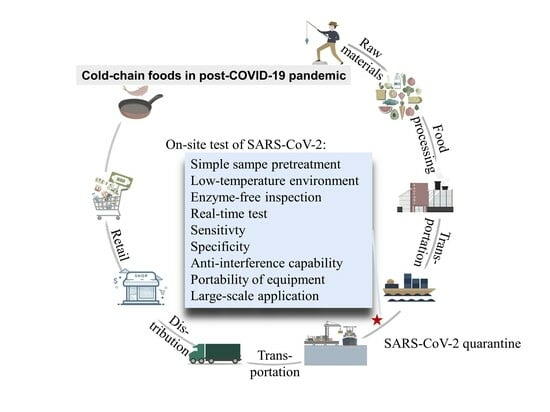
The Safety of Cold-Chain Food in Post-COVID-19 Pandemic: Precaution and Quarantine
Abstract
:1. Introduction
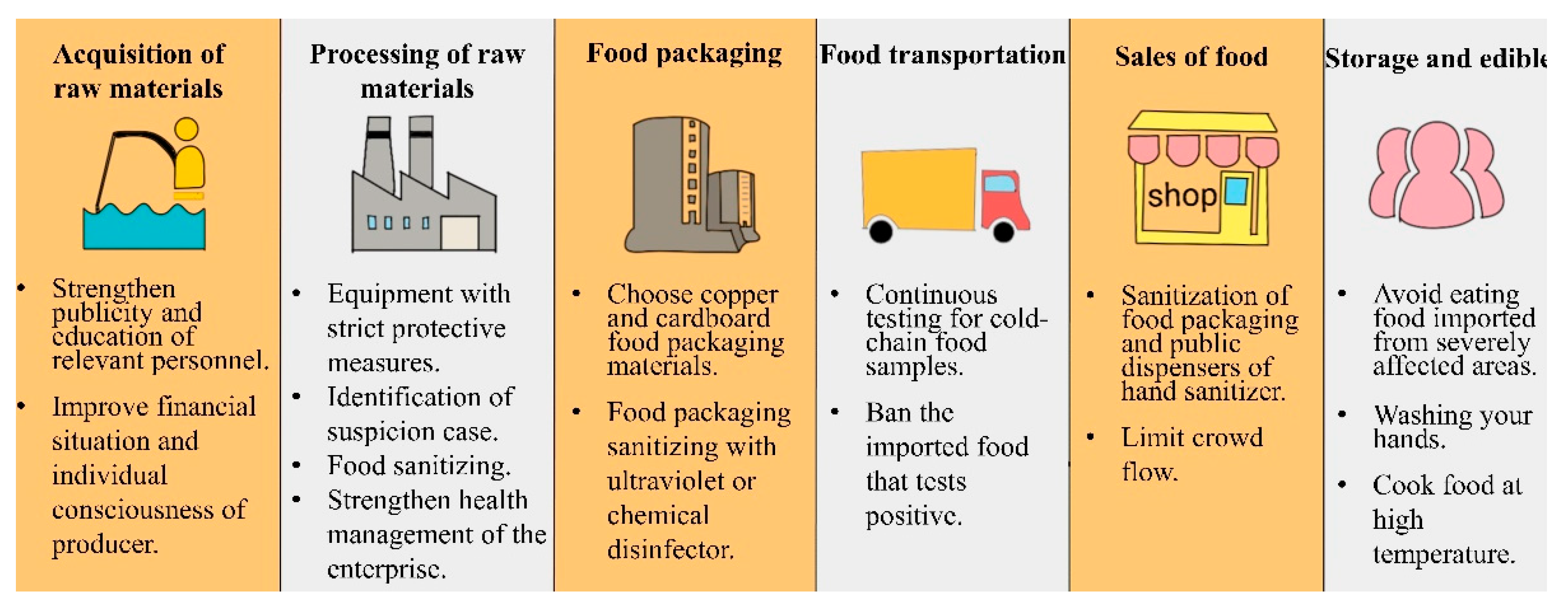
2. Safety Precautions in Cold-Chain Links
2.1. Acquisition of Food Raw Materials
2.2. Processing of Food Raw Materials
2.3. Food Packaging
2.4. Food Transportation
2.5. Sales of Food
2.6. Consumption
3. Characteristics for the Cold-Chain Food Quarantine
