Discrimination between Wild and Farmed Sea Bass by Using New Spectrometry and Spectroscopy Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Analytical Procedure
2.2.1. Extraction and Analysis of Fatty Acids
2.2.2. NIR Acquisition
2.3. Data Processing
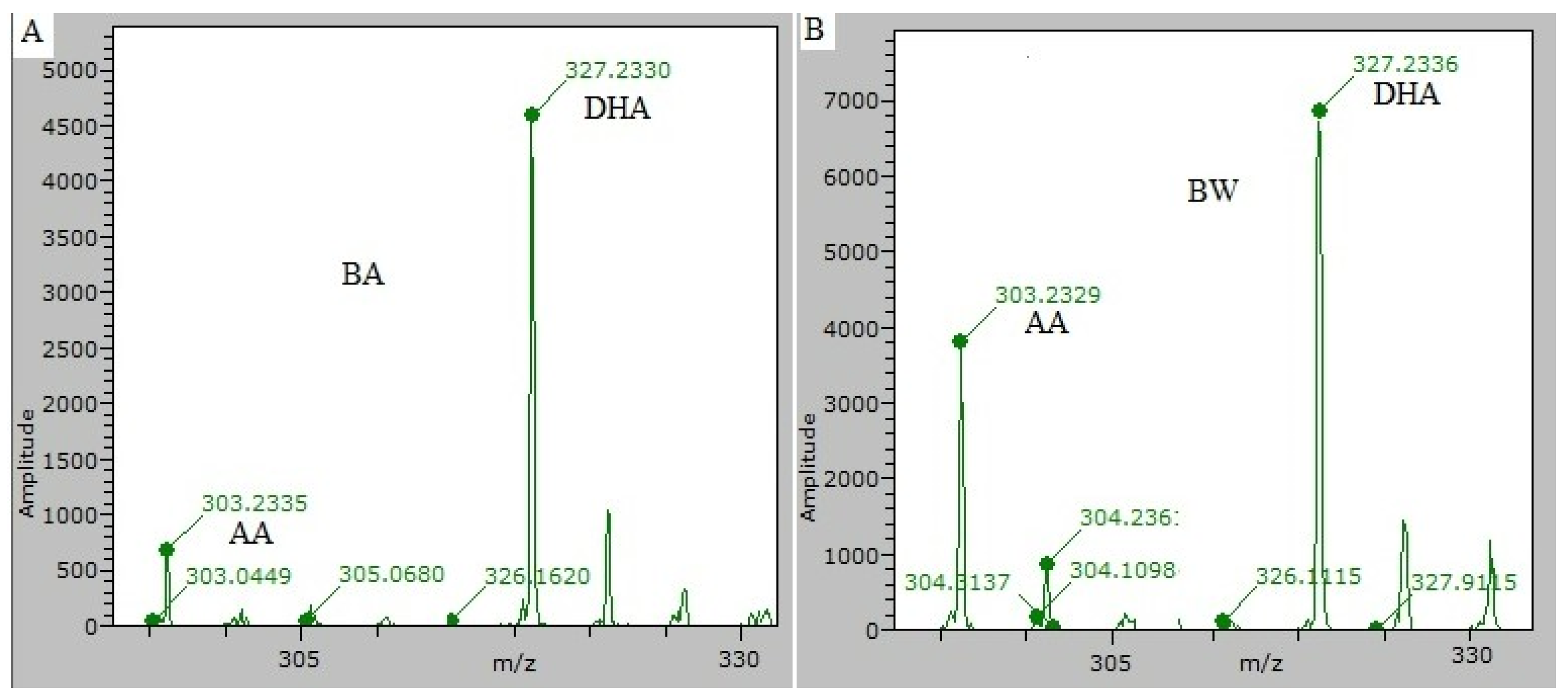
2.3.1. Mass Spectrometry
2.3.2. NIR Spectroscopy
- -
- Processed: log, average scan, 1st derivative and 2nd order polynomial, select range (nm), subtraction average.
- -
- Normalize: average scan, select range (nm), standard normal variate (SNV).
- -
- Processed & Normalize: log, average scan, 1st derivative and 2nd order polynomial, select range (nm), SNV.
