International Spread of Tet(X4)-Producing Escherichia coli Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. E. coli Genomes Collected
2.2. AMR Genotypes, Virulence Genotypes, and Plasmid Replicon Types Identification
2.3. Phylogenomic Analysis
2.4. Local Database Construction and BLAST+ Comparison
2.5. Data Analysis
3. Results
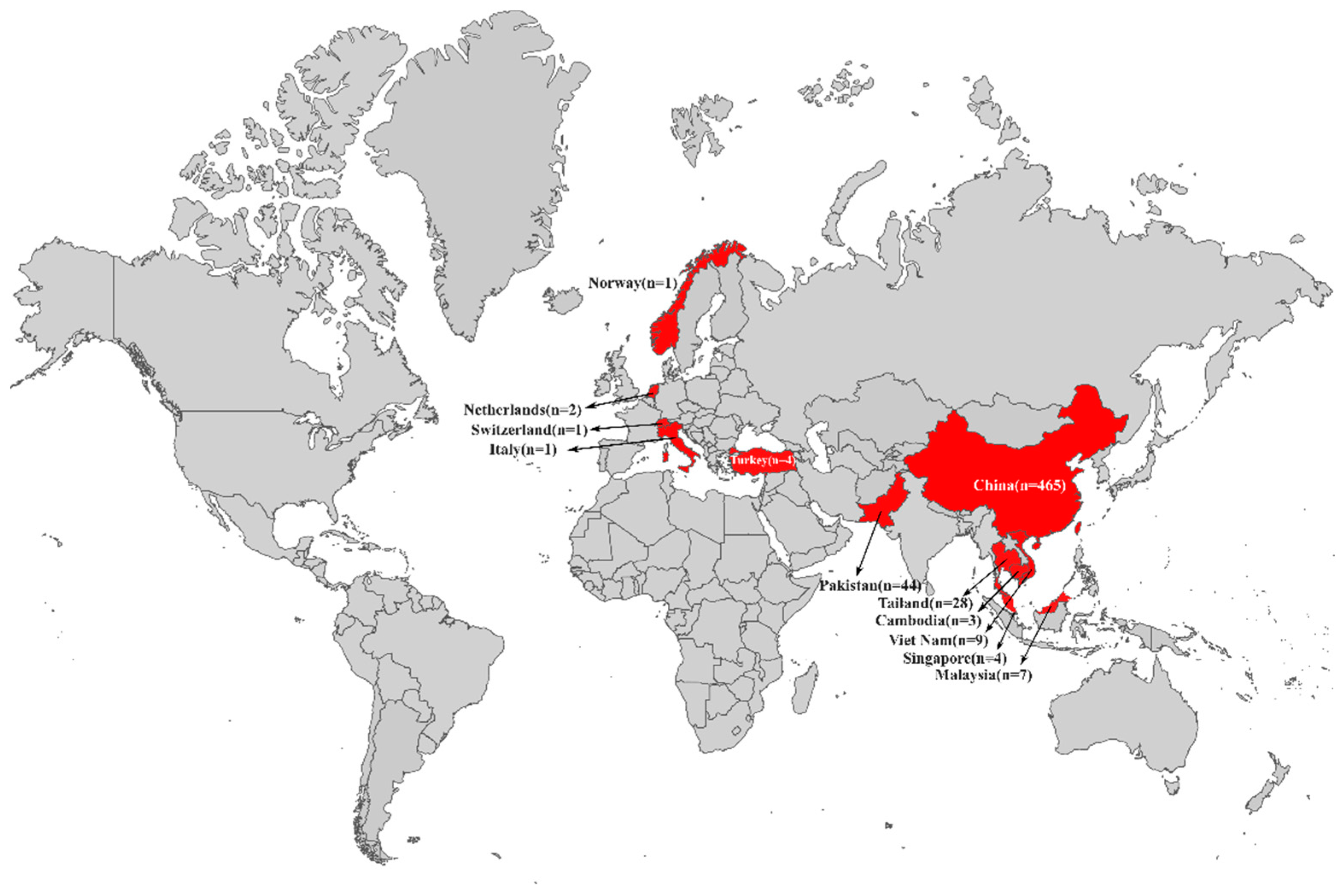
3.1. Dissemination of tet(X4)-Producing E. coli in Asia and Europe
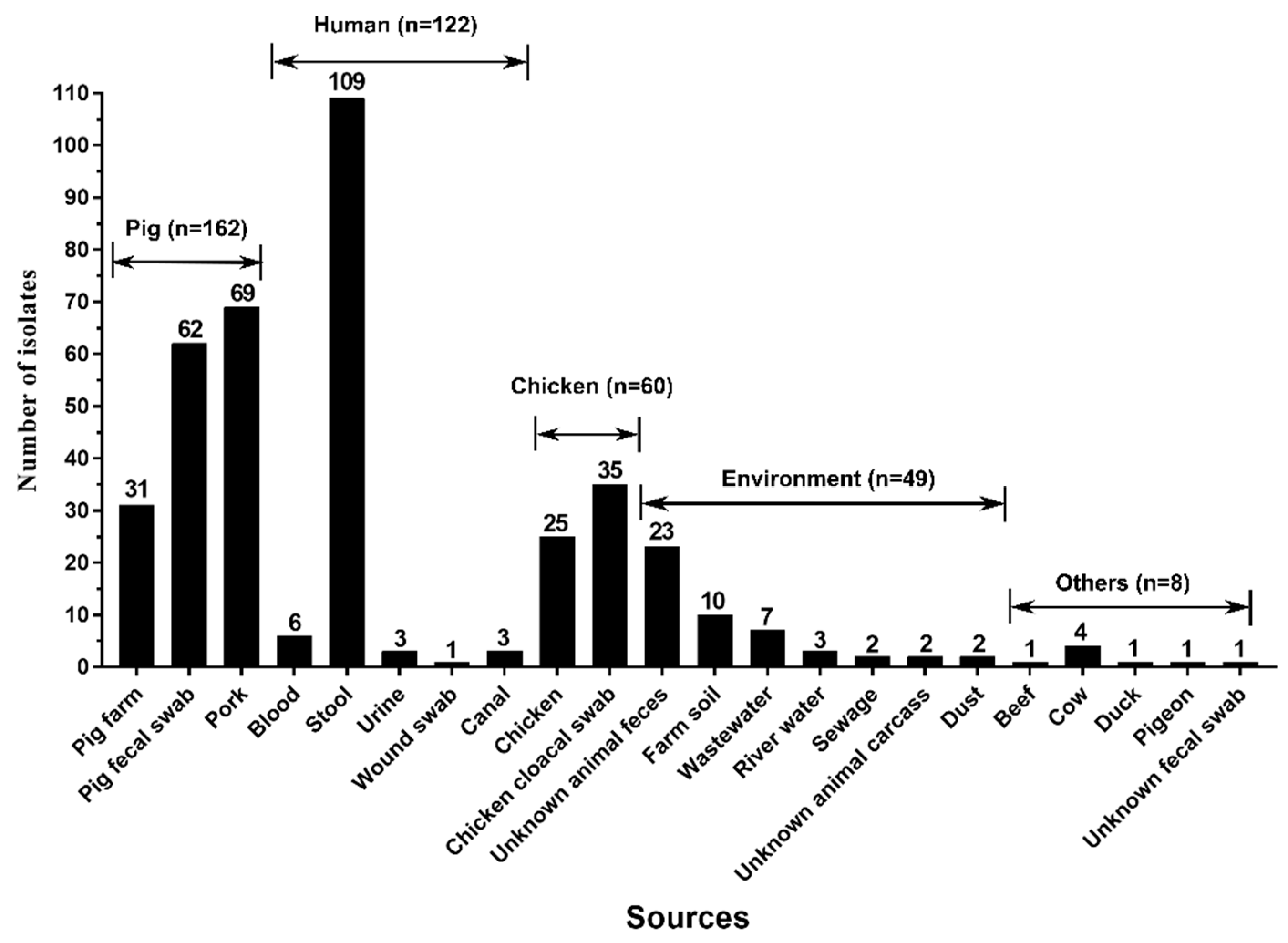
3.2. Pig and Its Products Were the Main Vehicles of tet(X4)-Producing E. coli
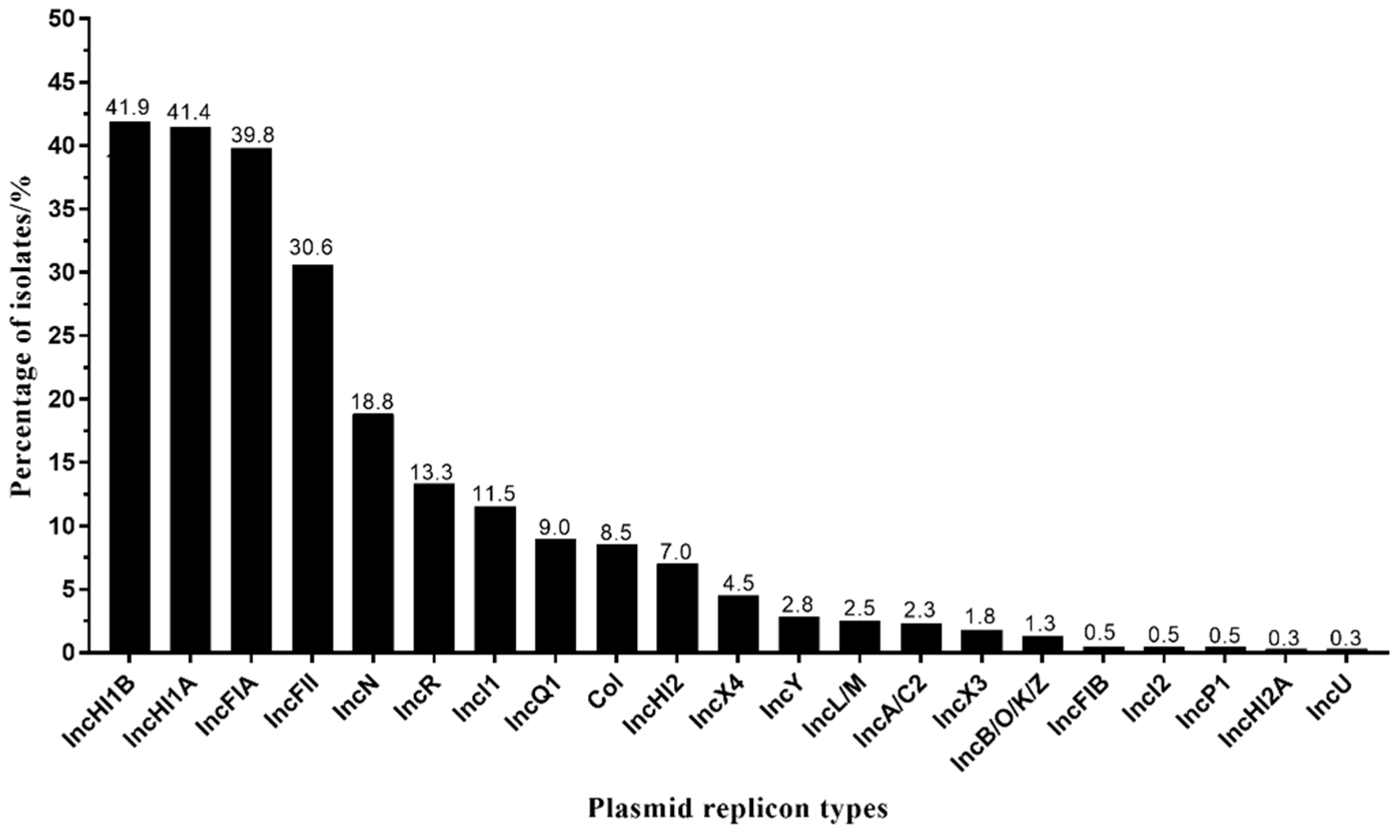
3.3. IncHI1A and IncHI1B Were the Dominant Plasmid Types in tet(X4)-Producing E. coli
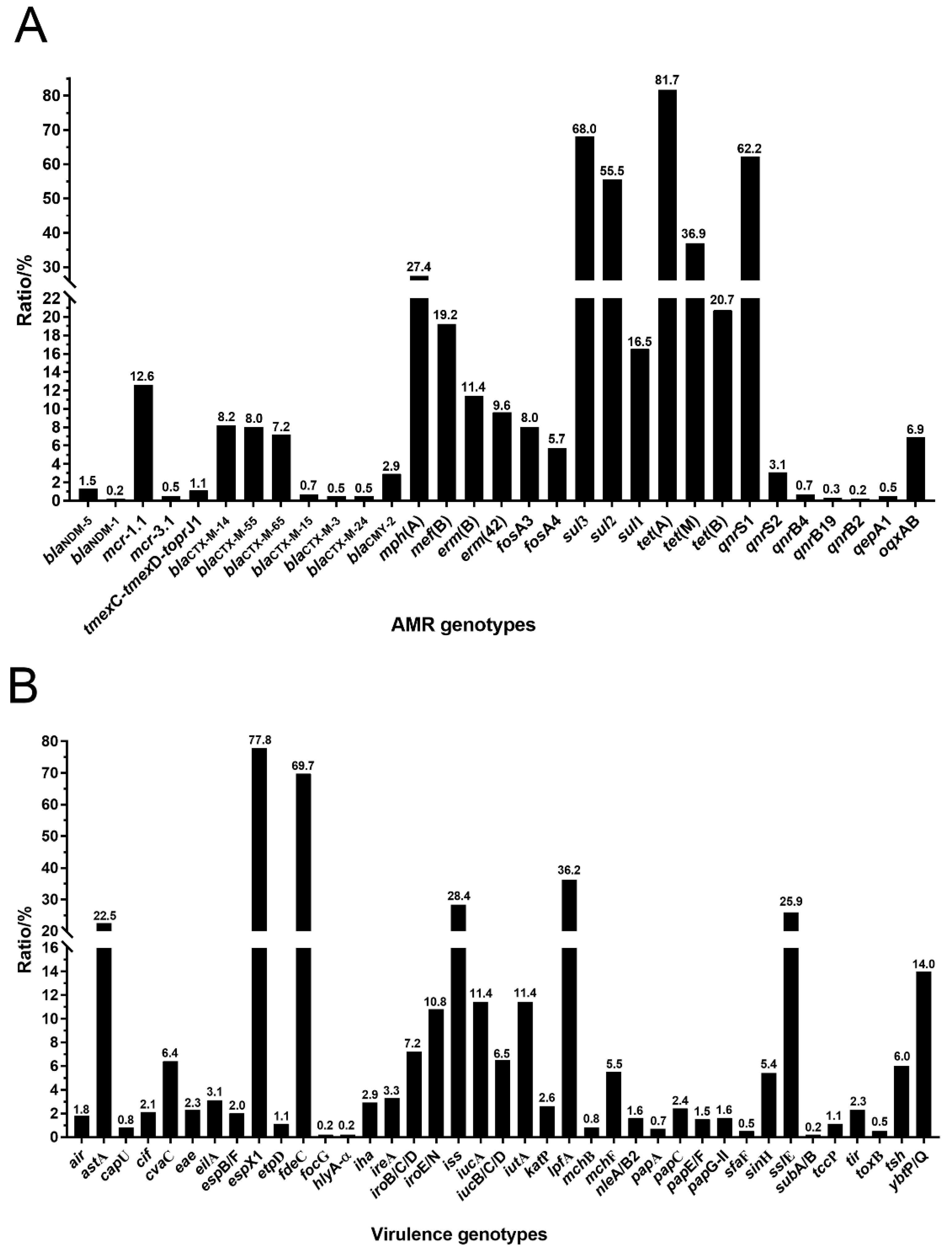
