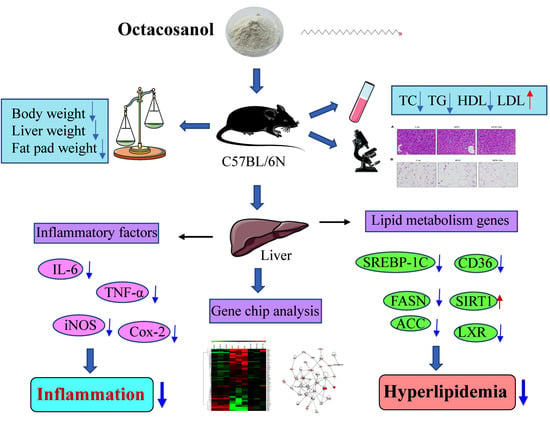
Octacosanol Modifies Obesity, Expression Profile and Inflammation Response of Hepatic Tissues in High-Fat Diet Mice
Abstract
:Highlights:
1. Introduction
2. Materials and Methods
2.1. Feed Formula of Mice
2.2. Octacosanol Treatment
2.3. Glucose Tolerance Testing
2.4. Analysis of Blood Biochemical Indicators
2.5. Histological Analysis
2.6. RNA Isolation and Array Processing
2.7. RT-PCR Analyzing mRNA Expressions
2.8. Western Blot Analysis
2.9. Statistical Analysis
3. Results
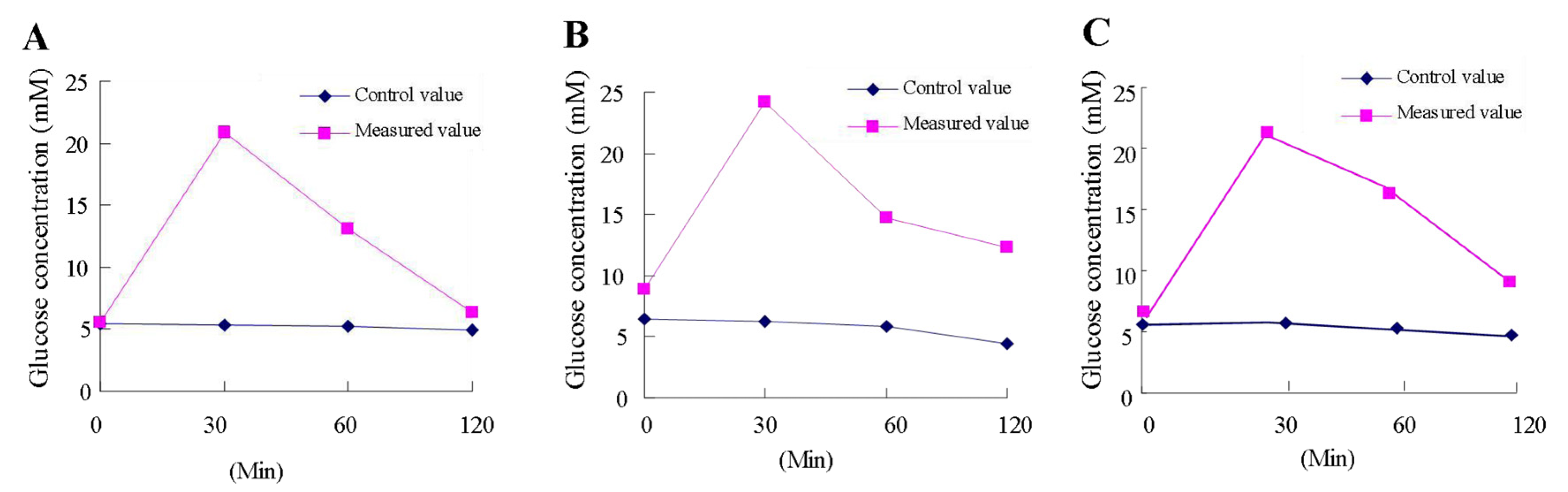
3.1. Octacosanol Enhances Glucose Tolerance of HFD Mice
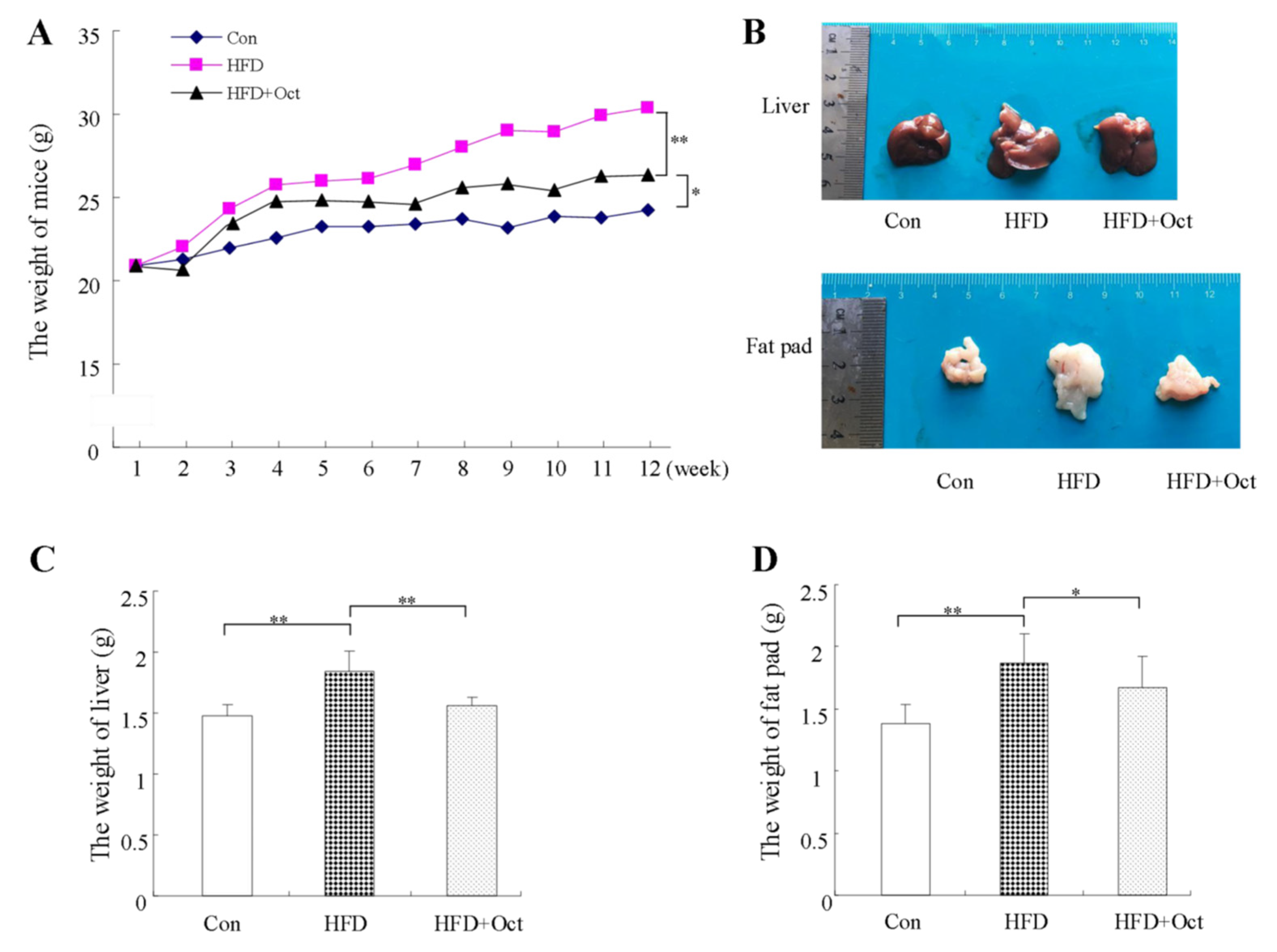
3.2. Octacosanol Ameliorate the Obesity Phenotype of HFD-Induced Mice
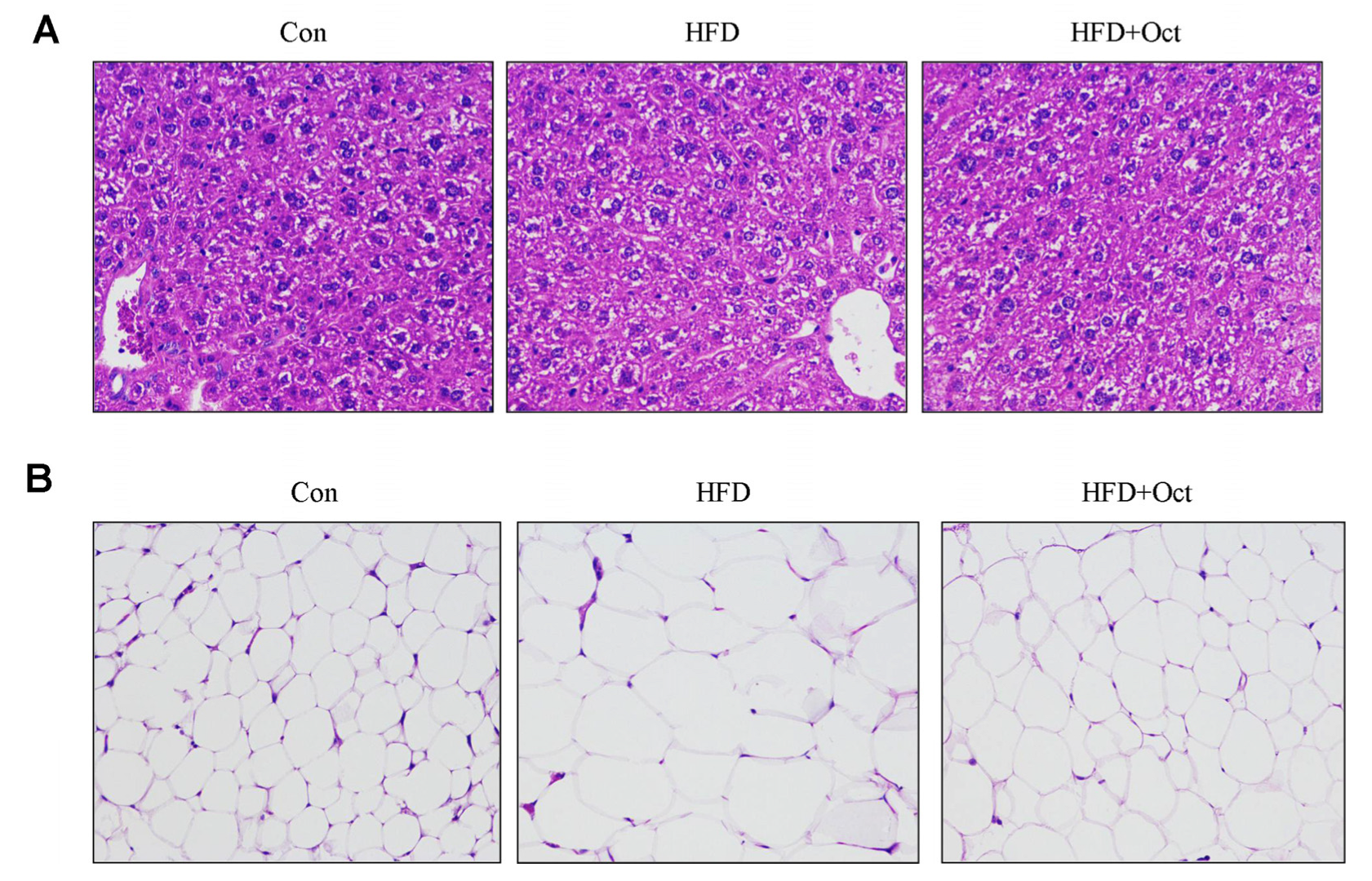
3.3. Histological Alterations of Liver Tissues and Epididymal Fat
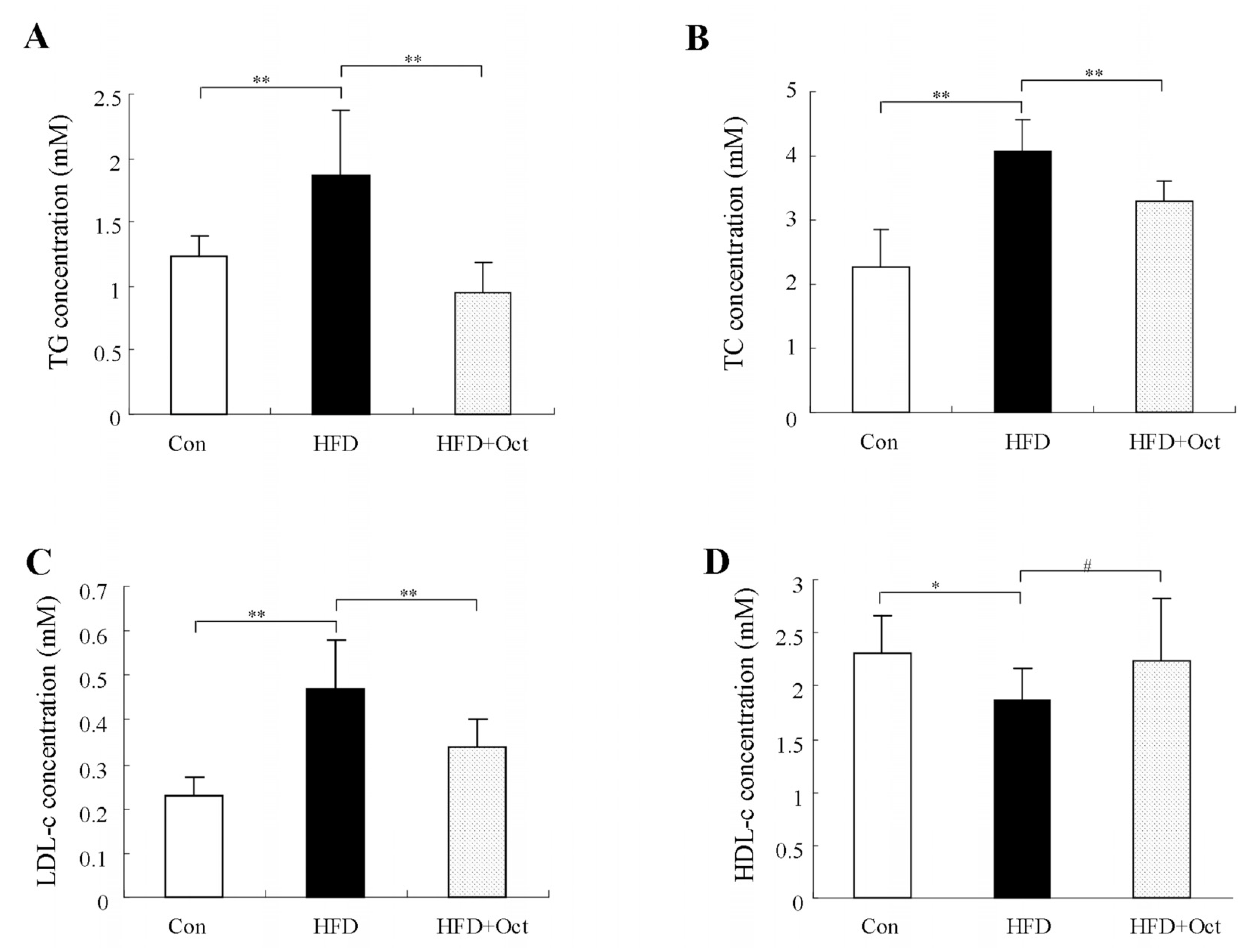
3.4. Octacosanol Improves the Plasma Lipid Level in High-Fat Diet Mice
3.5. Octacosanol Modulated the Gene Expression Profile of Liver Tissues
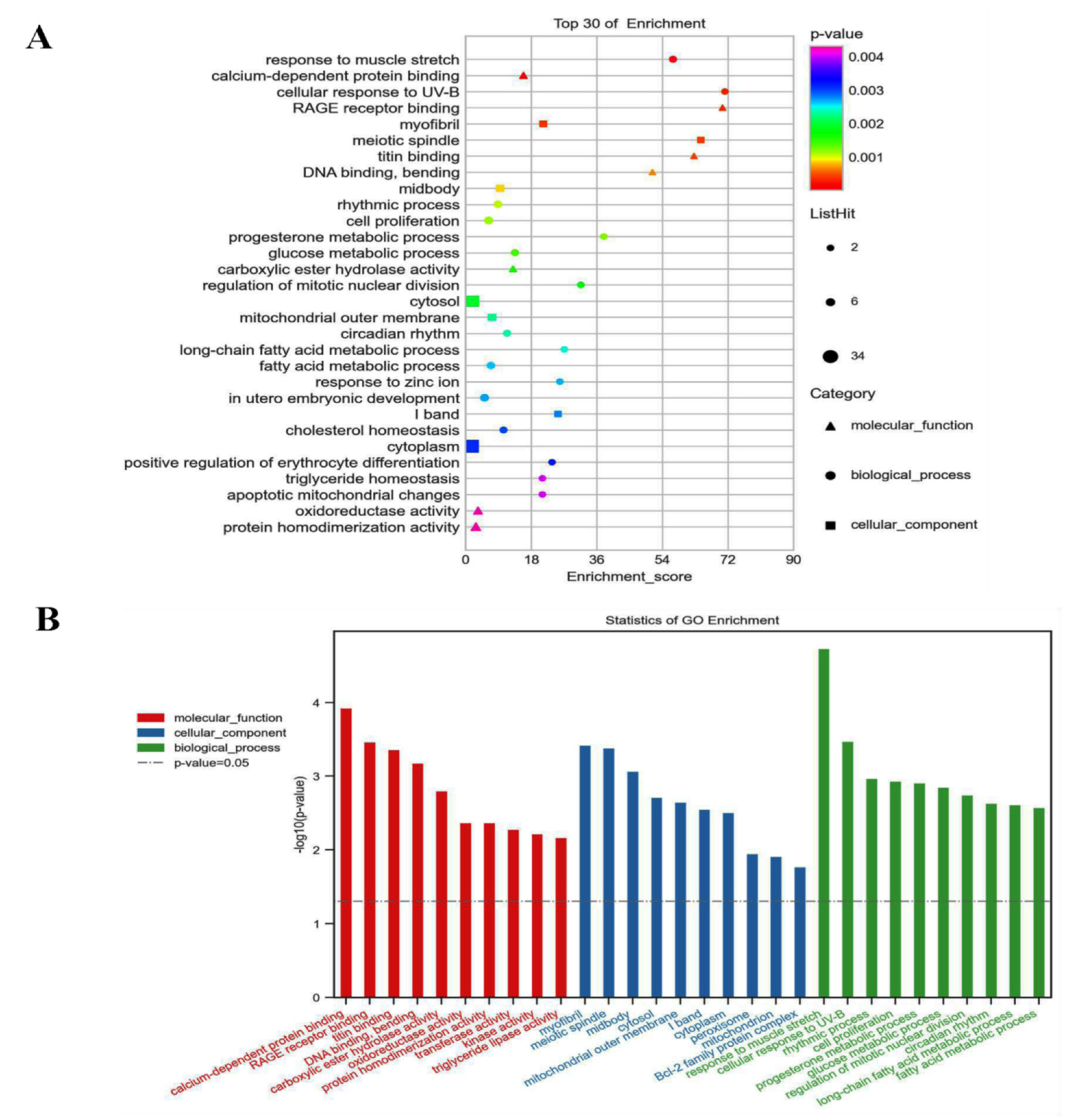
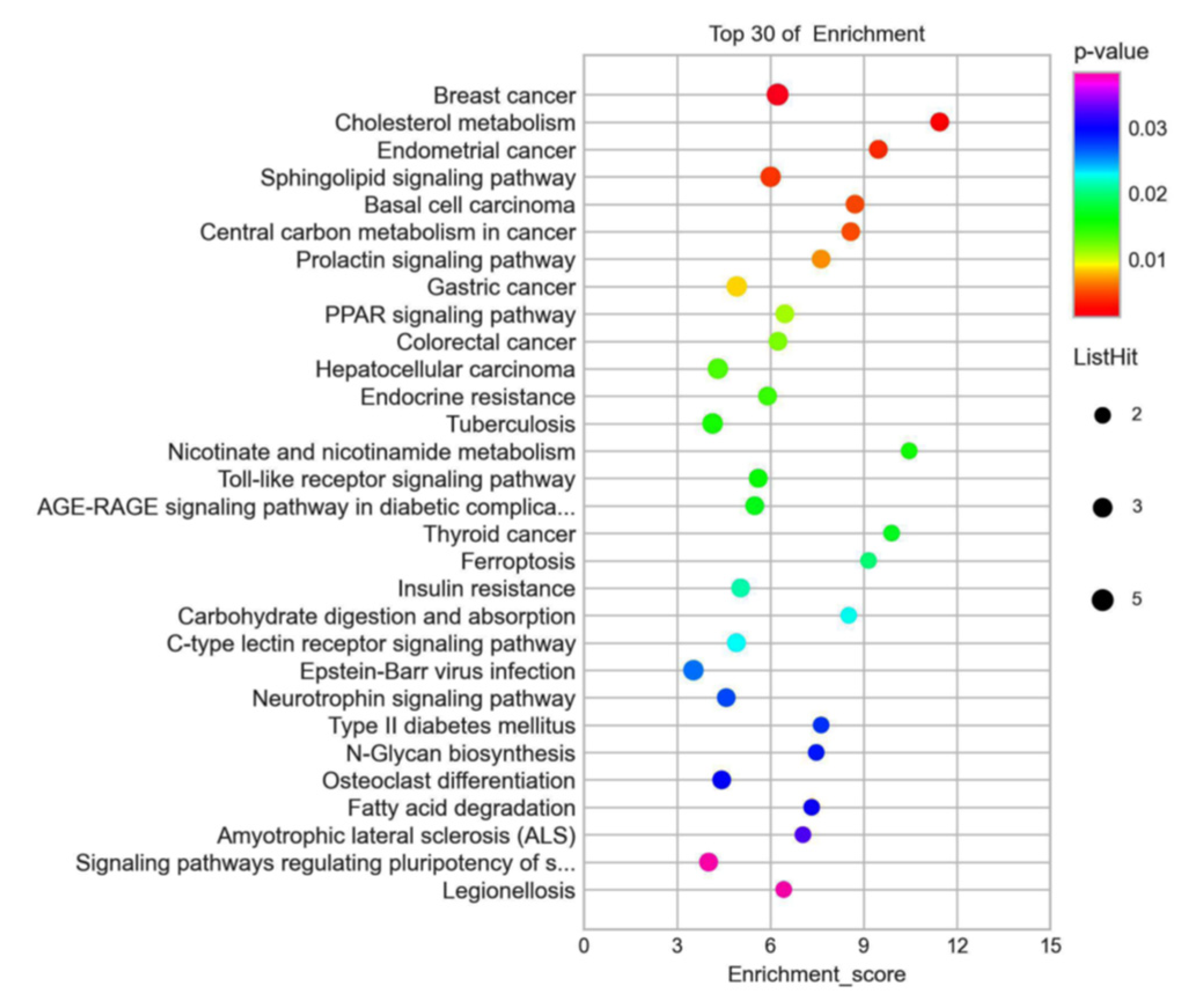
3.6. GO and KEGG Analyzed the 72 Different Expression Genes
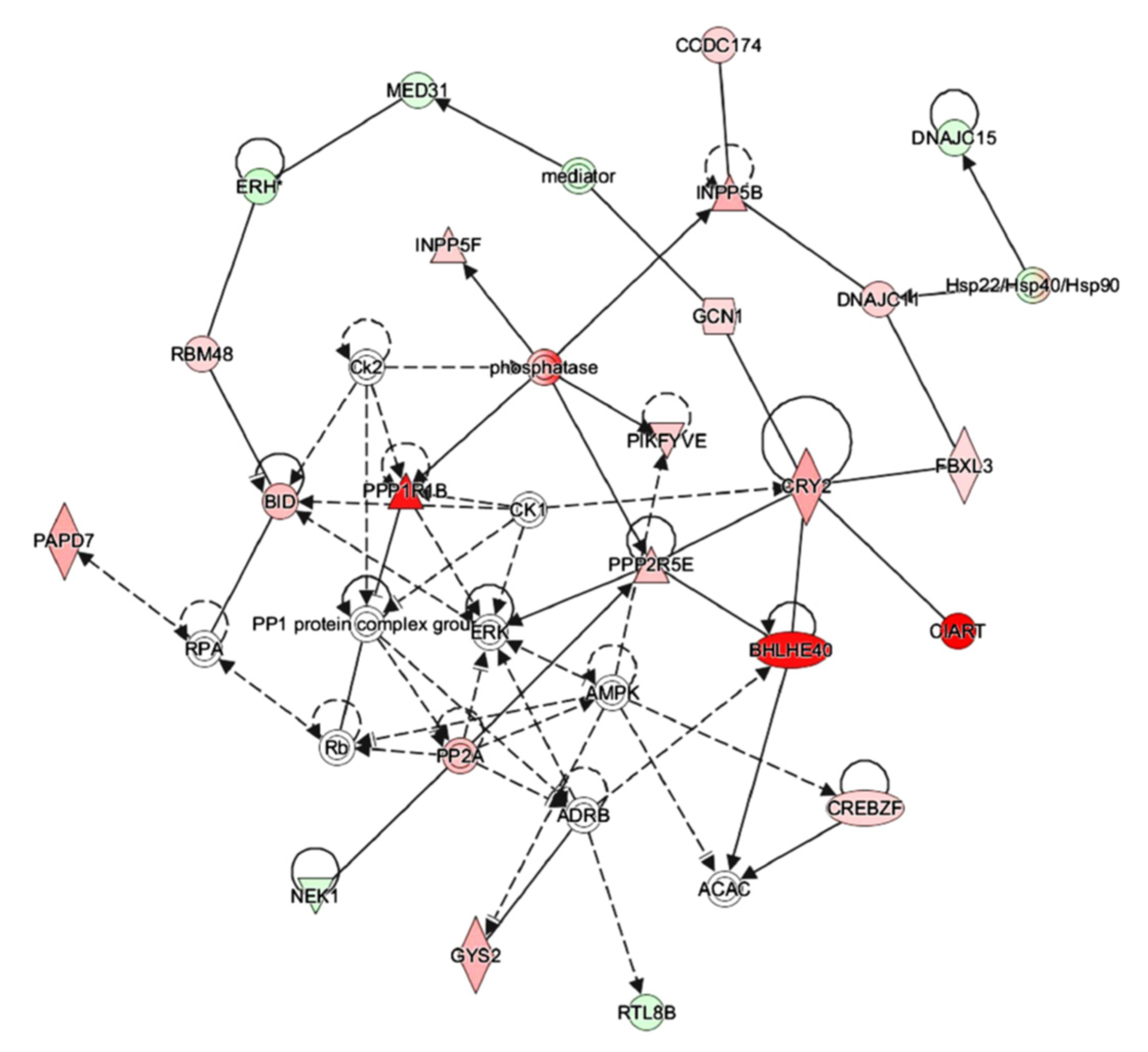
3.7. IPA Analysis Indicated That Octacosanol Modulated AMPK and ERK Pathways
3.8. Octacosanol Affected Lipid Metabolism-Related Gene Expressions
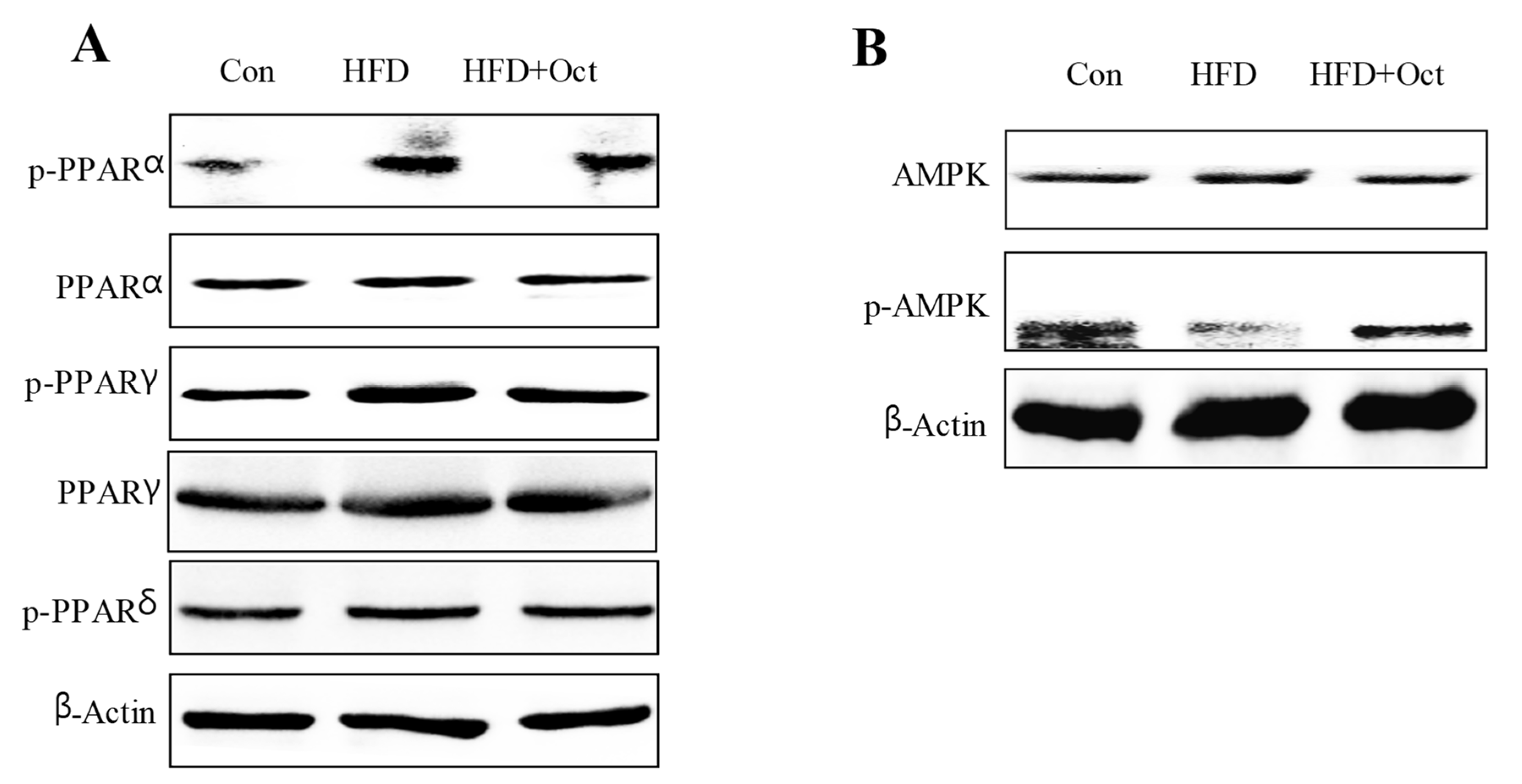
3.9. Octacosanol Affected PPAR and AMPK Signal Pathways
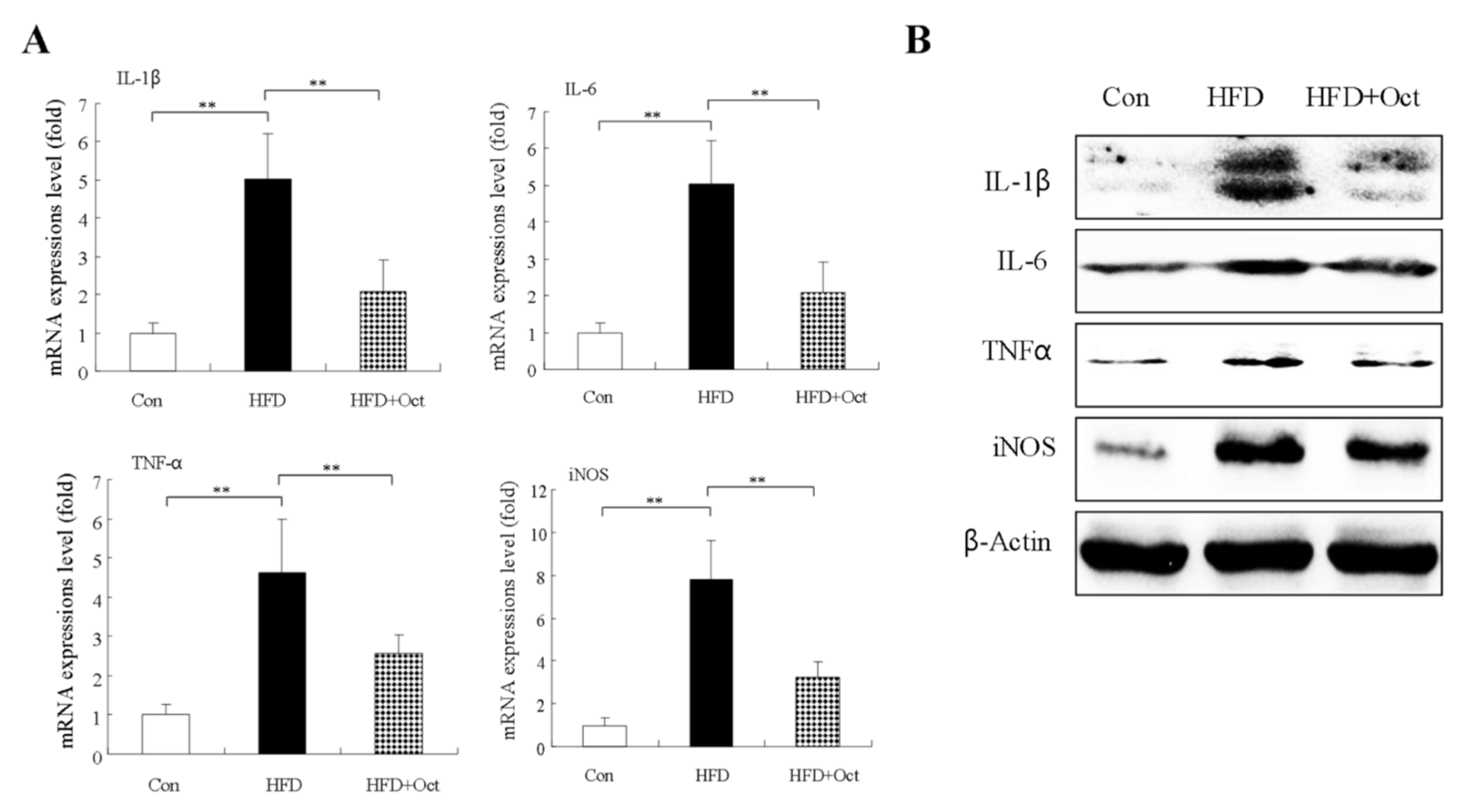
3.10. Octacosanol Decreases Inflammatory Factor Expressions
4. Discussions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barness, L.A.; Opitz, J.M.; Gilbert-Barness, E. Obesity: Genetic, molecular, and environmental aspects. Am. J. Med. Genet. 2007, 143, 3016–3034. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Stone, N.J.; Ballantyne, C.; Bittner, V.; Criqui, M.H.; Ginsberg, H.N.; Goldberg, A.C.; Howard, W.J. Triglycerides and cardiovascular disease: A scientific statement from the American Heart Association. Circulation 2011, 123, 2292–2333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toth, P.P. Triglyceride-rich lipoproteins as a causal factor for cardiovascular disease. Vasc. Health Risk Manag. 2016, 12, 171–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarp, J.B.; Jensen, A.S.; Engstrøm, T.; Holstein-Rathlou, N.H.; Søndergaard, L. Cyanotic congenital heart disease and atherosclerosis. Heart 2017, 103, 897–900. [Google Scholar] [CrossRef]
- Panahi, Y.; Ahmadi, Y.; Teymouri, M.; Johnston, T.P.; Sahebkar, A. Curcumin as a potential candidate for treating hyperlipidemia: A review of cellular and metabolic mechanisms. J. Cell. Physiol. 2018, 233, 141–152. [Google Scholar] [CrossRef]
- Beppu, F.; Li, H.; Yoshinaga, K.; Nagai, T.; Yoshinda, A.; Kubo, A. Dietary starfish oil prevents hepatic steatosis and hyperlipidemia in C57BL/6N mice fed high-fat diet. J. Oleo Sci. 2017, 66, 761–769. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Lin, Q.; Li, X.; Nie, Y.; Wang, L.; Shi, L.; Xu, W.; Hu, T. Octacosanol attenuates inflammation in both RAW264.7 macrophages and a mouse model of colitis. J. Agric. Food Chem. 2017, 65, 3647–3658. [Google Scholar] [CrossRef]
- Shen, J.J.; Luo, F.J.; Lin, Q.L. Policosanol: Extraction and biological functions. J. Funct. Foods 2019, 57, 351–353. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Cao, F.L.; Luo, F.J.; Li, Q.L. Octacosanol and health benefits: Biological functions and mechanisms of action. Food Biosci. 2022, 47, 101632. [Google Scholar] [CrossRef]
- Sharma, R.; Matsuzaka, T.; Kaushik, M.K.; Sugasawa, T.; Ohno, H.; Wang, Y.; Motomura, K.; Shimura, T. Octacosanol and policosanol prevent high-fat diet-induced obesity and metabolic disorders by activating brown adipose tissue and improving liver metabolism. Sci. Rep. 2019, 9, 5169–5181. [Google Scholar] [CrossRef]