- -
- (log)R & Normalize: log, average scan, select range (nm), 2nd derivative and 2nd order polynomial, SNV.
3. Results and Discussion
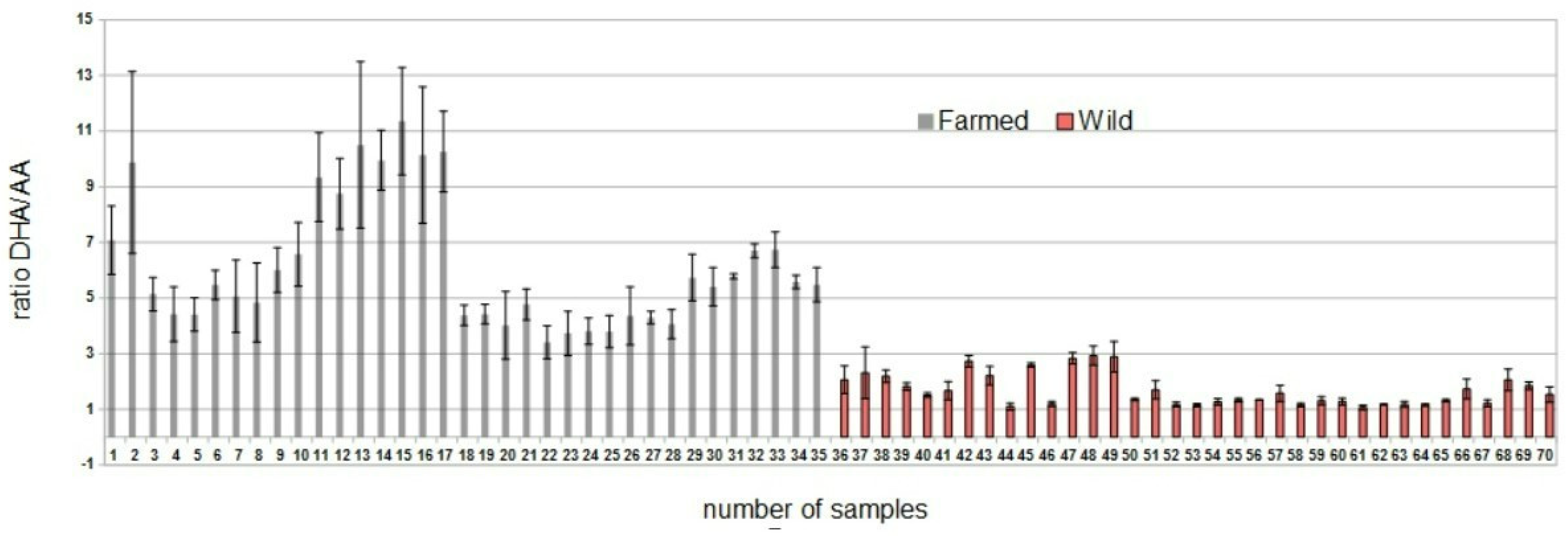
3.1. Fatty Acids Ratio
3.2. Statistical Analysis
3.2.1. Mass Spectrometry
3.2.2. NIR Spectroscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Jayachandran, M.; Bai, W.; Xu, B. A Critical Review on the Health Benefits of Fish Consumption and Its Bioactive Constituents. Food Chem. 2022, 369, 130874. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture 2020. Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Bjørndal, T.; Guillen, J. Market Integration between Wild and Farmed Fish in Mediterranean Countries. Fish. Aquac. Circ. 2018, 1131, 1–98. [Google Scholar]
- Fernandes, T.J.R.; Amaral, J.S.; Mafra, I. DNA Barcode Markers Applied to Seafood Authentication: An Updated Review. Crit. Rev. Food Sci. Nutr. 2021, 61, 3904–3935. [Google Scholar] [CrossRef] [PubMed]
- Martinsohn, J.T.; Raymond, P.; Knott, T.; Glover, K.A.; Nielsen, E.E.; Eriksen, L.B.; Ogden, R.; Casey, J.; Guillen, J. DNA-Analysis to Monitor Fisheries and Aquaculture: Too Costly? Fish Fish. 2019, 20, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Grazina, L.; Rodrigues, P.J.; Igrejas, G.; Nunes, M.A.; Mafra, I.; Arlorio, M.; Oliveira, M.B.; Amaral, J.S. Machine Learning Approaches Applied to GC-FID Fatty Acid Profiles to Discriminate Wild from Farmed Salmon. Foods 2020, 9, 1622. [Google Scholar] [CrossRef]
- Alasalvar, C.; Taylor, K.D.A.; Zubcov, E.; Shahidi, F.; Alexis, M. Differentiation of Cultured and Wild Sea Bass (Dicentrarchus Labrax): Total Lipid Content, Fatty Acid and Trace Mineral Composition. Food Chem. 2002, 79, 145–150. [Google Scholar] [CrossRef]
- Arechavala-Lopez, P.; Fernandez-Jover, D.; Black, K.D.; Ladoukakis, E.; Bayle-Sempere, J.T.; Sanchez-Jerez, P.; Dempster, T. Differentiating the Wild or Farmed Origin of Mediterranean Fish: A Review of Tools for Sea Bream and Sea Bass. Rev. Aquac. 2013, 5, 137–157. [Google Scholar] [CrossRef]