4. Potential Cold-Chain Food Quarantine Techniques
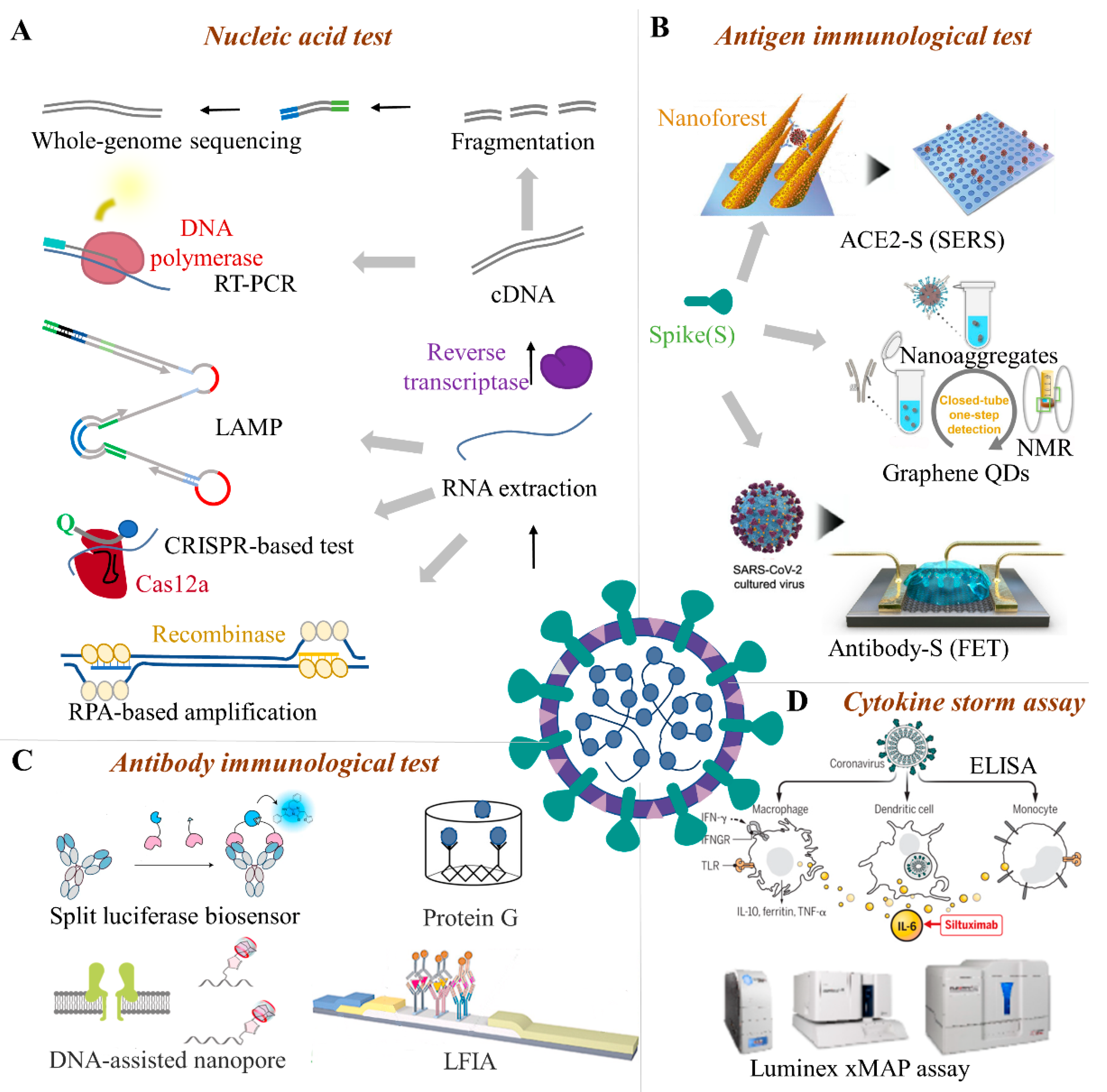
4.1. Nucleic Acid Test
4.1.1. PCR-Based Techniques
4.1.2. RT-LAMP
4.1.3. CRISPR-Based System
4.1.4. Microfluidic Biochip
4.1.5. Whole-Genome Sequencing
4.2. Immunological Methods
4.2.1. Antigen Immunological Test
4.2.2. Serum Antibody Immunological Test
4.2.3. Cytokine Storm Assay
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 20 February 2022).
