Simultaneous Determination of Nine Quinolones in Pure Milk Using PFSPE-HPLC-MS/MS with PS-PAN Nanofibers as a Sorbent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Equipment and HPLC-MS-MS Conditions
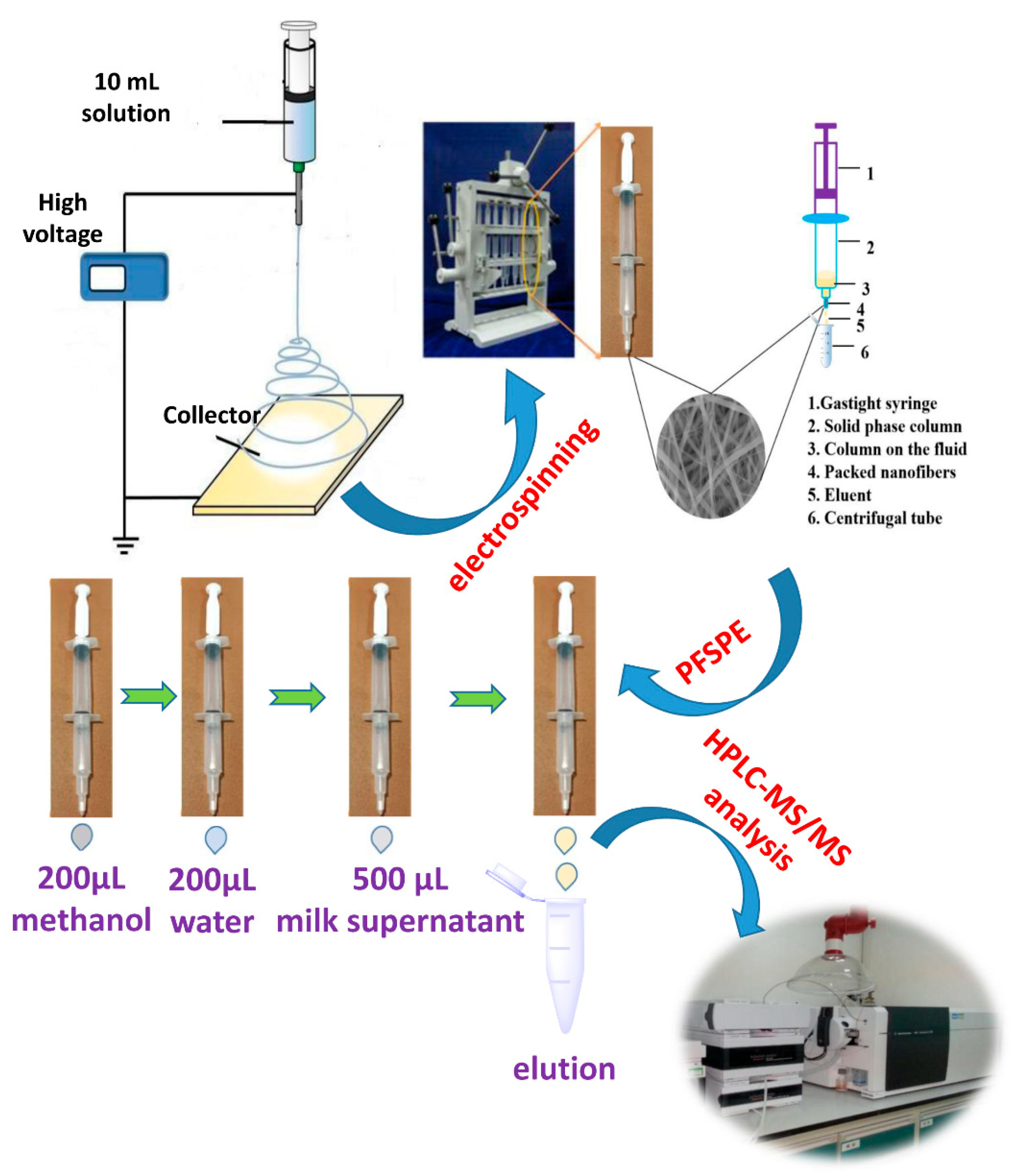
2.3. Preparation of PS-PAN Nanofibers
2.4. Characterization
2.5. Sample Pretreatment
2.6. Method Validation
3. Results and Discussion
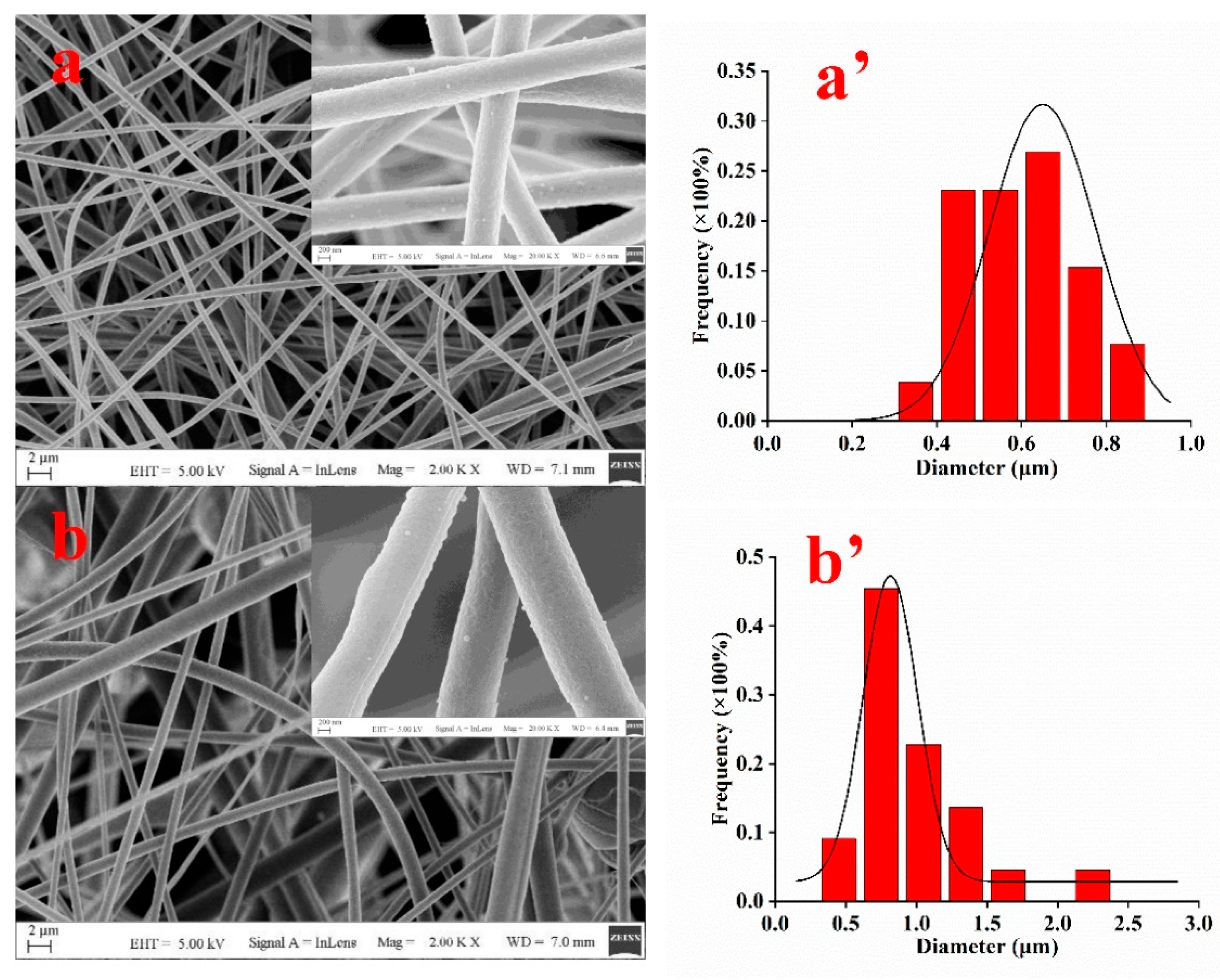
3.1. Characterization of Nanofibers
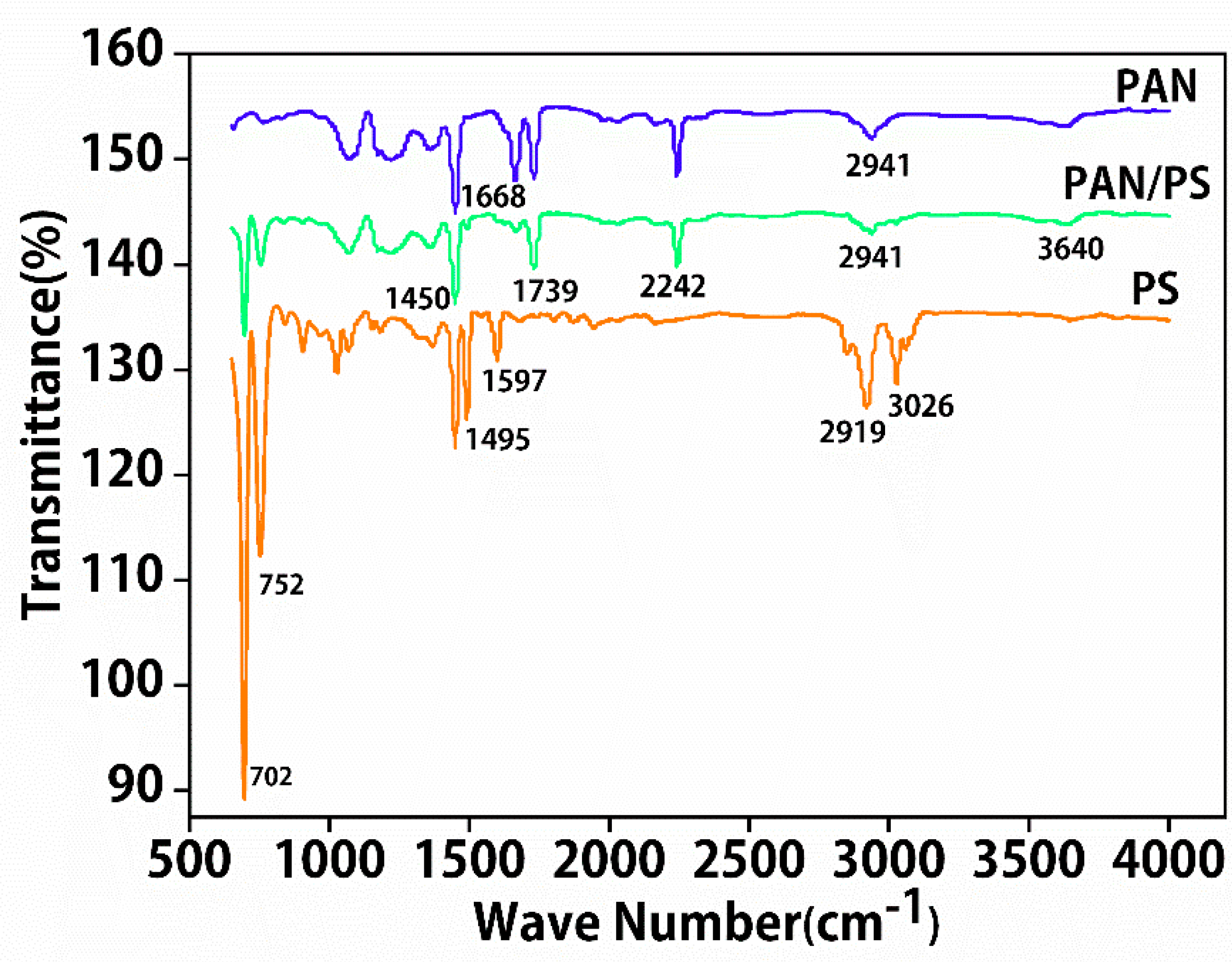
3.2. Molecular Interactions of Nanofibers
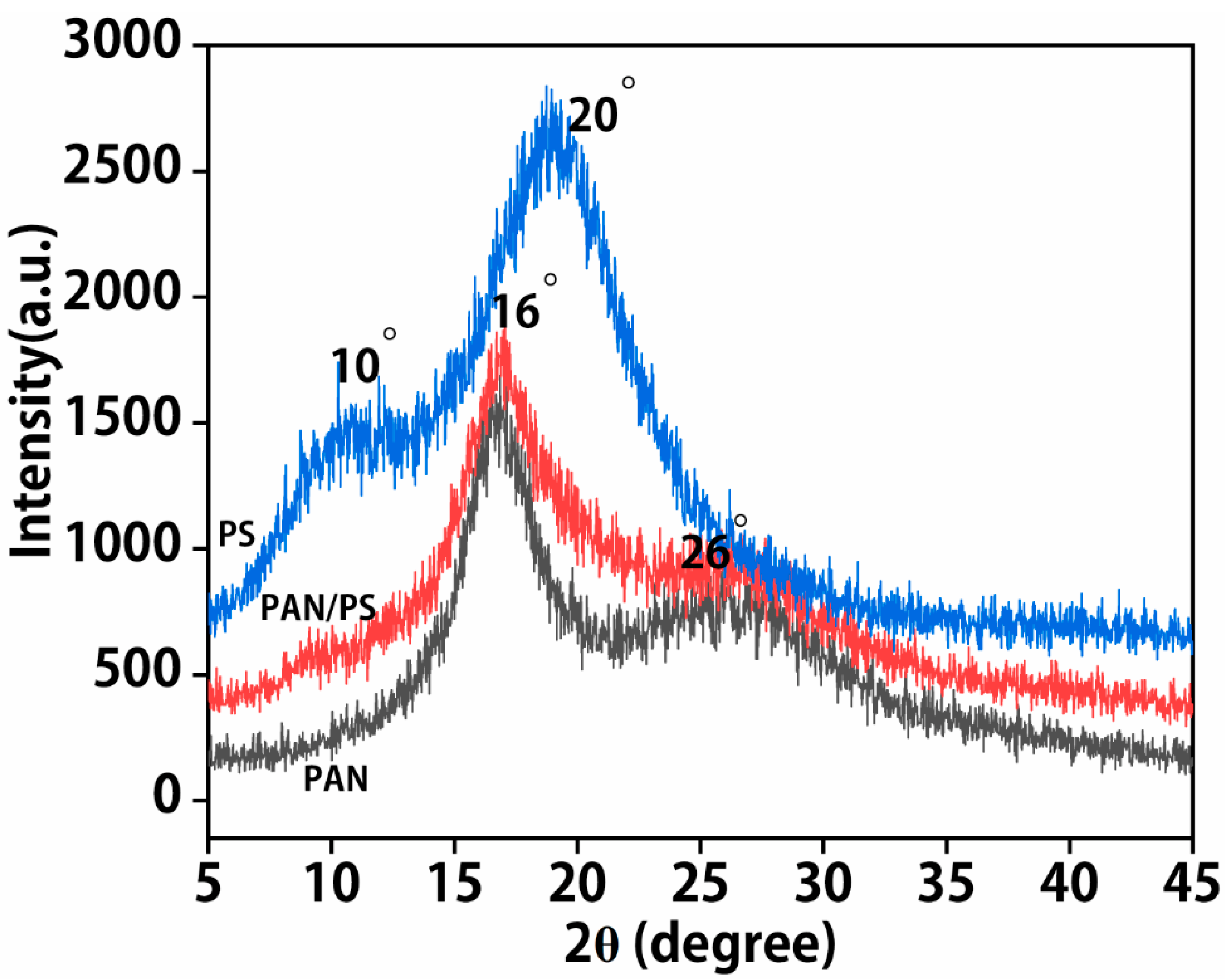
3.3. X-ray Diffraction Analysis (XRD)
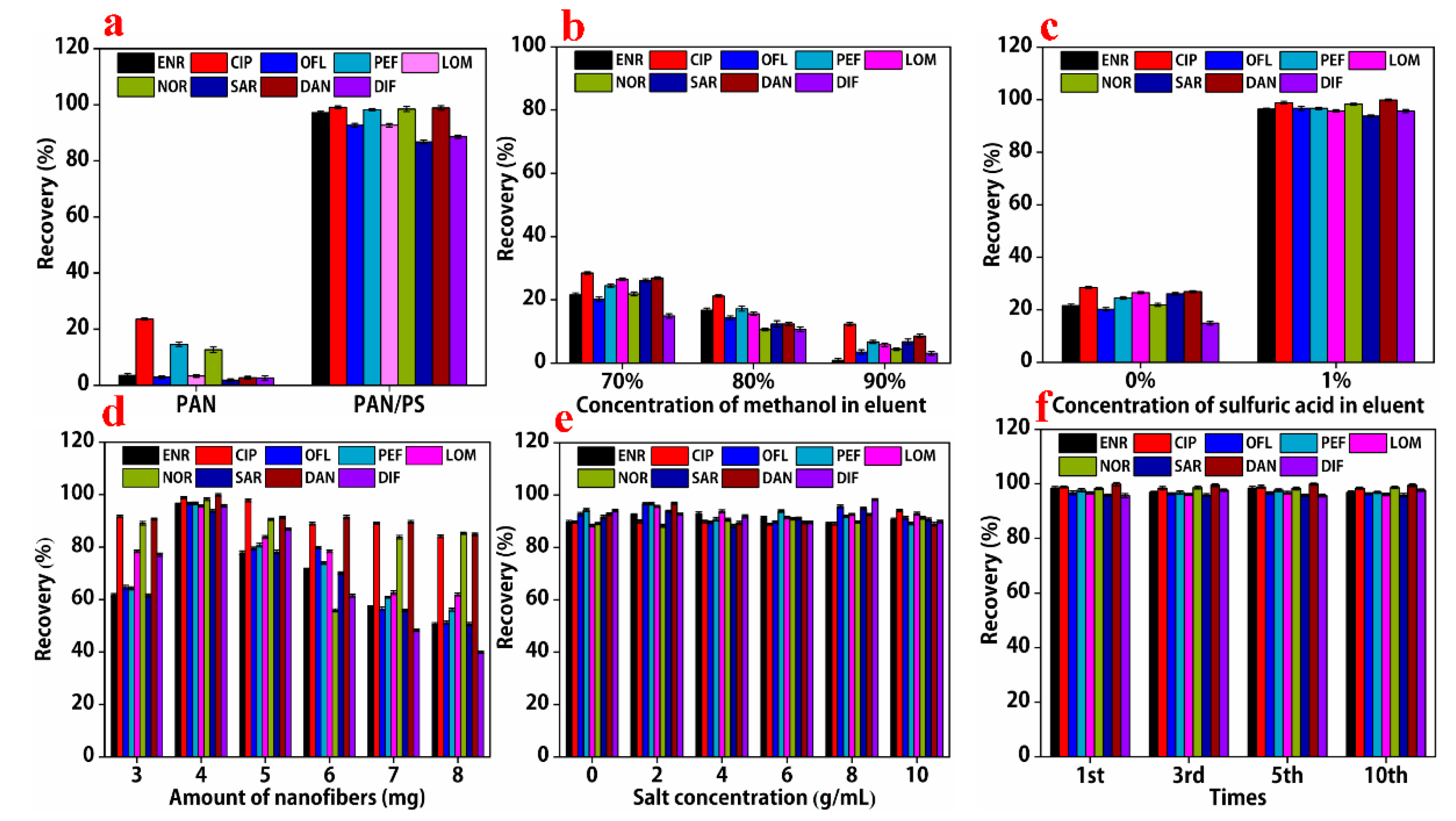
3.4. Comparison of Adsorption Efficiencies between PAN Nanofibers and PAN/PS Composite Nanofibers
3.5. Method Validation
3.6. Matrix Effect
3.7. Comparison of This Method with Other Measurement Methods
3.8. Application to Real Samples
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliveira, R.V.; Pietro, A.C.D.; Cass, Q.B. Quantification of cephalexin as residue levels in bovine milk by high-performance liquid chromatography with on-line sample cleanup. Talanta 2007, 71, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.X.; Gan, X.; Li, S.; Qin, W.D. A rapid CE-potential gradient detection method for determination of quinolones. Electrophoresis 2007, 28, 4101–4107. [Google Scholar] [CrossRef]
- Xu, J.; Li, X.; Li, C.; Xiao, Y. Hexafluoroisopropanolinduced salt-free catanionic surfactant coacervate extraction method for determination of fluoroquinolones in milk samples. Food Chem. 2018, 242, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Beucher, L.; Dervilly-Pinel, G.; Prévost, S.; Monteau, F.; Le, B. Determination of a large set of b-adrenergic agonists in animal matrices based on ion mobility and mass separations. Anal. Chem. 2015, 87, 9234–9242. [Google Scholar] [CrossRef] [PubMed]
