Specific Microbial Communities Are Selected in Minimally-Processed Fruit and Vegetables according to the Type of Product
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Culture-Dependent Microbiological Analysis
2.3. DNA Extraction and Amplicon Illumina Sequencing
2.4. Bioinformatics and Statistical Analysis
3. Results
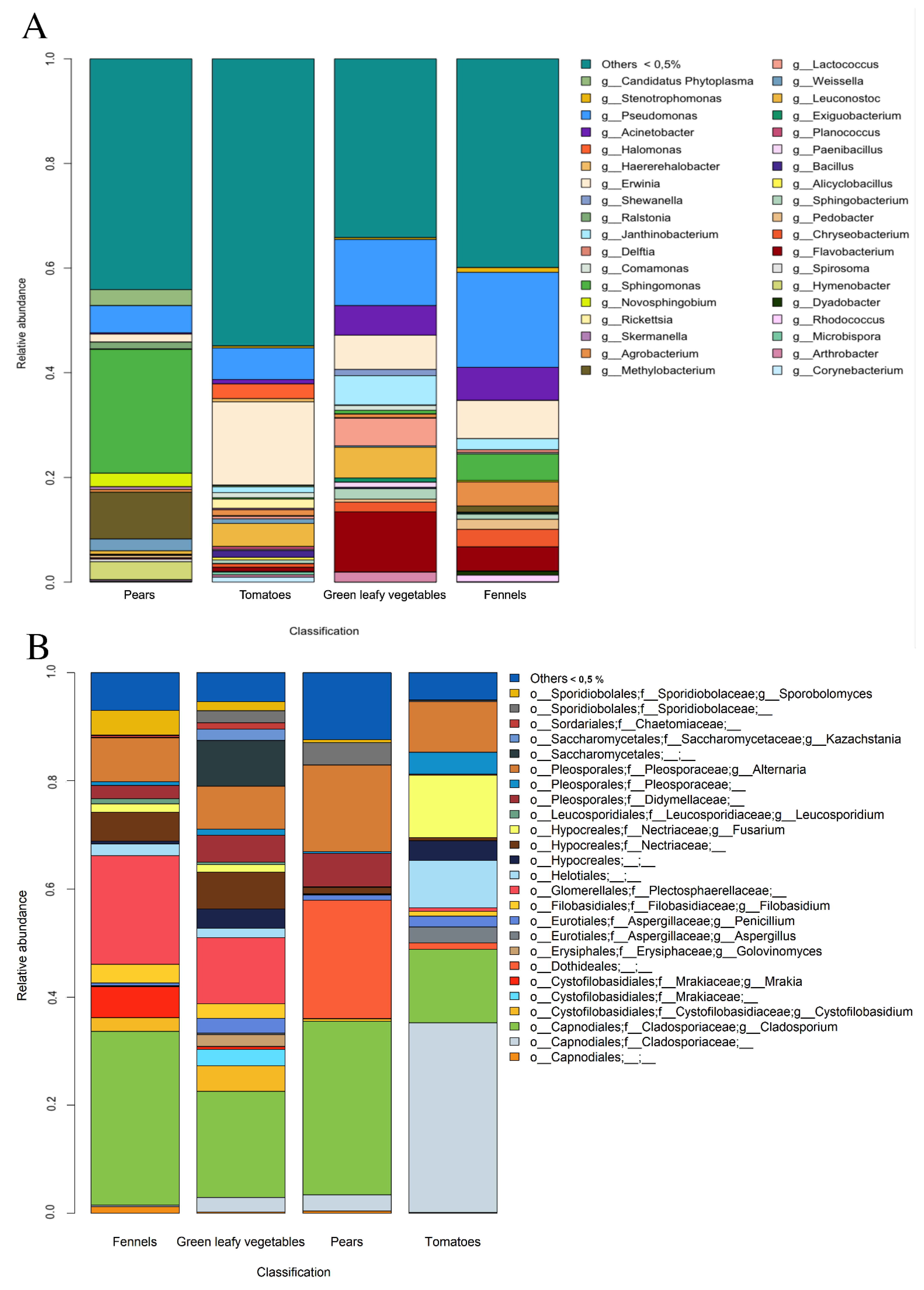
3.1. Culture-Dependent Analysis
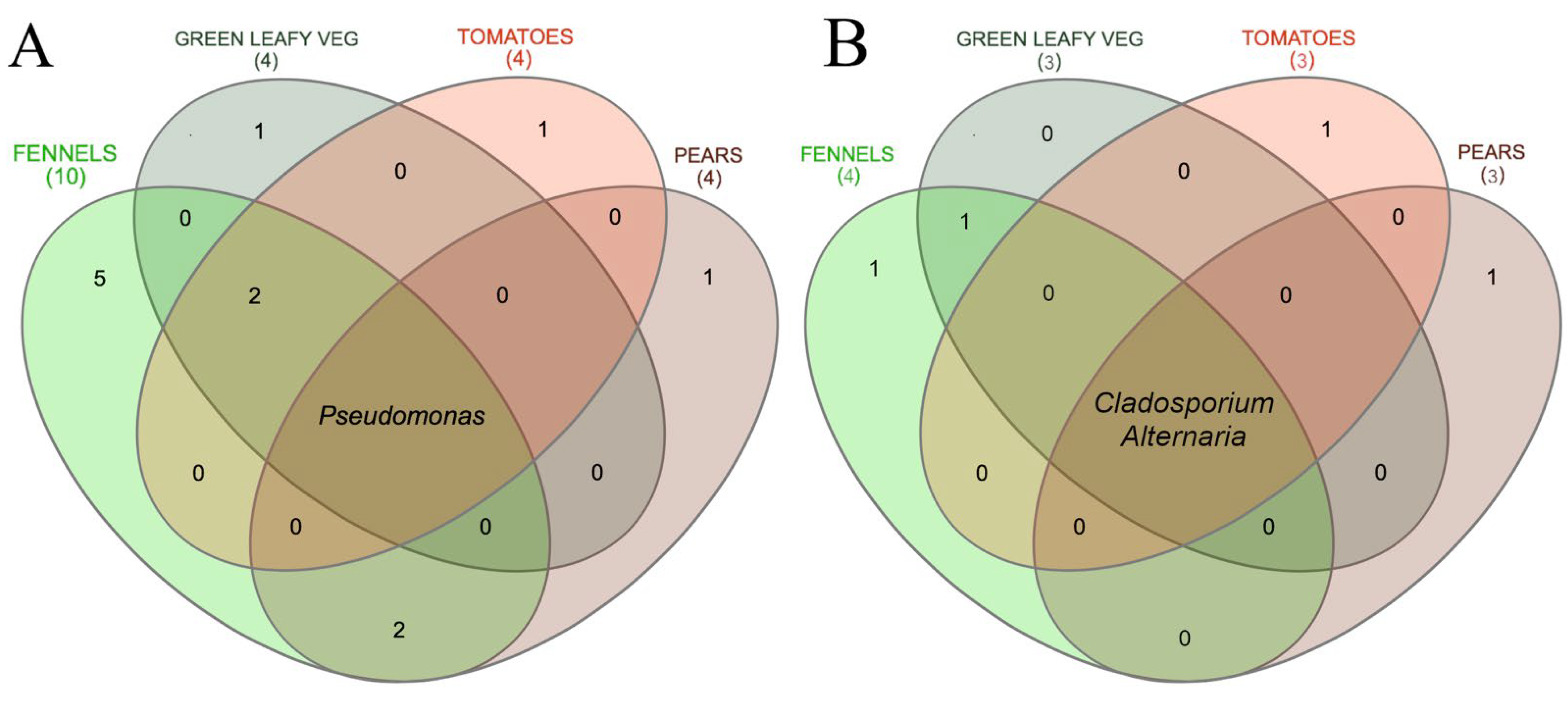
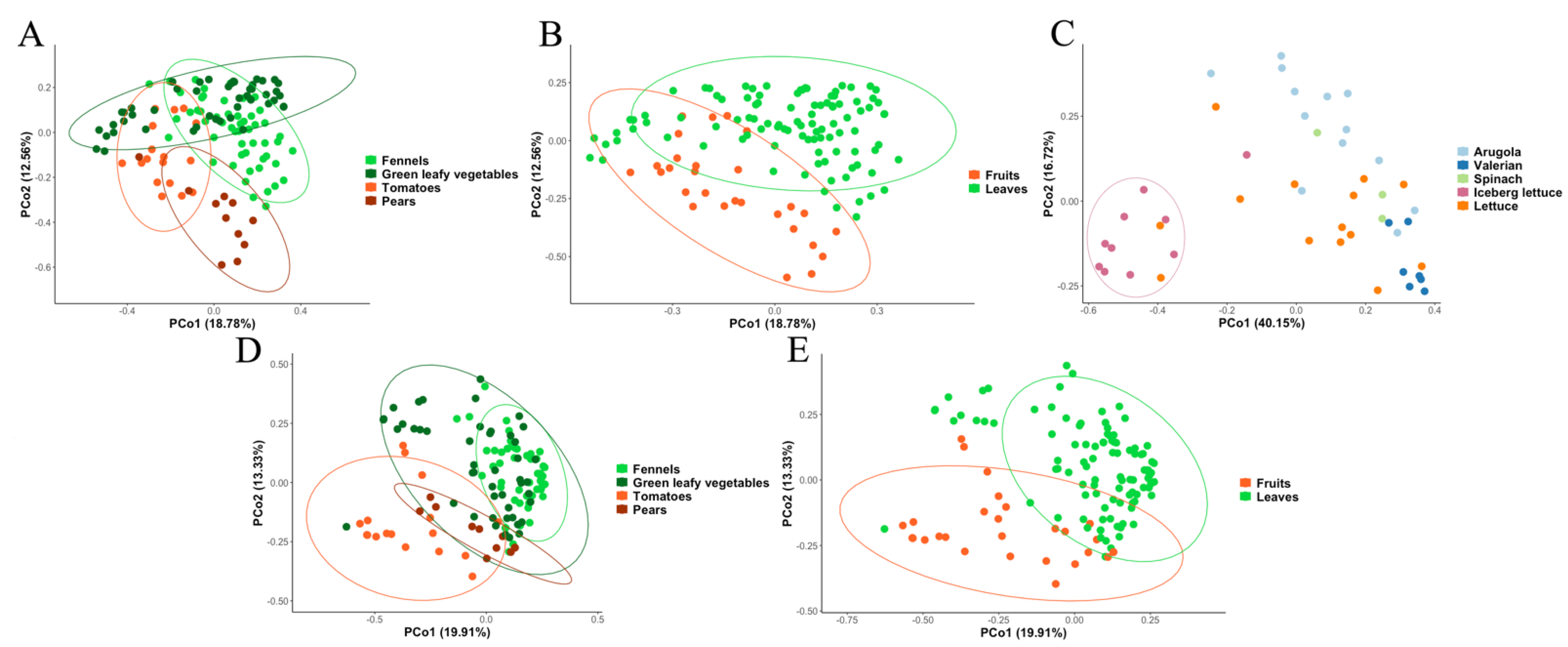
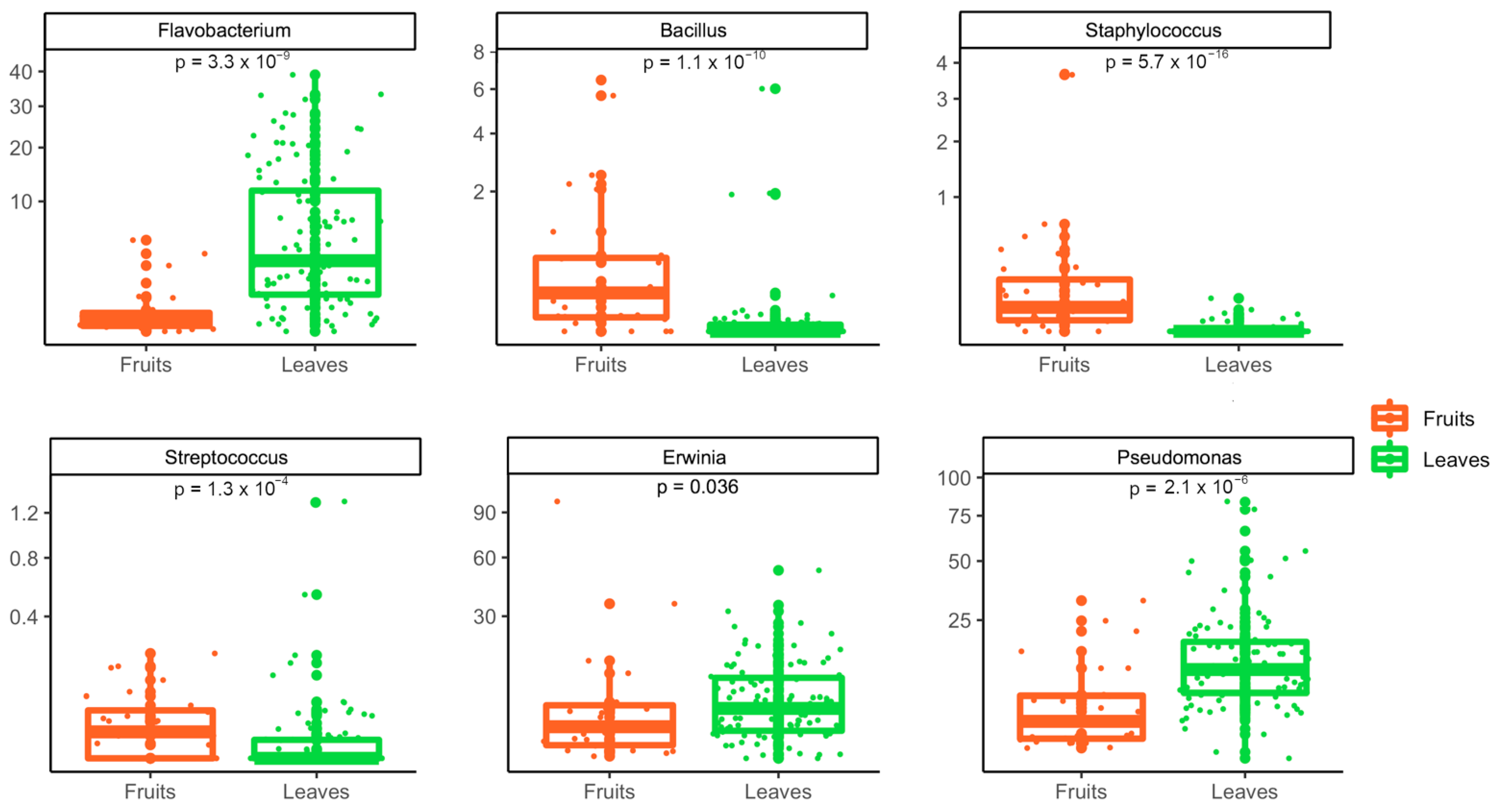
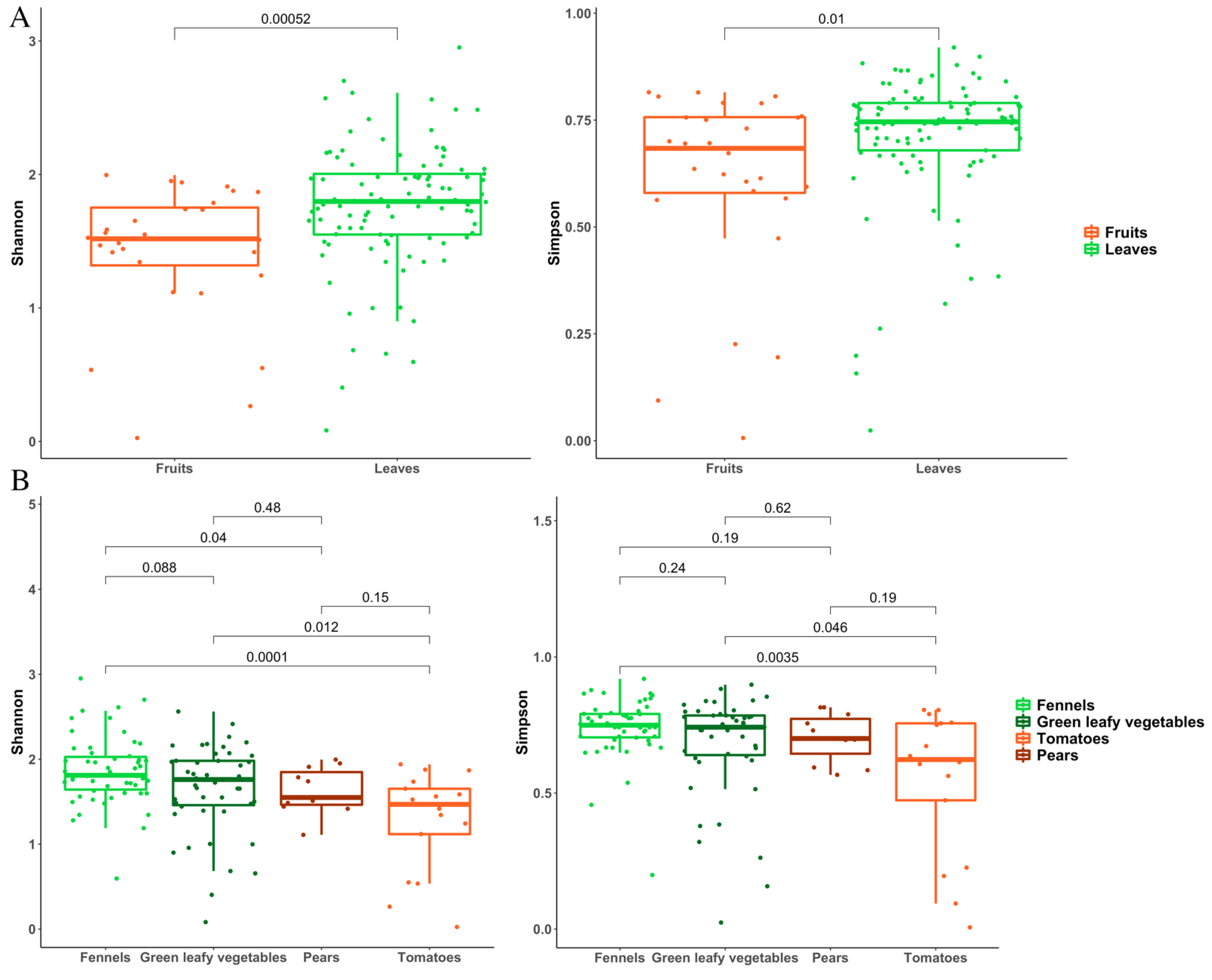
3.2. Metagenomic Analysis of Bacterial and Fungal Populations
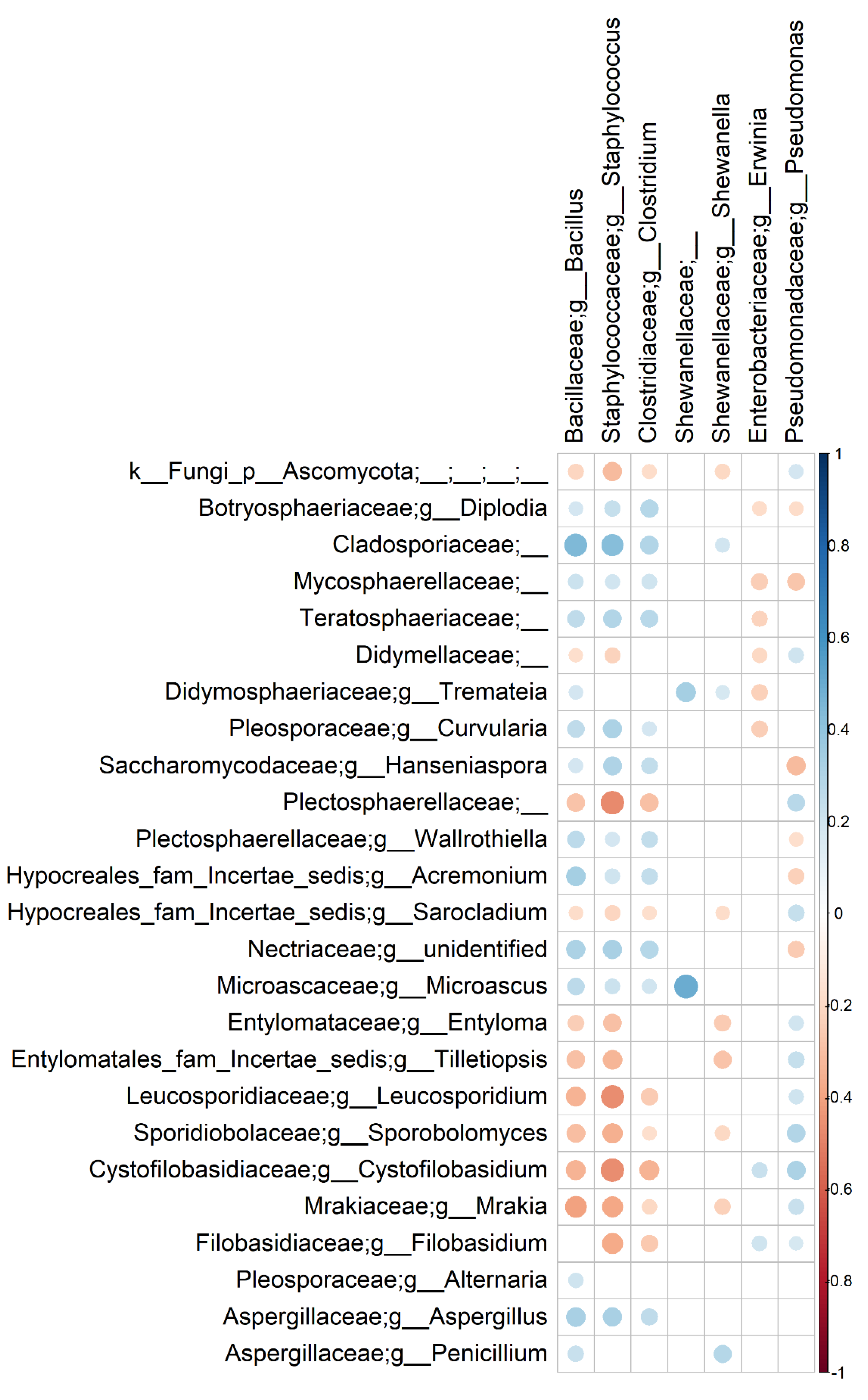
3.3. Bacterial and Fungal Communities Are Correlated
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Artés, F.; Allende, A. 26—Minimal fresh processing of vegetables, fruits and juices. In Emerging Technologies for Food Processing, 1st ed.; Sun., D.W., Ed.; Academic Press: Cambridge, MA, USA, 2005; Volume 1, pp. 677–716. [Google Scholar]
- Allende, A.; Jacxsens, L.; Devlieghere, F.; Debevere, J.; Artés, F. Effect of superatmopsheric oxygen packaging on sensorial quality, spoilage, and Listeria monocytogenes and Aeromonas caviae growth in fresh processed mixed salads. J. Food Prod. 2002, 65, 1565–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beuchat, L.R. Pathogenic microorganisms associated with fresh produce. J. Food Prod. 1996, 59, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407–415. [Google Scholar] [CrossRef]
- Callejon, R.M.; Rodríguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; Garcia-Parrilla, M.C.; Troncoso, A.M. Reported foodborne outbreaks due to fresh produce in the United States and European Union: Trends and causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef]
- Carstens, C.K.; Salazar, J.K.; Darkoh, C. Multistate outbreaks of foodborne illness in the United States associated with fresh produce from 2010 to 2017. Front. Microbiol. 2019, 10, 2667. [Google Scholar] [CrossRef] [Green Version]