- Nie, Y.; Luo, F.; Wang, L.; Yang, T.; Shi, L.; Li, X. Anti-hyperlipidemic effect of rice bran polysaccharide and its potential mechanism in high-fat diet mice. Food Funct. 2017, 8, 4028–4041. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, Q.; Yang, T.; Liang, Y.; Nie, Y.; Luo, Y.; Shen, J.; Fu, X. Oryzanol modifies high fat diet-induced obesity, liver gene expression profile, and inflammation response in mice. J. Agric. Food Chem. 2017, 65, 8374–8385. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, R.; Thurmond, D.C. Mechanisms by which skeletal muscle myokines ameliorate insulin resistance. Int. J. Mol. Sci. 2022, 23, 4636. [Google Scholar] [CrossRef] [PubMed]
- Mulvihill, E.E.; Burke, A.C.; Huff, M.W. Citrus flavonoids as regulators of lipoprotein metabolism and atherosclerosis. Annu. Rev. Nutr. 2016, 36, 275–295. [Google Scholar] [CrossRef] [PubMed]
- Hassen, W.; Gao, R.; Abdelkader, D.; Ding, L.; Wang, L.; Liu, J.; Shang, J. Reduced oxidative stress contributes to the lipid lowering effects of isoquercitrin in free fatty acids induced hepatocytes. Oxid. Med. Cell. Longev. 2014, 2014, 313602. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Zhang, H.B.; Yi, C.P.; Quan, K.; Lin, B. Chemical composition, structure and functional properties of rice bran dietary fiber modified by cellulase treatment. Food Chem. 2021, 342, 128352. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Hardie, D.G. AMPK: Sensing glucose as well as cellular energy status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Lin, Q.; Guo, T.; Yang, T.; Zhou, W.; Deng, X.; Yan, J.K.; Luo, Y. Polysaccharide isolated from Phellinus linteus mycelia exerts anti-inflammatory effects via MAPK and PPAR signaling pathways. Carbohydr. Polym. 2018, 200, 487–497. [Google Scholar] [CrossRef]
- Chang, H.C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014, 25, 138–145. [Google Scholar] [CrossRef]