- Cinquina, A.L.; Longo, F.; Anastasi, G.; Giannetti, L.; Cozzani, R. Validation of a high-performance liquid chromatography method for the determination of oxytetracycline, tetracycline, chlortetracycline and doxycycline in bovine milk and muscle. J. Chromatogr. A 2003, 987, 227–233. [Google Scholar] [CrossRef]
- Christodoulou, E.A.; Samanidou, V.F. Multiresidue HPLC analysis of ten quinolones in milk after solid phase extraction: Validation according to the European Union Decision 2002/657/EC. J. Sep. Sci. 2007, 30, 2421–2429. [Google Scholar] [CrossRef]
- Idowu, O.R.; Peggins, J.O. Simple, rapid determination of enrofloxacin and ciprofloxacin in bovine milk and plasma by high-performance liquid chromatography with fluorescence detection. J. Pharmaceut. Biomed. 2004, 35, 143–153. [Google Scholar] [CrossRef]
- Zheng, M.M.; Rui, G.; Xing, Z.; Feng, Y.Q. Selective sample pretreatment by molecularly imprinted polymer monolith for the analysis of fluoroquinolones from milk samples. J. Chromatogr. A 2010, 1217, 2075–2081. [Google Scholar] [CrossRef]
- Bogialli, S.; Curini, R.; Corcia, A.D.; Nazzari, M.; Sergi, M. Confirmatory analysis of sulfonamide antibacterials in bovine liver and kidney: Extraction with hot water and liquid chromatography coupled to a single- or triple-quadrupole mass spectrometer. Rapid Commun. Mass Spectrom. 2010, 17, 1146–1156. [Google Scholar] [CrossRef]
- Kantiani, L.; Farré, D.; Barceló, M. Rapid residue analysis of fluoroquinolones in raw bovine milk by online solid phase extraction followed by liquid chromatography coupled to tandem mass spectrometry. J. Chromatogr. A 2011, 1218, 9019–9027. [Google Scholar] [CrossRef]