- Iwu, C.D.; Okoh, A.I. Preharvest transmission routes of fresh produce associated bacterial pathogens with outbreak potentials: A review. Int. J. Environ. Res. Public Health 2019, 16, 4407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leff, J.W.; Fierer, N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef] [Green Version]
- Williams, T.R.; Moyne, A.L.; Harris, L.J.; Marco, M.L. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS ONE 2013, 8, e68642. [Google Scholar] [CrossRef]
- Kim, M.S.; Park, E.J. Postharvest-induced microbiota remodeling increases fungal diversity in the phyllosphere microbiota of broccoli florets. Postharvest Biol. Technol. 2021, 181, 111693. [Google Scholar] [CrossRef]
- Luna-Guevara, J.J.; Arenas-Hernandez, M.M.; Martinez de la Peña, C.; Silva, J.L.; Luna-Guevara, M.L. The role of pathogenic E. coli in fresh vegetables: Behavior, contamination factors, and preventive measures. Int. J. Microbiol. 2019, 2894328. [Google Scholar]
- Kusstatscher, P.; Zachow, C.; Harms, K.; Maier, J.; Eignes, H.; Berg, G.; Cernava, T. Microbiome-driven identification of microbial indicators for postharvest diseases of sugar beets. Microbiome 2019, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Barry-Ryan, C.; Pacussi, J.M.; O’beirne, D. Quality of shredded carrots as affected by packaging film and storage temperature. J. Food Sci. 2020, 65, 726–730. [Google Scholar] [CrossRef] [Green Version]
- Abdelfattah, A.; Whitehead, S.R.; Macarisin, D.; Liu, J.; Burchard, E.; Freilich, S.; Dardick, C.; Droby, S.; Wisniewski, M. Effect of washing, waxing and low-temperature storage on the postharvest microbiome of apple. Microorganisms 2020, 8, 944. [Google Scholar] [CrossRef]
- Parish, M.E.; Beuchat, L.R.; Suslow, T.V.; Harris, L.J.; Garrett, E.H.; Farber, J.N.; Busta, F.F. Methods to reduce/eliminate pathogens from fresh-cut produce. Compr. Rev. Food Sci. Food Saf. 2003, 2, 161–173. [Google Scholar] [CrossRef]