3.4. Antimicrobial Resistance (AMR) Genotypes and Virulence Genotypes in tet(X4)-Producing E. coli
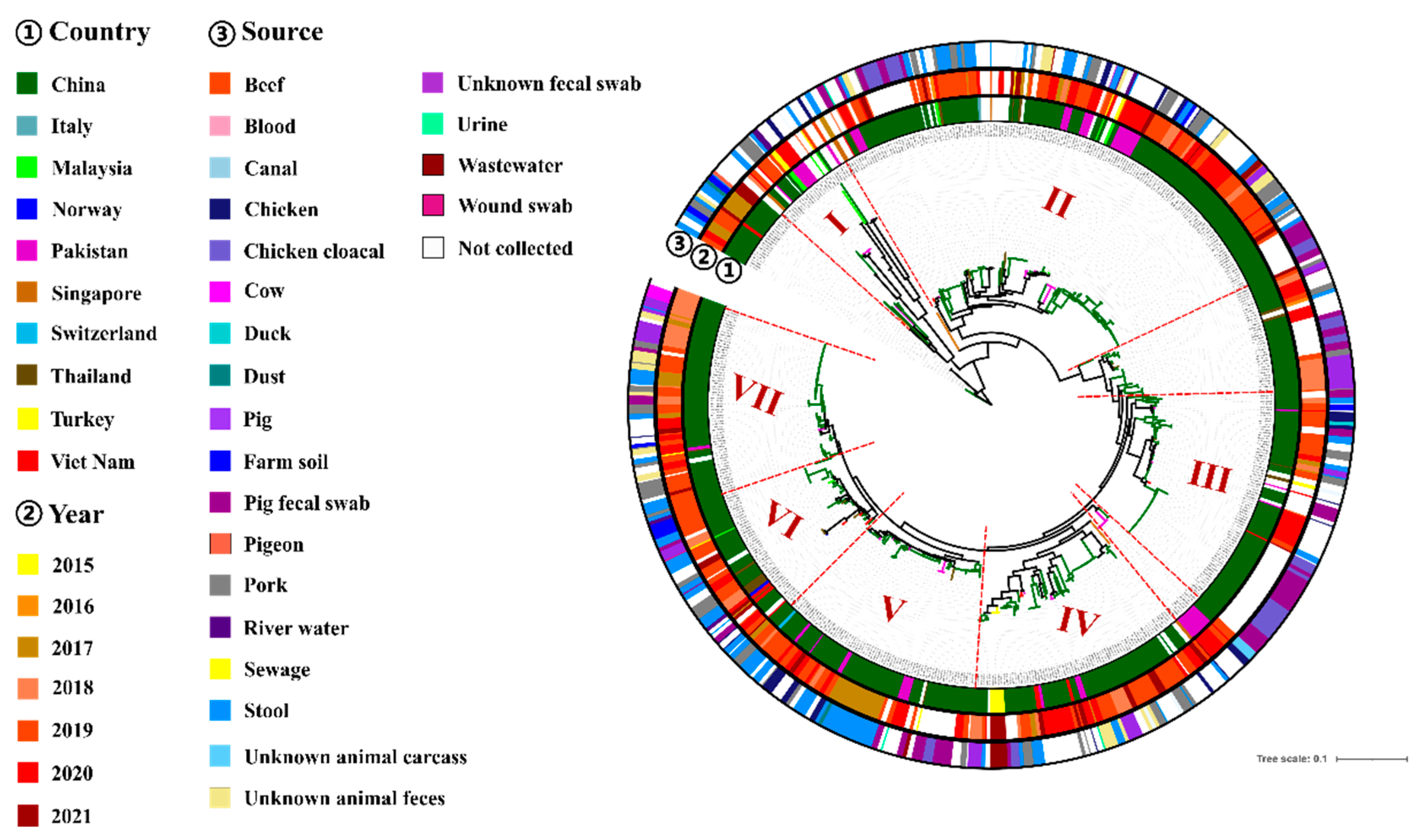
3.5. Phylogenomic Analysis of tet(X4)-Producing E. coli
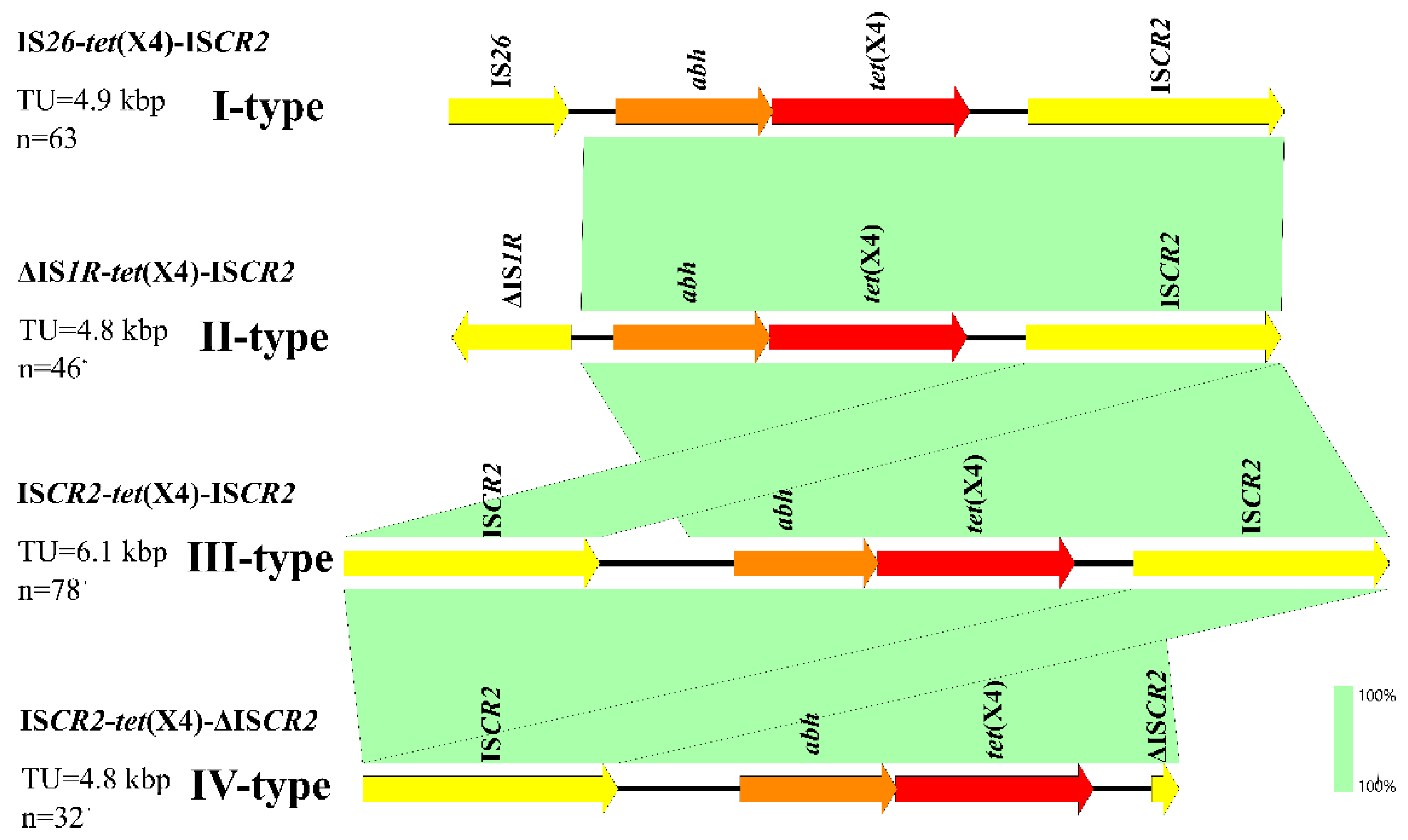
3.6. Genetic Environment of tet(X4) in E. coli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gerson, S.; Nowak, J.; Zander, E.; Ertel, J.; Wen, Y.; Krut, O.; Seifert, H.; Higgins, P.G. Diversity of Mutations in Regulatory Genes of Resistance-Nodulation-Cell Division Efflux Pumps in Association with Tigecycline Resistance in Acinetobacter Baumannii. J. Antimicrob. Chemother. 2018, 73, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Shi, Q.; Fu, Y.; Xu, J.; Yu, Y.; Du, X. Tigecycline Resistance Caused by RpsJ Evolution in a 59-Year-Old Male Patient Infected with KPC-Producing Klebsiella pneumoniae during Tigecycline Treatment. Infect. Genet. Evol. 2018, 66, 188–191. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of Plasmid-Mediated High-Level Tigecycline Resistance Genes in Animals and Humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, C.; Cui, C.-Y.; Zhang, Y.; Liu, X.; Cui, Z.-H.; Ma, X.-Y.; Feng, Y.; Fang, L.-X.; Lian, X.-L.; et al. Plasmid-Encoded tet(X) Genes That Confer High-Level Tigecycline Resistance in E. coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Q.; Peng, K.; Liu, Y.; Xiao, X.; Mohsin, M.; Li, R.; Wang, Z. Distribution and Genomic Characterization of Tigecycline-Resistant tet(X4)-Positive E. coli of Swine Farm Origin. Microb. Genom. 2021, 7, 000667. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.-Y.; Li, X.-J.; Chen, C.; Wu, X.-T.; He, Q.; Jia, Q.-L.; Zhang, X.-J.; Lin, Z.-Y.; Li, C.; Fang, L.-X.; et al. Comprehensive Analysis of Plasmid-Mediated tet(X4)-Positive E. coli Isolates from Clinical Settings Revealed a High Correlation with Animals and Environments-Derived Strains. Sci. Total Environ. 2022, 806, 150687. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Du, P.; Du, Y.; Sun, H.; Zhang, P.; Wan, Y.; Lin, Q.; Fanning, S.; Cui, S.; Wu, Y. Detection of Plasmid-Mediated Tigecycline-Resistant Gene tet(X4) in E. coli from Pork, Sichuan and Shandong Provinces, China, February 2019. Eurosurveillance 2019, 24, 