- Vidal, N.P.; Manzanos, M.J.; Goicoechea, E.; Guillén, M.D. Farmed and Wild Sea Bass (Dicentrarchus Labrax) Volatile Metabolites: A Comparative Study by SPME-GC/MS. J. Sci. Food Agric. 2016, 96, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Varrà, M.O.; Ghidini, S.; Zanardi, E.; Badiani, A.; Ianieri, A. Authentication of European Sea Bass According to Production Method and Geographical Origin by Light Stable Isotope Ratio and Rare Earth Elements Analyses Combined with Chemometrics. Ital. J. Food Saf. 2019, 8, 1. [Google Scholar] [CrossRef]
- Bell, J.G.; Preston, T.; Henderson, R.J.; Strachan, F.; Bron, J.E.; Cooper, K.; Morrison, D.J. Discrimination of Wild and Cultured European Sea Bass (Dicentrarchus Labrax) Using Chemical and Isotopic Analyses. J. Agric. Food Chem. 2007, 55, 5934–5941. [Google Scholar] [CrossRef] [PubMed]
- Sciuto, S.; Esposito, G.; Guglielmetti, C.; Mazza, M.; Cocco, C.; Ru, G.; Acutis, P.L. Development of a Novel Method for Rapid Discrimination between Wild and Farmed Sea Bream. J. Food Prot. 2019, 82, 1870–1873. [Google Scholar] [CrossRef] [PubMed]
- Ghidini, S.; Varrà, M.O.; Dall’Asta, C.; Badiani, A.; Ianieri, A.; Zanardi, E. Rapid Authentication of European Sea Bass (Dicentrarchus Labrax L.) According to Production Method, Farming System, and Geographical Origin by near Infrared Spectroscopy Coupled with Chemometrics. Food Chem. 2019, 280, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Ottavian, M.; Facco, P.; Fasolato, L.; Novelli, E.; Mirisola, M.; Perini, M.; Barolo, M. Use of Near-Infrared Spectroscopy for Fast Fraud Detection in Seafood: Application to the Authentication of Wild European Sea Bass (Dicentrarchus Labrax). J. Agric. Food Chem. 2012, 60, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Beć, K.B.; Grabska, J.; Huck, C.W. Miniaturized NIR Spectroscopy in Food Analysis and Quality Control: Promises, Challenges, and Perspectives. Foods 2022, 11, 1465. [Google Scholar] [CrossRef] [PubMed]
- The Lab–Consumer Physics. Available online: https://thelab.consumerphysics.com (accessed on 29 April 2022).
- Fan, J.; Gijbels, I. Local Polynomial Modelling and Its Applications: Monographs on Statistics and Applied Probability 66; Routledge: London, UK, 1996. [Google Scholar]
- Pepe, M.S. The Statistical Evaluation of Medical Tests for Classification and Prediction; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- Lenas, D.; Chatziantoniou, S.; Nathanailides, C.; Triantafillou, D. Comparison of Wild and Farmed Sea Bass (Dicentrarchus Labrax L) Lipid Quality. Procedia Food Sci. 2011, 1, 1139–1145. [Google Scholar] [CrossRef] [Green Version]
- Terova, G.; Moroni, F.; Antonini, M.; Bertacchi, S.; Pesciaroli, C.; Branduardi, P.; Labra, M.; Porro, D.; Ceccotti, C.; Rimoldi, S. Using Glycerol to Produce European Sea Bass Feed with Oleaginous Microbial Biomass: Effects on Growth Performance, Filet Fatty Acid Profile, and FADS2 Gene Expression. Front. Mar. Sci. 2021, 8, 1115. [Google Scholar] [CrossRef]
- Torrecillas, S.; Román, L.; Rivero-Ramírez, F.; Caballero, M.J.; Pascual, C.; Robaina, L.; Izquierdo, M.S.; Acosta, F.; Montero, D. Supplementation of Arachidonic Acid Rich Oil in European Sea Bass Juveniles (Dicentrarchus Labrax) Diets: Effects on Leucocytes and Plasma Fatty Acid Profiles, Selected Immune Parameters and Circulating Prostaglandins Levels. Fish Shellfish Immunol. 2017, 64, 437–445. [Google Scholar] [CrossRef] [PubMed]
| FARMED SEA BASS | WILD SEA BASS | |||||
|---|---|---|---|---|---|---|
| Geographical origin | Croatia | Greece | Oriental Mediterranean | Italy | Oriental Mediterranean | Italy |
| Weight (g) | 400–600 | 400 | 650 | 500–750 | 800–200 | 650–1700 |
| Number of samples | 15 | 8 | 15 | 7 | 15 | 30 |
| Model | Performance f1 | Predicted BA (%) | Predicted BW (%) |
|---|---|---|---|
| Processed | 0.88 | 86 | 91 |
| Normalize | 0.84 | 86 | 81 |
| Processed & Normalize | 0.83 | 84 | 86 |
| (log)R & Normalize | 0.80 | 76 | 85 |
| Model | Performance f1 | Class | Classification Rate per Class % | Overall Classification Rate % |
|---|---|---|---|---|
| Processed | 0.89 | BA | 70 (3/10) | 85 (17/20) |
| BW | 100 (10/10) | |||
| Normalize | 0.97 | BA | 90 (1/10) | 95 (19/20) |
| BW | 100 (10/10) | |||
| Processed & Normalize | 0.87 | BA | 70 (3/10) | 85 (17/20) |
| BW | 100 (10/10) | |||
| (log)R & Normalize | 0.97 | BA | 90 (1/10) | 95 (19/20) |
| BW | 100 (10/10) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, G.; Sciuto, S.; Guglielmetti, C.; Pastorino, P.; Ingravalle, F.; Ru, G.; Bozzetta, E.M.; Acutis, P.L. Discrimination between Wild and Farmed Sea Bass by Using New Spectrometry and Spectroscopy Methods. Foods 2022, 11, 1673. https://doi.org/10.3390/foods11121673
Esposito G, Sciuto S, Guglielmetti C, Pastorino P, Ingravalle F, Ru G, Bozzetta EM, Acutis PL. Discrimination between Wild and Farmed Sea Bass by Using New Spectrometry and Spectroscopy Methods. Foods. 2022; 11(12):1673. https://doi.org/10.3390/foods11121673
Chicago/Turabian StyleEsposito, Giovanna, Simona Sciuto, Chiara Guglielmetti, Paolo Pastorino, Francesco Ingravalle, Giuseppe Ru, Elena Maria Bozzetta, and Pier Luigi Acutis. 2022. "Discrimination between Wild and Farmed Sea Bass by Using New Spectrometry and Spectroscopy Methods" Foods 11, no. 12: 1673. https://doi.org/10.3390/foods11121673
APA StyleEsposito, G., Sciuto, S., Guglielmetti, C., Pastorino, P., Ingravalle, F., Ru, G., Bozzetta, E. M., & Acutis, P. L. (2022). Discrimination between Wild and Farmed Sea Bass by Using New Spectrometry and Spectroscopy Methods. Foods, 11(12), 1673. https://doi.org/10.3390/foods11121673