- Zhao, P.; Ndayambaje, J.P.; Liu, X.; Xia, X. Microbial spoilage of fruits: A review on causes and prevention methods. Food Rev. Intern. 2020, 1–20. [Google Scholar] [CrossRef]
- Buchholz, F.; Kostić, T.; Sessitsch, A.; Mitter, B. The potential of plant microbiota in reducing postharvest food loss. Microb. Biotechnol. 2018, 11, 971–975. [Google Scholar] [CrossRef] [Green Version]
- Leneveu-Jenvrin, C.; Charles, F.; Barba, F.J.; Remize, F. Role of biological control agents and physical treatments in maintaining the quality of fresh and minimally-processed fruit and vegetables. Crit. Rev. Food Sci. Nutr. 2020, 60, 2837–2855. [Google Scholar] [CrossRef]
- Tournas, V.H. Moulds and yeasts in fresh and minimally processed vegetables, and sprouts. Int. J. Food Microbiol. 2005, 99, 71–77. [Google Scholar] [CrossRef]
- Snyder, A.B.; Churey, J.J.; Worobo, R.W. Association of fungal genera from spoiled processed foods with physiochemical food properties and processing conditions. Food Microbiol. 2019, 83, 211–218. [Google Scholar] [CrossRef]
- Erkmen, O.; Bozoglu, T.F. Spoilage of vegetables and fruits. In Food Microbiology, Principles into Practice, 1st ed.; Erkmen, O., Bozoglu, T.F., Eds.; Wiley: Hoboken, NJ, USA, 2016; pp. 337–363. [Google Scholar]
- Oumer, O.J. Pectinase: Substrate, Production and their Biotechnological Applications. Int. J. Environ. Agric. Biotech. 2017, 2, 1007–1014. [Google Scholar] [CrossRef]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studied. Nucleic Acid. Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Laiola, M.; Paparo, L.; Calignano, A.; De Caro, C.; Coretti, L.; Chiarotti, L.; Gilbert, J.A.; et al. Specific signatures of the gut microbiota and increased levels of butyrate in children treated with fermented cow’s milk containing heat-killed Lactobacillus paracasei CBA L74. Appl. Environ. Microbiol. 2017, 83, e01206-17. [Google Scholar] [CrossRef] [Green Version]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved bacterial 16S rRNA Gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 2015, 1, e00009-15. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideut, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classification. Nucl. Acid. Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 2015, 16, 169. [Google Scholar] [CrossRef]
- Prasanna, V.; Prabha, T.N.; Tharanathan, R.N. Fruit ripening phenomena—An overview. Crit. Rev. Food Sci. Nutr. 2007, 47, 1–19. [Google Scholar] [CrossRef]
- Lee, Y.L.; Cesario, T.; Wang, Y.; Shanbrom, E.; Thrupp, L. Antibacterial activity of vegetables and juices. Nutrition 2003, 19, 994–996. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Kapoor, H.C. Antioxidants in fruits and vegetables—The millennium’s health. Int. J. Food Sci. 2001, 36, 703–725. [Google Scholar] [CrossRef]
- Beuchat, L.R. Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes Infect. 2002, 4, 413–423. [Google Scholar] [CrossRef]
- Hassan, S.A.; Altalhi, A.D.; Gherbawy, Y.A.; El-Deeb, B.A. Bacterial load of fresh vegetables and their resistance to the currently used antibiotics in Saudi Arabia. Foodborne Pathog. Dis. 2011, 8, 1011–1018. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Holley, R.A. Factors influencing the microbial safety of fresh produce: A review. Food Microbiol. 2012, 32, 1–19. [Google Scholar] [CrossRef]
- Uhlig, E.; Olsson, C.; He, J.; Stark, T.; Sadowska, Z.; Molin, G.; Ahrne, S.; Alsanius, B.; Hakansson, A. Effects of household washing on bacterial load and removal of Escherichia coli from lettuce and ready-to-eat salads. Food Sci. Nutr. 2017, 5, 1215–1220. [Google Scholar] [CrossRef]
- Tatsika, S.; Karamanoli, K.; Karayanni, H.; Genitsaris, S. Metagenomic characterization of bacterial communities on ready-to-eat vegetables and effects of household washing on their diversity and composition. Pathogens 2019, 8, 37. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Santoni, S.; Weber, A.; This, P.; Péros, J.P. Understanding the phyllosphere microbiome assemblage in grape species (Vitaceae) with amplicon sequence data structures. Sci. Rep. 2019, 9, 14294. [Google Scholar] [CrossRef] [Green Version]