- Lombardo-Agüí, M.; García-Campa, A.M.A.; Gámiz-Gracia, L.; Cruces-Blanco, C. Determination of quinolones of veterinary use in bee products by ultra-high performance liquid chromatography-tandem mass spectrometry using a QuEChERS extraction procedure. Talanta 2012, 93, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Lombardo-Agüí, M.; Gámiz-Gracia, L.; García-Campaña, A.M.; Cruces-Blanco, C. Sensitive determination of fluoroquinolone residues in waters by capillary electrophoresis with laser-induced fluorescence detection. Anal. Bioanal. Chem. 2009, 396, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- González-Gómez, L.; Morante-Zarcero, S.; Pérez-Quintanilla, D.; Sierra, I. Simultaneous Determination of Furanic Compounds and Acrylamide in Insect-Based Foods by HPLC-QqQ-MS/MS Employing a Functionalized Mesostructured Silica as Sorbent in Solid-Phase Extraction. Foods 2021, 10, 1557. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Zhang, Q.H.; Ma, K.; Li, H.M.; Guo, Z. Identification and determination of 34 water-soluble synthetic dyes in foodstuff by high performance liquid chromatography-diode array detection-ion trap time-of-flight tandem mass spectrometry. Food Chem. 2015, 182, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Oberson, J.M.; Meschiari, M.; Munari, C. Determination of 18 water-soluble artificial dyes by LC-MS in selected Matrices. Food Chem. 2016, 197, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Kawashima, A.; Okamoto, M.; Kanetsuki, K.; Makino, T.; Hamada, N. Fundamental study of a novel membrane filtration cleanup method for pesticide analysis in agricultural products. Food Control. 2016, 64, 1–9. [Google Scholar] [CrossRef]
- Chigome, S.; Torto, N. A review of opportunities for electrospun nanofibers in analytical chemistry. Anal. Chimi. Acta 2011, 706, 25–36. [Google Scholar] [CrossRef]