1900340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Cui, M.; Zhang, S.; Liu, D.; Fu, B.; Li, Z.; Bai, R.; Wang, Y.; Wang, H.; Song, L.; et al. Genomic Epidemiology of Animal-Derived Tigecycline-Resistant E. coli across China Reveals Recent Endemic Plasmid-Encoded tet(X4) Gene. Commun. Biol. 2020, 3, 412. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, M.; Hassan, B.; Martins, W.M.B.S.; Li, R.; Abdullah, S.; Sands, K.; Walsh, T.R. Emergence of Plasmid-Mediated Tigecycline Resistance tet(X4) Gene in E. coli Isolated from Poultry, Food and the Environment in South Asia. Sci. Total Environ. 2021, 787, 147613. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, H.; Mei, C.-Y.; Wang, Y.; Wang, Z.-Y.; Lu, M.-J.; Pan, Z.-M.; Jiao, X. Multiple Mechanisms of Tigecycline Resistance in Enterobacteriaceae from a Pig Farm, China. Microbiol. Spectr. 2021, 9, e00416-21. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xiao, X.; Liu, Y.; Li, R.; Wang, Z. Emerging Opportunity and Destiny of mcr-1- and tet(X4)-Coharboring Plasmids in E. coli. Microbiol. Spectr. 2021, 9, e01520-21. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhai, W.; Fu, Y.; Li, R.; Du, P.; Bai, L. Co-Occurrence of Plasmid-Mediated Resistance Genes tet(X4) and blaNDM-5 in a Multidrug-Resistant E. coli Isolate Recovered from Chicken in China. J. Glob. Antimicrob. Res. 2021, 24, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J.; et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio 2020, 11, e02930-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zhan, Z.; Shi, C. International Spread of Tet(X4)-Producing Escherichia coli Isolates. Foods 2022, 11, 2010. https://doi.org/10.3390/foods11142010
Zhang Z, Zhan Z, Shi C. International Spread of Tet(X4)-Producing Escherichia coli Isolates. Foods. 2022; 11(14):2010. https://doi.org/10.3390/foods11142010
Chicago/Turabian StyleZhang, Zengfeng, Zeqiang Zhan, and Chunlei Shi. 2022. "International Spread of Tet(X4)-Producing Escherichia coli Isolates" Foods 11, no. 14: 2010. https://doi.org/10.3390/foods11142010
APA StyleZhang, Z., Zhan, Z., & Shi, C. (2022). International Spread of Tet(X4)-Producing Escherichia coli Isolates. Foods, 11(14), 2010. https://doi.org/10.3390/foods11142010