- Jia, T.; Yao, Y.; Guo, T.; Wang, R.; Chai, B. Effects of Plant and Soil Characteristics on Phyllosphere and Rhizosphere Fungal Communities during Plant Development in a Copper Tailing Dam. Front. Microbiol. 2020, 11, 556002. [Google Scholar] [CrossRef]
- Stanevičienė, R.; Lukša, J.; Strazdaitė-Žielienė, Z.; Ravoitytė, B.; Losinska-Sičiūnienė, R.; Mozūraitis, R.; Servienė, E. Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts. Microorganisms 2021, 9, 1423. [Google Scholar] [CrossRef]
- Kokaeva, L.; Chudinova, E.; Berezov, A.; Yarmeeva, M.; Balabko, P.; Belosokhov, A.; Elansky, S. Fungal diversity in tomato (Solanum lycopersicum) leaves and fruits in Russia. J. Cent. Eur. Agric. 2020, 21, 809–816. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J.; Tech, J.J. Grapevine microbiomics: Bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. In International Symposium on Biological Control of Postharvest Diseases: Challenges and Opportunities; ISHS: Leesburg, VA, USA, 2011; pp. 31–42. [Google Scholar]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Velasco, G.; Welbaum, G.E.; Boyer, R.R.; Mane, S.P.; Ponder, M.A. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J. Appl. Microbiol. 2011, 110, 1203–1214. [Google Scholar] [CrossRef]
- Kim, M.S.; Bae, J.W.; Park, E.J. Geographic and host-associated variations in bacterial communities on the floret surfaces of field-grown broccoli. Appl. Environ. Microbiol. 2018, 84, e02837-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.T.; Lin, Y.T.; Wan, T.W.; Wang, D.Y.; Lin, H.Y.; Lin, C.Y.; Chen, Y.C.; Teng, L.J. Distribution of antibiotic resistance genes among Staphylococcus species isolated from ready-to-eat foods. J. Food Drug Anal. 2019, 27, 841–848. [Google Scholar] [CrossRef] [Green Version]