- Chigome, S.; Torto, N. Electrospun nanofiber-based solid-phase extraction. Trends Anal. Chem. 2012, 38, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Chigome, S.; Darko, G.; Torto, N. Electrospun nanofibers as sorbent material for solid phase extraction. Analyst 2011, 136, 2879–2889. [Google Scholar] [CrossRef] [Green Version]
- Qi, F.; Qian, L.; Liu, J.; Li, X.; Lu, L.; Xu, Q. A high-throughput nanofibers mat based micro-solid phase extraction for the determination of cationic dyes in wastewater. J. Chromatogr. A 2016, 1460, 24–32. [Google Scholar] [CrossRef]
- Qi, F.; Jian, N.; Qian, L.; Cao, W.; Xu, Q.; Li, J. Development and optimization of a novel sample preparation method cored on functionalized nanofibers mat-solid phase extraction for the simultaneous efficient extraction of illegal anionic and cationic dyes in foods. Anal. Bioanal. Chem. 2017, 409, 5697–5709. [Google Scholar] [CrossRef] [PubMed]
- Jian, N.G.; Zhao, M.; Liang, S.H.; Cao, J.K.; Wang, C.M.; Xu, Q.; Li, J. High-Throughput and High-Efficient Micro-solid Phase Extraction Based on Sulfonated-Polyaniline/Polyacrylonitrile Nanofiber Mats for Determination of Fluoroquinolones in Animal-Origin Foods. J. Agric. Food Chem. 2019, 67, 6892–6901. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.L.; Song, Y.; Liu, P.; Kang, X.J. Polystyrene nanofibers capped with copper nanoparticles for selective extraction of glutathione prior to its determination by HPLC. Microchim. Acta 2018, 185, 321. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Y.; Xu, X.; Zhang, L. Metal-organic framework-derived three-dimensional porous graphitic octahedron carbon cages-encapsulated copper nanoparticles hybrids as highly efficient enrichment material for simultaneous determination of four fluoroquinolones. J. Chromatogr. A 2018, 1533, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kopf, T.; Schmitz, G. Analysis of non-esterified fatty acids in human samples by solidphase-extraction and gas chromatography/mass spectrometry. J. Chromatogr. B 2013, 938, 22–26. [Google Scholar] [CrossRef] [PubMed]