- Rossi, G.A.; Sacco, O.; Mancino, E.; Cristiani, L.; Midulla, F. Differences and similarities between SARS-CoV and SARS-CoV-2: Spike receptor-binding domain recognition and host cell infection with support of cellular serine proteases. Infection 2020, 48, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Aboubakr, H.A.; Sharafeldin, T.A.; Goyal, S.M. Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the influence of climatic conditions: A review. Transbound. Emerg. Dis. 2021, 68, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Krammer, F. SARS-CoV-2 vaccines: Status report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, J.V.; Ratzan, S.C.; Palayew, A.; Gostin, L.O.; Larson, H.J.; Rabin, K.; Kimball, S.; El-Mohandes, A. A global survey of potential acceptance of a COVID-19 vaccine. Nat. Med. 2021, 27, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Sharun, K.; Tiwari, R.; Dadar, M.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W. COVID-19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vacc. Immunother. 2020, 16, 1232–1238. [Google Scholar] [CrossRef]
- Waltenburg, M.A.; Victoroff, T.; Rose, C.E.; Butterfield, M.; Jervis, R.H.; Fedak, K.M.; Gabel, J.A.; Feldpausch, A.; Dunne, E.M.; Austin, C.; et al. Update: COVID-19 among workers in meat and poultry processing facilities―United States, April–May 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 887. [Google Scholar] [CrossRef]
- Fisher, D.; Reilly, A.; Zheng, A.K.E.; Cook, A.R.; Anderson, D. Seeding of outbreaks of COVID-19 by contaminated fresh and frozen food. bioRxiv 2020, 17, 255166. [Google Scholar]
- Desai, A.N.; Aronoff, D.M. Food Safety and COVID-19. JAMA 2020, 323, 1982. [Google Scholar] [CrossRef]
- Li, Z.; Liu, F.; Cui, J.; Peng, Z.; Chang, Z.; Lai, S.; Chen, Q.; Wang, L.; Gao, G.; Feng, Z. Comprehensive large-scale nucleic acid–testing strategies support China’s sustained containment of COVID-19. Nat. Med. 2021, 27, 740–742. [Google Scholar] [CrossRef]
- Ma, H.; Wang, Z.; Zhao, X.; Han, J.; Zhang, Y.; Wang, H.; Chen, C.; Wang, J.; Jiang, F.; Lei, J.; et al. Long Distance Transmission of SARS-CoV-2 from Contaminated Cold Chain Products to Humans-Qingdao City, Shandong Province, China, September 2020. China CDC Wkly. 2021, 3, 637–644. [Google Scholar] [CrossRef]
- Tebas, P.; Yang, S.P.; Boyer, J.D.; Reuschel, E.L.; Patel, A.; Christensen-Quick, A.; Andrade, V.M.; Morrow, M.P.; Kraynyak, K.; Agnes, J.; et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. eClinicalMedicine 2021, 31, 100689. [Google Scholar] [CrossRef]
- Pang, X.; Ren, L.; Wu, S.; Ma, W.; Yang, J.; Di, L.; Li, J.; Xiao, Y.; Kang, L.; Du, S.; et al. Cold-chain food contamination as the possible origin of COVID-19 resurgence in Beijing. Natl. Sci. Rev. 2020, 7, 1861–1864. [Google Scholar] [CrossRef]
- Lu, L.-C.; Quintela, I.; Lin, C.-H.; Lin, T.-C.; Lin, C.-H.; Wu, V.C.H.; Lin, C.-S. A review of epidemic investigation on cold-chain food-mediated SARS-CoV-2 transmission and food safety consideration during COVID-19 pandemic. J. Food Saf. 2021, 41, e12932. [Google Scholar] [CrossRef]
- Mercier, S.; Villeneuve, S.; Mondor, M.; Uysal, I. Time–Temperature Management Along the Food Cold Chain: A Review of Recent Developments. Compr. Rev. Food Sci. Food Saf. 2017, 16, 647–667. [Google Scholar] [CrossRef]