- Li, X. SIRT1 and energy metabolism. Acta Biochim. Biophys. Sin. 2013, 45, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Saha, A.K.; Ruderman, N.B. Malonyl-CoA and AMP-activated protein kinase: An expanding partnership. Mol. Cell. Biochem. 2003, 253, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Munday, M.R.; Hemingway, C.J. The regulation of acetyl-CoA carboxylase--a potential target for the action of hypolipidemic agents. Adv. Enzym. Regul. 1999, 39, 205–234. [Google Scholar] [CrossRef]
- Bi, L.; Jiang, Z.; Zhou, J. The role of lipin-1 in the pathogenesis of alcoholic fatty liver. Alcohol Alcohol. 2015, 50, 146–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Lu, W.; Jin, Z.; Yu, J.; Shi, B. Carbenoxolone ameliorates hepatic lipid metabolism and inflammation in obese mice induced by high fat diet via regulating the JAK2/STAT3 signaling pathway. Int. Immunopharmacol. 2019, 74, 105498. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog. Lipid Res. 2013, 52, 175–191. [Google Scholar] [CrossRef] [Green Version]
- Bełtowski, J.; Jamroz-Wiśniewska, A.; Widomska, S. Adiponectin and its role in cardiovascular diseases. Cardiovasc. Hematol. Disord. Drug Targets 2008, 8, 7–46. [Google Scholar] [CrossRef]
- Wagner, N.; Wagner, K.D. PPAR Beta/delta and the hallmarks of cancer. Cells 2020, 9, 1133. [Google Scholar] [CrossRef]
- Shi, L.; Lin, Q.; Li, X.; Nie, Y.; Sun, S.; Deng, X.; Wang, L.; Lu, J. Alliin, a garlic organosulfur compound, ameliorates gut inflammation through MAPK-NF-kappaB/AP-1/STAT-1 inactivation and PPAR-gamma activation. Mol. Nutr. Food Res. 2017, 61, 160101. [Google Scholar] [CrossRef]
- Shi, L.; Lin, Q.; Yang, T.; Nie, Y.; Li, X.; Liu, B.; Shen, J.; Liang, Y. Oral administration of Lentinus edodes β-glucans ameliorates DSS-induced ulcerative colitis in mice via MAPK-Elk-1 and MAPK-PPARγ pathways. Food Funct. 2016, 7, 4614–4627. [Google Scholar] [CrossRef]