- National Standard Method. Determination of 14 Quinolones Residues in Animal-original Food by HPLC-MS/MS. 2007. Available online: https://max.book118.com/html/2019/0617/5044013312002044.shtm (accessed on 10 May 2022).
- Hong, J.W.; Liu, X.M.; Yang, X.Y.; Wang, Y.S. Ionic liquid-based dispersive liquid-liquid microextraction followed by magnetic solid-phase extraction for determination of quinolones. Microchimi. Acta 2022, 153, 022003. [Google Scholar] [CrossRef]
- Kergaravat, S.V.; Nagel, O.G.; Althaus, R.L.; Silvia, R.H. Detection of quinolones in milk and groundwater samples using an indirect immunofluorescent magneto assay. Int. J. Environl. Anal. Chem. 2020, 1–18. [Google Scholar] [CrossRef]
- Ye, Z.; Huang, Y.; Luo, Q.; Wang, L.; Huang, X. Preparation of highly fluorinated and boron-rich adsorbent for magnetic solid-phase extraction of fluoroquinolones in water and milk samples. J. Chromatogr. A 2019, 1601, 86–94. [Google Scholar] [CrossRef]
- Wang, G.N.; Yang, K.; Liu, H.Z.; Feng, M.X.; Wang, J.P. Molecularly imprinted polymer-based solid phase extraction combined high performance liquid chromatography for determination of fluoroquinolones in milk. Anal. Methods 2016, 8, 5511–5518. [Google Scholar] [CrossRef]
- Liu, C.; Yang, L.Y.; Zhang, W.; Li, D.W.; Li, L.A.; Wang, H.Y.; Ma, Y.; Li, C. A magnetic nanoparticle-based lateral flow immunochromatography assay for the rapid detection of fluoroquinolones in milk. Eur. Food Res. Technol. 2021, 247, 2645–2656. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Yang, L.; Zhang, W.; Lin, J.; Li, C. Determination of eight quinolones in milk using immunoaffinity microextraction in a packed syringe and liquid chromatography with fluorescence detection. J. Chromatogr. B 2017, 1064, 68–74. [Google Scholar] [CrossRef] [PubMed]