- Quintieri, L.; Fanelli, F.; Caputo, L. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: An underestimated risk and the control strategies. Foods 2019, 8, 372. [Google Scholar] [CrossRef] [Green Version]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: A need for quantitative risk assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Schroth, M.N.; Cho, J.J.; Green, S.K.; Kominos, S.D. Epidemiology of Pseudomonas aeruginosa in agricultural areas. J. Med. Microbiol. 2018, 67, 1191–1201. [Google Scholar] [CrossRef]
- Ruiz-Roldán, L.; Rojo-Bezares, B.; Lozano, C.; López, M.; Chichón, G.; Torres, C.; Sáenz, Y. Occurrence of Pseudomonas spp. in raw vegetables: Molecular and phenotypical analysis of their antimicrobial resistance and virulence-related traits. Int. J. Mol. Sci. 2021, 22, 12626. [Google Scholar] [CrossRef]
- Pinto, L.; Ippolito, A.; Baruzzi, F. Control of spoiler Pseudomonas spp. on fresh cut vegetables by neutral electrolyzed water. Food Microbiol. 2015, 50, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Sanzani, S.M.; Reverberi, M.; Geisen, R. Mycotoxins in harvested fruits and vegetables: Insights in producing fungi, biological role, conducive conditions, and tools to manage postharvest contamination. Postharvest Biol. Technol. 2016, 122, 95–105. [Google Scholar] [CrossRef]
- Amiri, A.; Bompeix, G. Diversity and population dynamics of Penicillium spp. on apples in pre- and postharvest environments: Consequences for decay development. Plant Pathol. 2005, 54, 74–81. [Google Scholar] [CrossRef]
- Logrieco, A.; Ferracane, R.; Haidukowsky, M.; Cozzi, G.; Visconti, A.; Ritieni, A. Fumonisin B2 production by Aspergillus niger from grapes and natural occurrence in must. Food Addit. Contamin 2009, 26, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Barkai-Golan, R. Aspergillus mycotoxins. In Mycotoxins in Fruits and Vegetables, 1st ed.; Barkai-Golan, R., Paster, N., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 115–151. [Google Scholar]