- Wang, Y.; Nakajima, T.; Gonzalez, F.J.; Tanaka, N. PPARs as metabolic regulators in the liver: Lessons from liver-specific PPAR-null mice. Int. J. Mol. Sci. 2020, 21, 2061. [Google Scholar] [CrossRef] [Green Version]
- Kyriakis, J.M.; Avruch, J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol. Rev. 2012, 92, 689–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.J.; Yang, T.; Xu, Y.; Luo, Y.; Zhong, X.; Shi, L.; Hu, T.; Guo, T. δ-Tocotrienol, isolated from rice bran, exerts an anti-Inflammatory effect via MAPKs and PPARs signaling pathways in lipopolysaccharide-stimulated macrophages. Int. J. Mol. Sci. 2018, 19, 3022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morello, S.; Ito, K.; Yamamura, S.; Lee, K.Y.; Jazrawi, E.; Desouza, P.; Barnes, P.; Cicala, C. IL-1 beta and TNF-alpha regulation of the adenosine receptor (A2A) expression: Differential requirement for NF-kappa B binding to the proximal promoter. J. Immunol. 2006, 177, 7173–7183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beetz, A.; Peter, R.U.; Oppel, T.; Kaffenberger, W.; Rupec, R.A.; Meyer, M.; Beuningen, D.; Kind, P. NF-kappaB and AP-1 are responsible for inducibility of the IL-6 promoter by ionizing radiation in HeLa cells. Int. J. Radiat. Biol. 2000, 76, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Steer, J.H.; Kroeger, K.M.; Abraham, L.J.; Joyce, D.A. Glucocorticoids suppress tumor necrosis factor-alpha expression by human monocytic THP-1 cells by suppressing transactivation through adjacent NF-kappa B and c-Jun-activating transcription factor-2 binding sites in the promoter. J. Biol. Chem. 2000, 275, 18432–18440. [Google Scholar] [CrossRef] [Green Version]
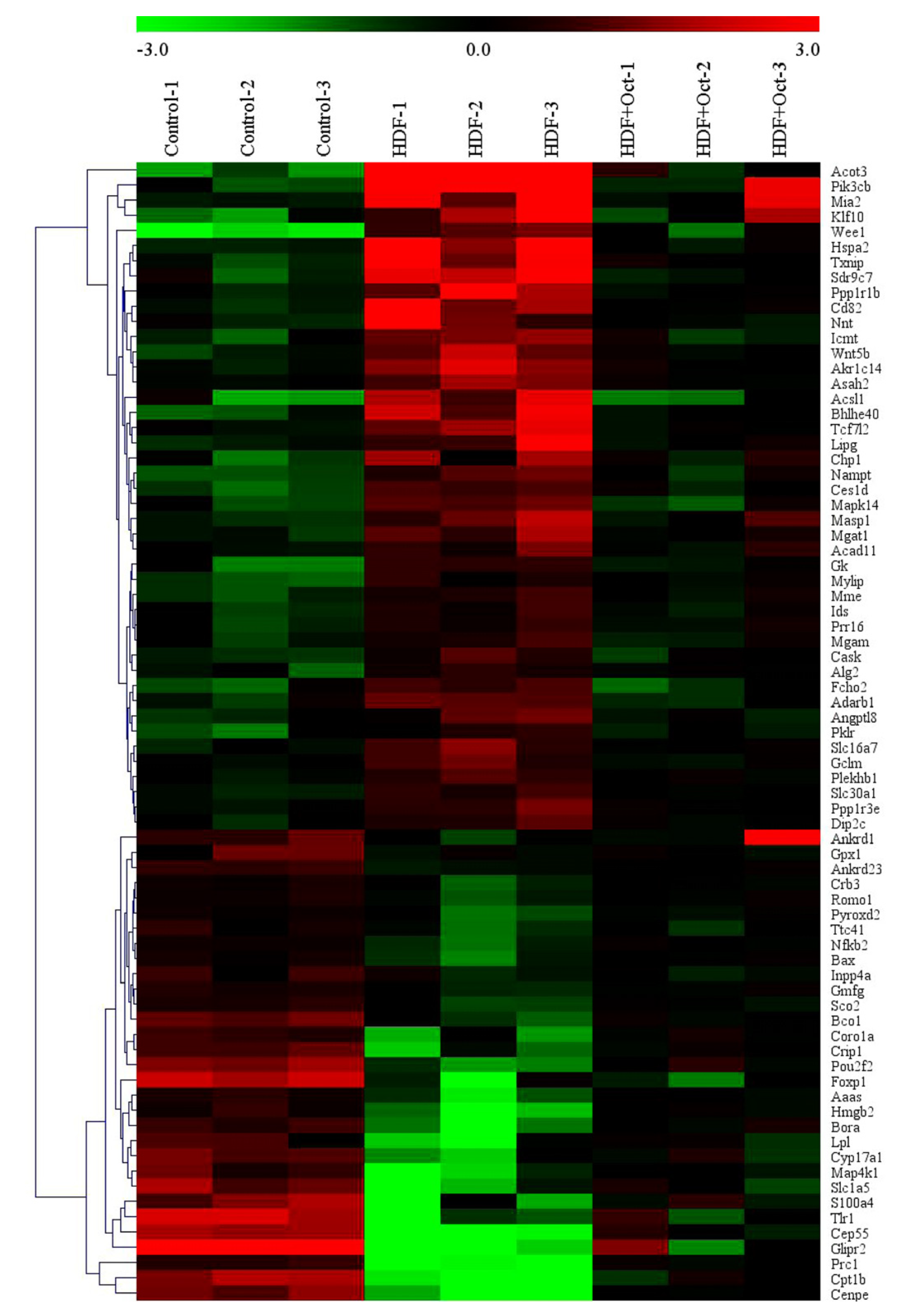

| ID | Gene Name | HFD/Con Fold Change | HFD/Con p-Value | HFD/HFD+Oct Fold Change | HFD/HFD+Oct p-Value |
|---|---|---|---|---|---|
| ENSMUSG00000026113.17 | Inpp4a | 0.53 | 0.036 | 0.53 | 0.019 |
| ENSMUSG00000060791.15 | Gmfg | 0.51 | 0.014 | 0.66 | 0.022 |