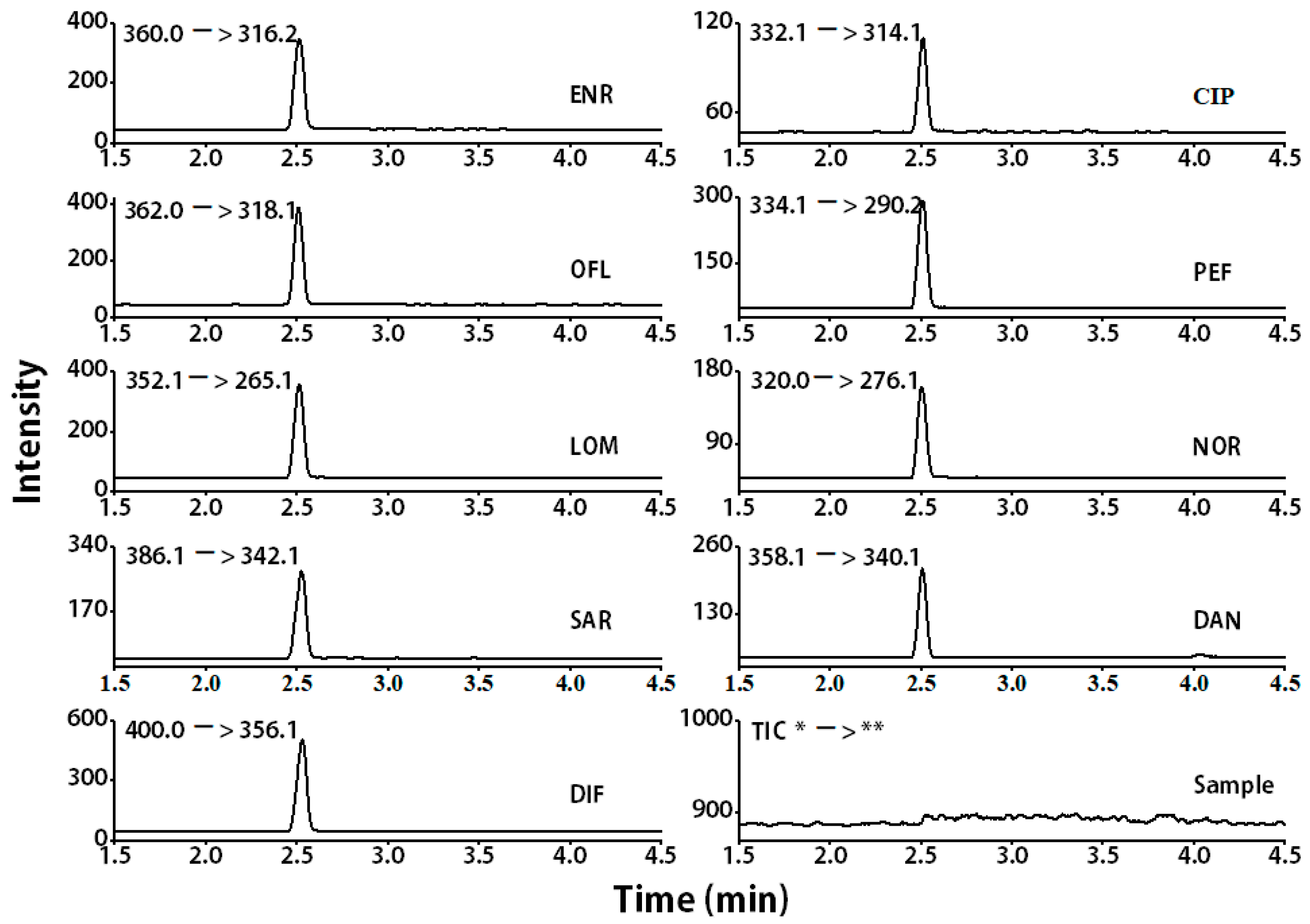
| Analyte | Structure | Formula | Precursor ion (m/z) | Product ions (m/z) | Cone Voltage (V) | Collision Energy (V) |
|---|---|---|---|---|---|---|
| Enrofloxacin (ENR) | 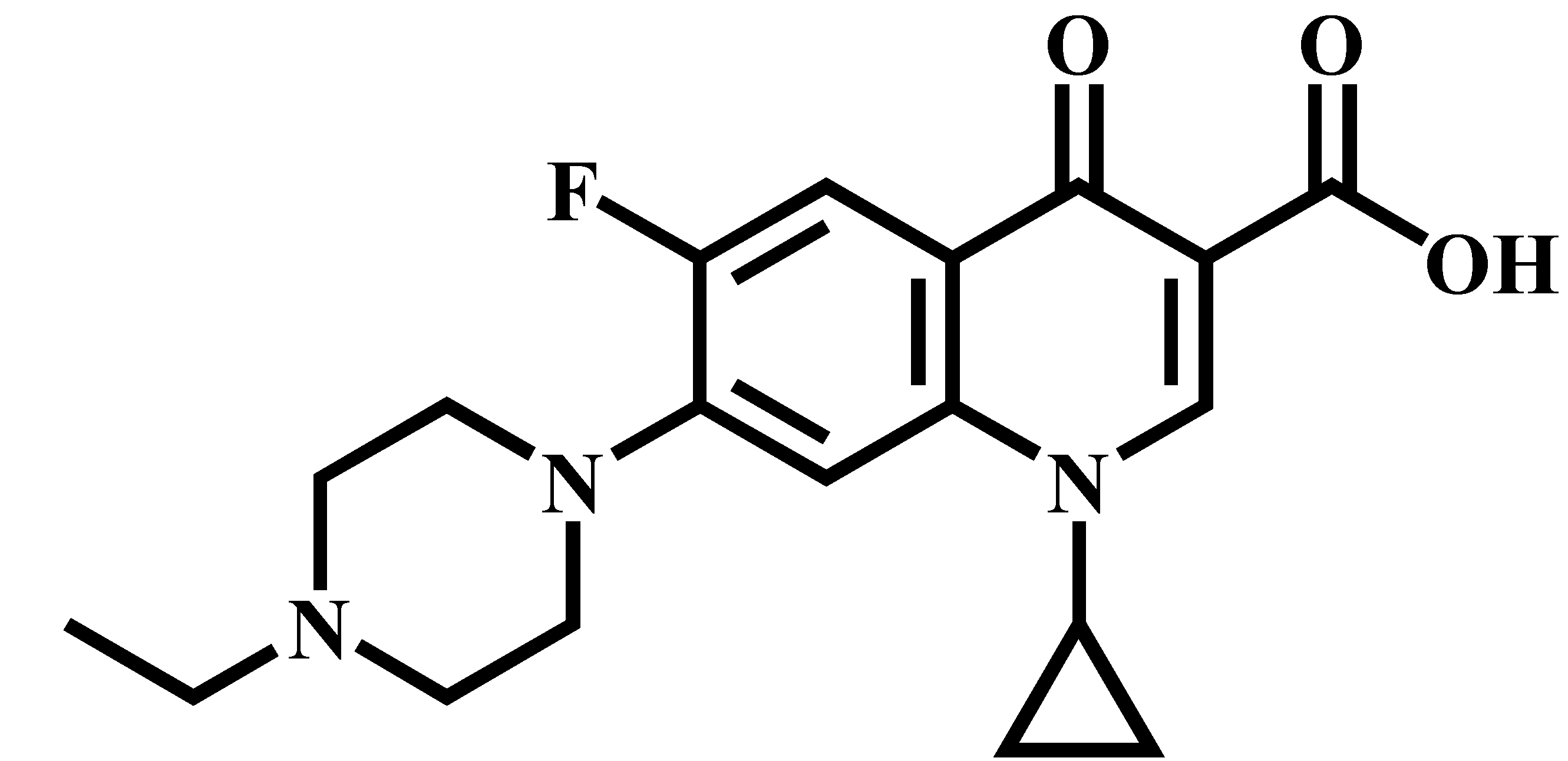 | C19H22FN3O3 | 360.0 | 316.2 * | 120 | 20 |
| 342.1 | 20 | |||||
| 244.9 | 40 | |||||
| Ciprofloxacin (CIP) | 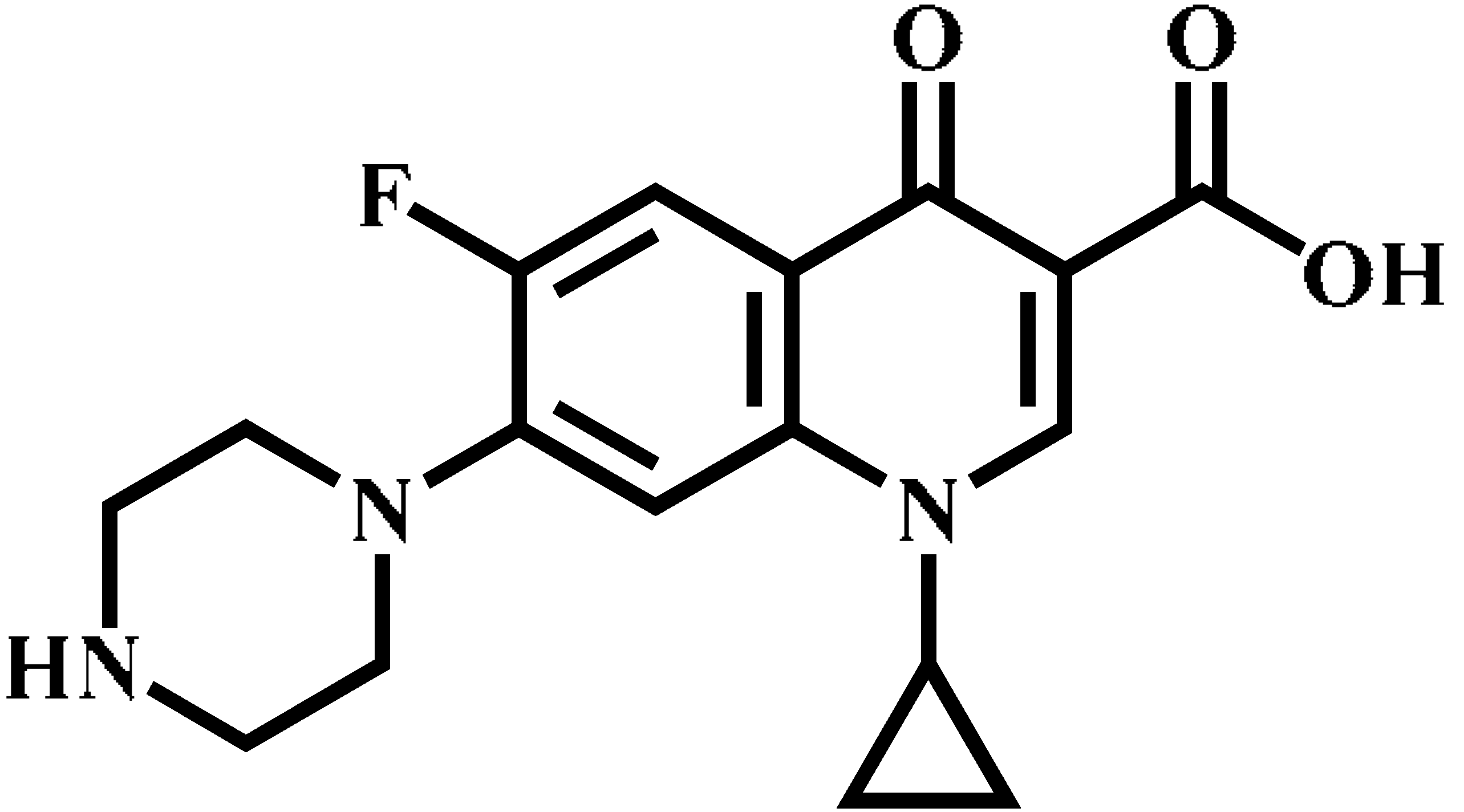 | C17H18FN3O3 | 332.1 | 314.1 * | 135 | 20 |
| 231.0 | 42 | |||||
| Ofloxacin (OFL) | 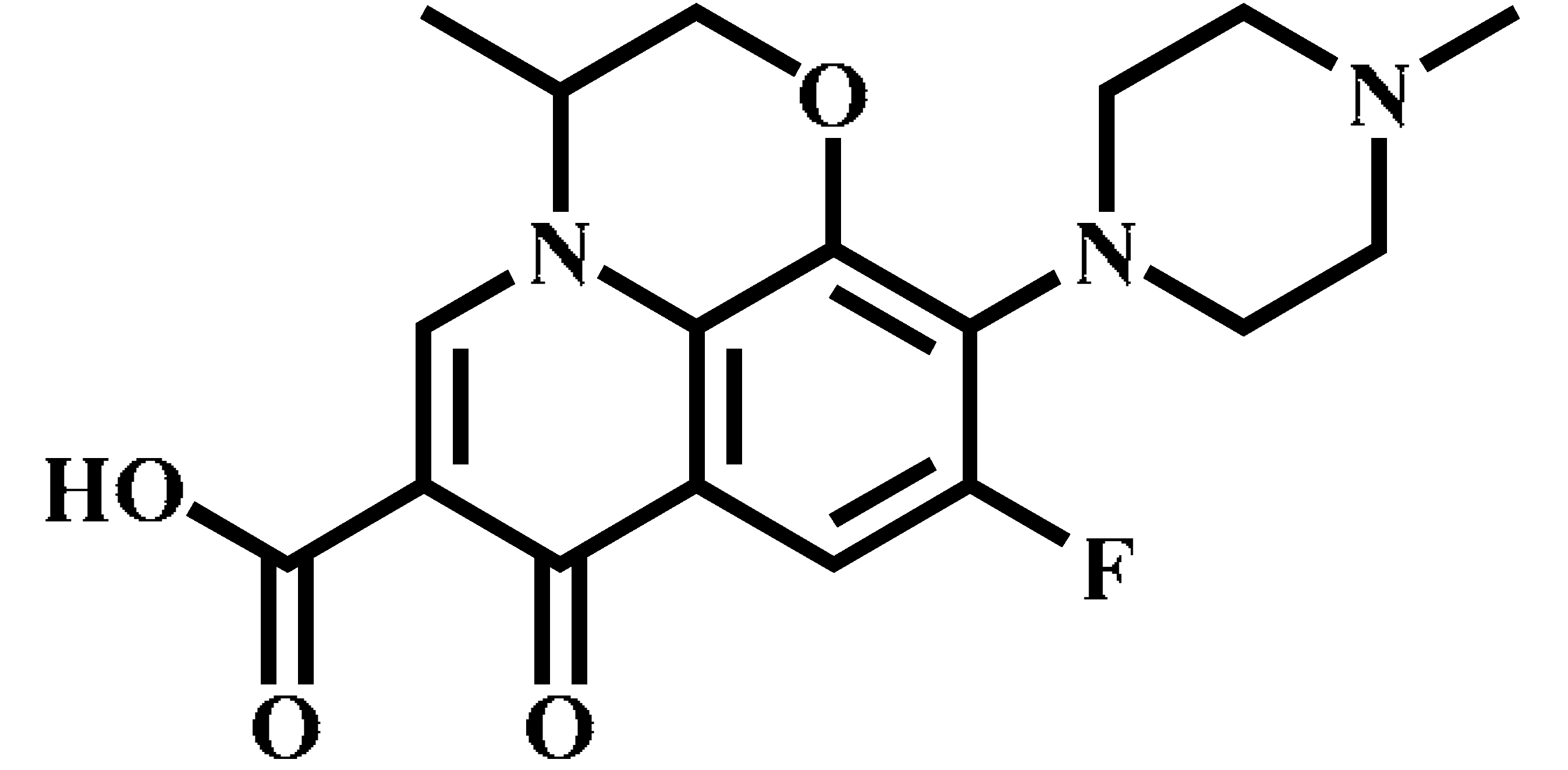 | C18H20FN3O4 | 362.0 | 318.1 * | 130 | 15 |
| 261.1 | 26 | |||||
| Pefloxacin (PEF) |  | C17H20FN3O3 | 334.1 | 290.2 * | 130 | 16 |
| 316.2 | 20 | |||||
| Lomefloxacin (LOM) |  | C17H19F2N3O3 | 352.1 | 265.1 * | 130 | 20 |
| 308.1 | 10 | |||||
| Norfloxacin (NOR) | 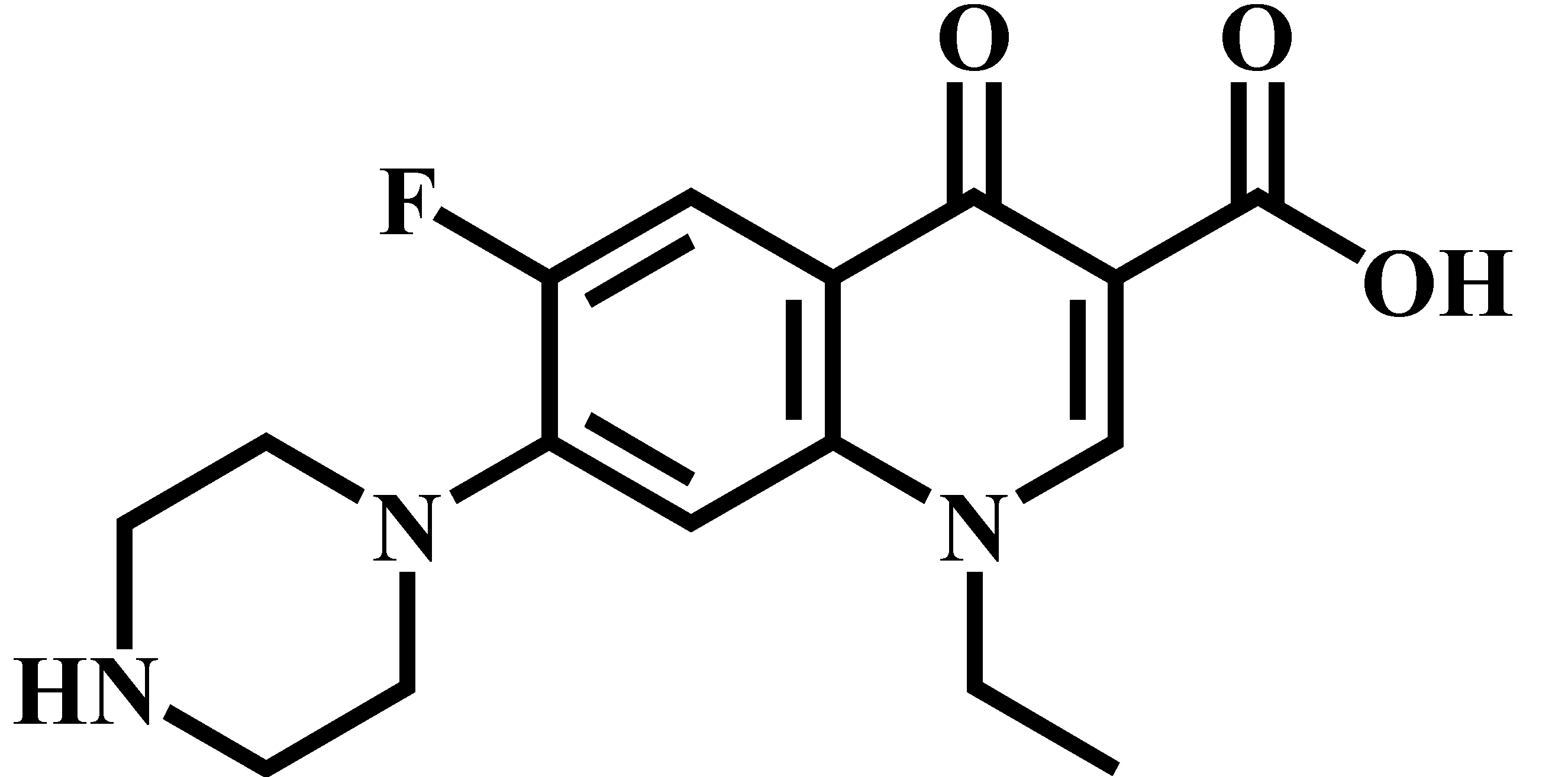 | C16H18FN3O3 | 320.0 | 276.1 * | 130 | 15 |
| 302.1 | 20 | |||||
| Sarafloxacin (SAR) |  | C20H17F2N3O3 | 386.1 | 342.1 * | 130 | 15 |
| 368.1 | 20 | |||||
| Danfloxacin (DAN) | 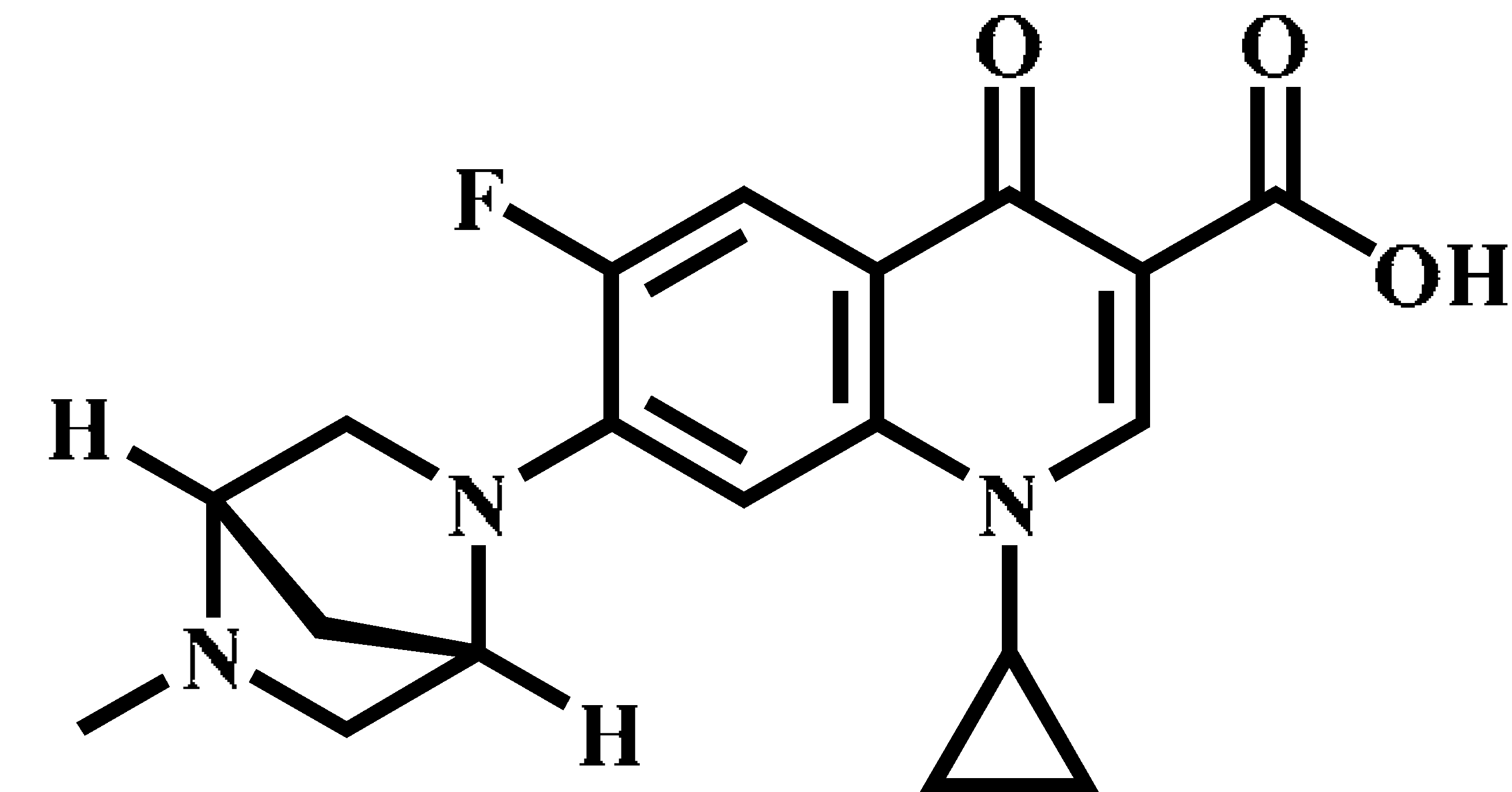 | C19H20FN3O3 | 358.1 | 340.1 * | 140 | 25 |
| 255.0 | 46 | |||||
| Difloxacin (DIF) |  | C21H19F2N3O3 | 400.0 | 356.1 * | 140 | 20 |
| 382.1 | 20 |
| Analyte | Linearity Range (ng/mL) | Correlation Coefficient (R2) | LOD (ng/mL) | LOQ (ng/mL) |
|---|---|---|---|---|
| ENR | 1.0–100 | 0.9999 | 0.18 | 0.59 |
| CIP | 0.9994 | 0.36 | 1.20 | |
| OFL | 0.9996 | 0.23 | 0.76 | |
| PEF | 0.9996 | 0.16 | 0.53 | |
| LOM | 0.9997 | 0.23 | 0.75 | |
| NOR | 0.9994 | 0.39 | 1.29 | |
| SAR | 0.9996 | 0.27 | 0.91 | |
| DAN | 0.9992 | 0.18 | 0.59 | |
| DIF | 0.9998 | 0.36 | 1.19 |
| Analyte | Spiked Concentration (ng/mL) | Recovery (%) ± RSD (n = 3) | |