- Punja, Z.K.; Rodriguez, G.; Tirajoh, A.; Formby, S. Role of fruit surface mycoflora, wounding and storage conditions on post-harvest disease development on greenhouse tomatoes. Can. J. Plant Pathol. 2016, 38, 448–459. [Google Scholar] [CrossRef]
- Vilanova, L.; Vall-Llaura, N.; Torres, R.; Usall, J.; Teixidò, N.; Larrigaudière, C.; Giné-Bordonaba, J. Penicillium expansum (compatible) and Penicillium digitatum (non-host) pathogen infection differentially alter ethylene biosynthesis in apple fruit. Plant Physiol. Biochem. 2017, 120, 132–143. [Google Scholar] [CrossRef]
- Rooney-Latham, S.; Lutz, M.; Blomquist, C.L.; Romberg, M.K.; Scheck, H.J.; Piątek, M. Entyloma helianthi: Identification and characterization of the causal agent of sunflower white leaf smut. Mycologia 2017, 109, 520–528. [Google Scholar] [CrossRef]
- Ortega, S.F.; Ferrocino, I.; Adams, I.; Silvestri, S.; Spadaro, D.; Gullino, M.L.; Boonham, N. Monitoring and surveillance of aerial mycobiota of rice paddy through DNA metabarcoding and qPCR. J. Fungi 2020, 6, 372. [Google Scholar] [CrossRef]
- Liu, D. Sporobolomyces. In Molecular Detection of Human Fungal Pathogens, 1st ed.; Liu, D., Ed.; CRC Press: Boca Raton, FL, USA, 2011; pp. 677–680. [Google Scholar]
- Arendrup, M.M.; Boekhout, T.; Akova, M.; Meis, J.F.; Cornely, O.A.; Lorholary, O.; European Society of Clinical Microbiology and Infectious Diseases Fungal Infection Study Group; European Confederation of Medical Mycology. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin. Microbiol. Infect. 2014, 20 (Suppl. S3), 76–98. [Google Scholar] [CrossRef] [Green Version]
- Kadke, U.B.; Kadke, H.U. Incidence of post-harvest disease and airborne fungal spores in a vegetable market. Acta Bot. Croat. 2012, 71, 147–157. [Google Scholar]
- Anees-Hill, S.; Douglas, P.; Pashley, C.H.; Hansell, A.; Marczylo, E.L. A systematic review of outdoor airborne fungal spore seasonality across Europe and the implications for health. Sci. Total Environ. 2021, 818, 151716. [Google Scholar] [CrossRef] [PubMed]