| ENSMUSG00000067847.13 | Romo1 | 0.50 | 0.035 | 0.75 | 0.001 |
| ENSMUSG00000060224.3 | Pyroxd2 | 0.47 | 0.048 | 0.72 | 0.028 |
| ENSMUSG00000067653.12 | Ankrd23 | 0.46 | 0.001 | 0.57 | 0.000 |
| ENSMUSG00000044937.14 | Ttc41 | 0.46 | 0.007 | 0.60 | 0.018 |
| ENSMUSG00000025225.14 | Nfkb2 | 0.45 | 0.022 | 0.78 | 0.031 |
| ENSMUSG00000091780.2 | Sco2 | 0.45 | 0.039 | 0.59 | 0.044 |
| ENSMUSG00000003873.11 | Bax | 0.44 | 0.014 | 0.77 | 0.035 |
| ENSMUSG00000024803.8 | Ankrd1 | 0.44 | 0.022 | 0.46 | 0.008 |
| ENSMUSG00000031845.15 | Bco1 | 0.34 | 0.021 | 0.44 | 0.005 |
| ENSMUSG00000030707.15 | Coro1a | 0.33 | 0.044 | 0.59 | 0.043 |
| ENSMUSG00000036678.7 | Aaas | 0.33 | 0.046 | 0.63 | 0.013 |
| ENSMUSG00000006360.11 | Crip1 | 0.30 | 0.030 | 0.49 | 0.015 |
| ENSMUSG00000003555.7 | Cyp17a1 | 0.28 | 0.033 | 0.42 | 0.050 |
| ENSMUSG00000054717.7 | Hmgb2 | 0.26 | 0.036 | 0.66 | 0.018 |
| ENSMUSG00000008496.19 | Pou2f2 | 0.25 | 0.006 | 0.42 | 0.033 |
| ENSMUSG00000037337.11 | Map4k1 | 0.24 | 0.049 | 0.49 | 0.032 |
| ENSMUSG00000022070.6 | Bora | 0.24 | 0.028 | 0.58 | 0.018 |
| ENSMUSG00000030067.17 | Foxp1 | 0.23 | 0.032 | 0.26 | 0.006 |
| ENSMUSG00000001918.17 | Slc1a5 | 0.22 | 0.045 | 0.39 | 0.031 |
| ENSMUSG00000001020.8 | S100a4 | 0.22 | 0.043 | 0.40 | 0.043 |
| ENSMUSG00000044827.10 | Tlr1 | 0.18 | 0.044 | 0.30 | 0.037 |
| ENSMUSG00000024989.14 | Cep55 | 0.14 | 0.016 | 0.36 | 0.017 |
| ENSMUSG00000078937.8 | Cpt1b | 0.14 | 0.022 | 0.34 | 0.006 |
| ENSMUSG00000038943.16 | Prc1 | 0.14 | 0.026 | 0.58 | 0.022 |
| ENSMUSG00000028480.14 | Glipr2 | 0.13 | 0.004 | 0.21 | 0.024 |
| ENSMUSG00000045328.11 | Cenpe | 0.11 | 0.048 | 0.41 | 0.004 |
| ID | Gene Name | HFD/Con Fold Change | HFD/Con p-Value | HFD/HFD+Oct Fold Change | HFD/HFD+Oct p-Value |
|---|---|---|---|---|---|
| ENSMUSG00000048264.15 | Dip2c | 2.05 | 0.026 | 1.66 | 0.047 |
| ENSMUSG00000039740.6 | Alg2 | 2.09 | 0.043 | 1.36 | 0.005 |