|---|---|---|---|
| Intra−Day | Inter−Day | ||
| ENR | 2 | 92.51 ± 1.71 | 90.64 ± 5.02 |
| 10 | 90.94 ± 2.61 | 88.68 ± 3.10 | |
| 25 | 95.39 ± 4.41 | 93.25 ± 4.67 | |
| CIP | 2 | 97.02 ± 1.56 | 96.72 ± 3.92 |
| 10 | 97.27 ± 5.38 | 95.59 ± 6.07 | |
| 25 | 93.56 ± 2.67 | 91.30 ± 4.16 | |
| OFL | 2 | 97.93 ± 2.74 | 97.64 ± 3.15 |
| 10 | 95.53 ± 3.15 | 94.20 ± 2.61 | |
| 25 | 94.92 ± 2.27 | 95.94 ± 2.01 | |
| PEF | 2 | 93.51 ± 1.60 | 93.12 ± 3.34 |
| 10 | 92.60 ± 1.08 | 90.74 ± 1.10 | |
| 25 | 89.86 ± 1.53 | 89.10 ± 3.50 | |
| LOM | 2 | 94.11 ± 2.36 | 91.12 ± 5.01 |
| 10 | 95.78 ± 1.68 | 93.30 ± 4.22 | |
| 25 | 92.22 ± 2.46 | 92.84 ± 2.96 | |
| NOR | 2 | 91.81 ± 1.20 | 91.49 ± 2.30 |
| 10 | 92.28 ± 2.66 | 92.12 ± 3.94 | |
| 25 | 95.26 ± 1.28 | 92.86 ± 2.41 | |
| SAR | 2 | 90.98 ± 2.36 | 90.86 ± 2.06 |
| 10 | 92.06 ± 3.77 | 91.72 ± 2.37 | |
| 25 | 91.49 ± 2.55 | 92.04 ± 2.93 | |
| DAN | 2 | 96.00 ± 2.66 | 94.39 ± 4.29 |
| 10 | 91.32 ± 1.25 | 90.49 ± 3.06 | |
| 25 | 97.63 ± 2.40 | 94.96 ± 3.09 | |
| DIF | 2 | 95.33 ± 2.04 | 94.11 ± 4.35 |
| 10 | 94.61 ± 3.05 | 94.31 ± 2.72 | |
| 25 | 95.98 ± 4.92 | 95.04 ± 5.58 | |
| Analyte | Matrix | Slope | Slope Matrix/Solvent | %ME |
|---|---|---|---|---|
| ENR | Water | 2040.14239 | 0.9826 | 98.3% |
| Milk | 2004.60237 | |||
| CIP | Water | 1018.39131 | 0.9661 | 96.6% |
| Milk | 983.82133 | |||
| OFL | Water | 1586.97198 | 0.9782 | 97.8% |
| Milk | 1552.37639 | |||
| PEF | Water | 2363.71296 | 0.9502 | 95.0% |
| Milk | 2246.1182 | |||
| LOM | Water | 1654.79423 | 0.9532 | 95.3% |
| Milk | 1577.41885 | |||
| NOR | Water | 934.71011 | 0.9793 | 97.9% |
| Milk | 915.32412 | |||
| SAR | Water | 1347.2961 | 0.9671 | 96.7% |
| Milk | 1303.02713 | |||
| DAN | Water | 2089.56314 | 0.9528 | 95.3% |
| Milk | 1991.0296 | |||
| DIF | Water | 1053.1916 | 0.9416 | 94.2% |
| Milk | 991.72708 |