- López Couso, V.P.; Tortajada-Girbés, M.; Rodriguez Gil, D.; Martínez Quesada, J.; Palacios Pelaez, R. Fungi Sensitization in Spain: Importance of Alternaria alternata species and its major allergen Alt a 1 in the allergenicity. J. Fungi 2021, 7, 631. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Yeasts and Molds (28 °C) | Total Psychrotrophic Anaerobic Counts (20 °C) | Total Psychrotrophic Aerobic Counts (20 °C) | Enterobacteriaceae (37 °C) | |
|---|---|---|---|---|
| Green leafy vegetables (n = 47) | 4.20 ± 0.82 a | 7.27 ± 0.96 a | 7.56 ± 0.48 a | 4.53 ± 0.89 a |
| Tomatoes (n = 17) | 2.99 ± 0.72 b | 4.13 ± 0.96 b | 4.38 ± 1.14 b | 2.30 ± 1.16 b |
| Fennels (n = 54) | 3.91 ± 1.66 a | 6.47 ± 1.23 a | 6.74 ± 0.82 a | 3.49 ± 1.20 b |
| Pears (n = 11) | 3.38 ± 0.72 a,b | 4.20 ± 0.88 b | 4.29 ± 0.83 b | 1.32 ± 1.94 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sequino, G.; Valentino, V.; Torrieri, E.; De Filippis, F. Specific Microbial Communities Are Selected in Minimally-Processed Fruit and Vegetables according to the Type of Product. Foods 2022, 11, 2164. https://doi.org/10.3390/foods11142164
Sequino G, Valentino V, Torrieri E, De Filippis F. Specific Microbial Communities Are Selected in Minimally-Processed Fruit and Vegetables according to the Type of Product. Foods. 2022; 11(14):2164. https://doi.org/10.3390/foods11142164
Chicago/Turabian StyleSequino, Giuseppina, Vincenzo Valentino, Elena Torrieri, and Francesca De Filippis. 2022. "Specific Microbial Communities Are Selected in Minimally-Processed Fruit and Vegetables according to the Type of Product" Foods 11, no. 14: 2164. https://doi.org/10.3390/foods11142164
APA StyleSequino, G., Valentino, V., Torrieri, E., & De Filippis, F. (2022). Specific Microbial Communities Are Selected in Minimally-Processed Fruit and Vegetables according to the Type of Product. Foods, 11(14), 2164. https://doi.org/10.3390/foods11142164