| ENSMUSG00000030701.17 | Plekhb1 | 2.10 | 0.035 | 1.69 | 0.005 |
| ENSMUSG00000028124.15 | Gclm | 2.11 | 0.037 | 2.21 | 0.048 |
| ENSMUSG00000068587.9 | Mgam | 2.11 | 0.042 | 1.79 | 0.005 |
| ENSMUSG00000073565.4 | Prr16 | 2.13 | 0.028 | 1.43 | 0.015 |
| ENSMUSG00000090150.8 | Acad11 | 2.14 | 0.048 | 1.60 | 0.002 |
| ENSMUSG00000035847.15 | Ids | 2.17 | 0.034 | 1.57 | 0.000 |
| ENSMUSG00000072494.7 | Ppp1r3e | 2.20 | 0.010 | 1.70 | 0.048 |
| ENSMUSG00000037434.7 | Slc30a1 | 2.25 | 0.009 | 1.55 | 0.010 |
| ENSMUSG00000041237.12 | Pklr | 2.33 | 0.039 | 1.56 | 0.034 |
| ENSMUSG00000031012.17 | Cask | 2.43 | 0.026 | 1.70 | 0.015 |
| ENSMUSG00000020102.15 | Slc16a7 | 2.44 | 0.010 | 1.79 | 0.038 |
| ENSMUSG00000047822.7 | Angptl8 | 2.45 | 0.017 | 2.04 | 0.047 |
| ENSMUSG00000027820.12 | Mme | 2.53 | 0.005 | 1.36 | 0.023 |
| ENSMUSG00000038175.9 | Mylip | 2.56 | 0.007 | 1.24 | 0.043 |
| ENSMUSG00000024887.9 | Asah2 | 2.57 | 0.014 | 2.23 | 0.029 |
| ENSMUSG00000020262.15 | Adarb1 | 2.59 | 0.025 | 2.96 | 0.012 |
| ENSMUSG00000041685.15 | Fcho2 | 2.72 | 0.023 | 2.61 | 0.023 |
| ENSMUSG00000025059.16 | Gk | 2.79 | 0.033 | 1.60 | 0.017 |
| ENSMUSG00000020346.16 | Mgat1 | 2.81 | 0.040 | 1.73 | 0.017 |
| ENSMUSG00000053436.14 | Mapk14 | 2.93 | 0.018 | 2.59 | 0.020 |
| ENSMUSG00000014077.13 | Chp1 | 3.00 | 0.016 | 1.57 | 0.027 |
| ENSMUSG00000022887.8 | Masp1 | 3.05 | 0.031 | 1.61 | 0.004 |
| ENSMUSG00000056973.6 | Ces1d | 3.08 | 0.002 | 1.61 | 0.015 |
| ENSMUSG00000053846.4 | Lipg | 3.08 | 0.032 | 2.05 | 0.045 |
| ENSMUSG00000024985.18 | Tcf7l2 | 3.12 | 0.026 | 2.69 | 0.014 |
| ENSMUSG00000039662.16 | Icmt | 3.13 | 0.013 | 2.44 | 0.030 |
| ENSMUSG00000030170.14 | Wnt5b | 3.14 | 0.013 | 1.94 | 0.042 |
| ENSMUSG00000020572.7 | Nampt | 3.14 | 0.004 | 1.65 | 0.043 |
| ENSMUSG00000025453.16 | Nnt | 3.16 | 0.019 | 2.69 | 0.035 |
| ENSMUSG00000033715.14 | Akr1c14 | 3.22 | 0.020 | 2.24 | 0.024 |
| ENSMUSG00000061718.12 | Ppp1r1b | 3.61 | 0.048 | 2.76 | 0.035 |