| Extraction Method | Detection Method | Target | Recovery Rate (%) | LOD | LOQ | Linearity Range | References |
|---|---|---|---|---|---|---|---|
| HLB-SPE | LC-MS/MS | 14 | 79.0~119.9 | 0.5~1.5 μg/kg | 2.0~5.0 μg/kg | 2.5~100.0 μg/L | [26] |
| IL-DLLME-MSPE | HPLC | 3 | 81.2~109.0 | 1.5 μg/L | 4.0~8.0 μg/L | 4~1000 μg/L | [27] |
| 96-well-based | IFA | 1 | 90.0~100.0 | 4.0 μg/L | / | 0.01~400 μg/L | [28] |
| MSPE | HPLC-DAD | 7 | 78.9~119.0 | 0.010~0.046 μg/kg | / | 0.05~200.0 μg/kg | [29] |
| MIP-SPE | HPLC | 4 | 76.8~97.7 | 10.0~20.0 ng/mL | 20.0~50.0 ng/mL | 20~1000 ng/mL | [30] |
| MNP | LFIA | 10 | 16.47~83.67 | 1.0~2.0 ng/mL | / | 0.2~10 µg/m | [31] |
| IA-MEPS | HPLC | 8 | 53.9~90.6 | 0.05~0.1 ng/g | 0.15~0.3 ng/g | 0.1~100.0 µg/mL | [32] |
| PFSPE | LC-MS/MS | 9 | 80.64~95.26 | 0.31~0.91 ng/mL | 1.03~3.03 ng/mL | 1.0~100.0 ng/mL | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, L.; Chen, Y.; Shao, D.; Li, J. Simultaneous Determination of Nine Quinolones in Pure Milk Using PFSPE-HPLC-MS/MS with PS-PAN Nanofibers as a Sorbent. Foods 2022, 11, 1843. https://doi.org/10.3390/foods11131843
Wei L, Chen Y, Shao D, Li J. Simultaneous Determination of Nine Quinolones in Pure Milk Using PFSPE-HPLC-MS/MS with PS-PAN Nanofibers as a Sorbent. Foods. 2022; 11(13):1843. https://doi.org/10.3390/foods11131843
Chicago/Turabian StyleWei, Lanlan, Yanan Chen, Dongliang Shao, and Jingjun Li. 2022. "Simultaneous Determination of Nine Quinolones in Pure Milk Using PFSPE-HPLC-MS/MS with PS-PAN Nanofibers as a Sorbent" Foods 11, no. 13: 1843. https://doi.org/10.3390/foods11131843
APA StyleWei, L., Chen, Y., Shao, D., & Li, J. (2022). Simultaneous Determination of Nine Quinolones in Pure Milk Using PFSPE-HPLC-MS/MS with PS-PAN Nanofibers as a Sorbent. Foods, 11(13), 1843. https://doi.org/10.3390/foods11131843





