer and Chemometrics for Olive Oil Applications. Foods 2022, 11, 2265. https://doi.org/10.3390/foods11152265
Ciaccheri L, Adinolfi B, Mencaglia AA, Mignani AG. Bluetooth-Connected Pocket Spectrometer and Chemometrics for Olive Oil Applications. Foods. 2022; 11(15):2265. https://doi.org/10.3390/foods11152265
Chicago/Turabian StyleCiaccheri, Leonardo, Barbara Adinolfi, Andrea Azelio Mencaglia, and Anna Grazia Mignani. 2022. "Bluetooth-Connected Pocket Spectrometer and Chemometrics for Olive Oil Applications" Foods 11, no. 15: 2265. https://doi.org/10.3390/foods11152265
APA StyleCiaccheri, L., Adinolfi, B., Mencaglia, A. A., & Mignani, A. G. (2022). Bluetooth-Connected Pocket Spectrometer and Chemometrics for Olive Oil Applications. Foods, 11(15), 2265. https://doi.org/10.3390/foods11152265