| ENSMUSG00000027215.13 | Cd82 | 3.69 | 0.011 | 2.33 | 0.032 |
| ENSMUSG00000018796.13 | Acsl1 | 3.93 | 0.032 | 3.48 | 0.007 |
| ENSMUSG00000030103.11 | Bhlhe40 | 3.99 | 0.008 | 2.23 | 0.028 |
| ENSMUSG00000040127.13 | Sdr9c7 | 4.10 | 0.009 | 3.47 | 0.003 |
| ENSMUSG00000038393.14 | Txnip | 4.15 | 0.006 | 2.33 | 0.022 |
| ENSMUSG00000037465.9 | Klf10 | 4.27 | 0.014 | 1.82 | 0.000 |
| ENSMUSG00000035349.5 | Mia2 | 4.43 | 0.043 | 2.00 | 0.032 |
| ENSMUSG00000059970.6 | Hspa2 | 4.44 | 0.015 | 3.03 | 0.011 |
| ENSMUSG00000031016.9 | Wee1 | 4.96 | 0.002 | 1.47 | 0.049 |
| ENSMUSG00000032462.14 | Pik3cb | 6.10 | 0.008 | 2.78 | 0.047 |
| ENSMUSG00000021228.14 | Acot3 | 10.43 | 0.003 | 4.28 | 0.007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, J.; Yang, T.; Zhou, Y.; Xu, W.; Han, S.; Guo, T.; Zhu, L.; Qin, D.; Luo, Y.; Hu, Z.; et al. Octacosanol Modifies Obesity, Expression Profile and Inflammation Response of Hepatic Tissues in High-Fat Diet Mice. Foods 2022, 11, 1606. https://doi.org/10.3390/foods11111606
Bai J, Yang T, Zhou Y, Xu W, Han S, Guo T, Zhu L, Qin D, Luo Y, Hu Z, et al. Octacosanol Modifies Obesity, Expression Profile and Inflammation Response of Hepatic Tissues in High-Fat Diet Mice. Foods. 2022; 11(11):1606. https://doi.org/10.3390/foods11111606
Chicago/Turabian StyleBai, Jie, Tao Yang, Yaping Zhou, Wei Xu, Shuai Han, Tianyi Guo, Lingfeng Zhu, Dandan Qin, Yi Luo, Zuomin Hu, and et al. 2022. "Octacosanol Modifies Obesity, Expression Profile and Inflammation Response of Hepatic Tissues in High-Fat Diet Mice" Foods 11, no. 11: 1606. https://doi.org/10.3390/foods11111606
APA StyleBai, J., Yang, T., Zhou, Y., Xu, W., Han, S., Guo, T., Zhu, L., Qin, D., Luo, Y., Hu, Z., Wu, X., Luo, F., Liu, B., & Lin, Q. (2022). Octacosanol Modifies Obesity, Expression Profile and Inflammation Response of Hepatic Tissues in High-Fat Diet Mice. Foods, 11(11), 1606. https://doi.org/10.3390/foods11111606