- Oliva, F.; Revetria, R. A system dynamic model to support cold chain management in food supply chain. In Proceedings of the WSEAS International Conference on Mathematics and Computers in Science and Engineering, Heraklion, Greece, 22–24 July 2008. [Google Scholar]
- Ferioli, M.; Cisternino, C.; Leo, V.; Pisani, L.; Palange, P.; Nava, S. Protecting healthcare workers from SARS-CoV-2 infection: Practical indications. Eur. Respir. Rev. 2020, 29, 200068. [Google Scholar] [CrossRef]
- Liu, L.; Hu, J.; Hou, Y.; Tao, Z.; Chen, Z.; Chen, K. Pit latrines may be a potential risk in rural China and low-income countries when dealing with COVID-19. Sci. Total Environ. 2021, 761, 143283. [Google Scholar]
- Shahbaz, M.; Bilal, M.; Moiz, A.; Zubair, S.; Iqbal, H. Food safety and COVID-19: Precautionary measures to limit the spread of coronavirus at food service and retail sector. J. Pure Appl. Microbiol. 2020, 14, 749–756. [Google Scholar] [CrossRef]
- Goli, M. Review of novel human β-coronavirus (2019-nCoV or SARS-CoV-2) from the food industry perspective—Appropriate approaches to food production technology. Food Sci. Nutr. 2020, 8, 5228–5237. [Google Scholar]
- Lee, A.C.Y.; Zhang, A.J.; Chan, J.F.W.; Li, C.; Fan, Z.; Liu, F.; Chen, Y.; Liang, R.; Sridhar, S.; Cai, J.-P.; et al. Oral SARS-CoV-2 inoculation establishes subclinical respiratory infection with virus shedding in golden Syrian hamsters. Cell Rep. Med. 2020, 1, 100121. [Google Scholar] [CrossRef]
- Azuma, K.; Yanagi, U.; Kagi, N.; Kim, H.; Ogata, M.; Hayashi, M. Environmental factors involved in SARS-CoV-2 transmission: Effect and role of indoor environmental quality in the strategy for COVID-19 infection control. Environ. Health Prev. Med. 2020, 25, 66. [Google Scholar] [CrossRef]
- Chan, K.-H.; Peiris, J.M.; Lam, S.; Poon, L.; Yuen, K.; Seto, W.H. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv. Virol. 2011, 2021, 734690. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fang, X.; Mao, Y.; Qi, H.; Wu, J.; Liu, X.; You, F.; Zhao, W.; Chen, Y.; Zheng, L. Real-time, selective, and low-cost detection of trace level SARS-CoV-2 spike-protein for cold-chain food quarantine. Npj Sci. Food 2021, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Zhang, J.; Wu, J.; Fang, X.; You, F.; Zhao, W.; Liu, X.; Chen, Y.; Zheng, L. Real-time Sensing of Trace Biomarkers from Viruses with a Microfluidic Immunosensor: A Case Study of SARS-CoV-2 Detection in Cold-chain Food. ChemRxiv 2020, 1, 12. [Google Scholar] [CrossRef]
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef]
- Oliveira, T.C.; Abranches, M.V.; Lana, R.M. (In) Segurança alimentar no contexto da pandemia por SARS-CoV-2. Cad. Saude Publica 2020, 36, e00055220. [Google Scholar] [CrossRef]
- Baležentis, T.; Morkūnas, M.; Žičkienė, A.; Volkov, A.; Ribašauskienė, E.; Štreimikienė, D. Policies for Rapid Mitigation of the Crisis’ Effects on Agricultural Supply Chains: A Multi-Criteria Decision Support System with Monte Carlo Simulation. Sustainability 2021, 13–21, 11899. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; Perera, M.R.A.; Hui, K.P.Y.; Yen, H.L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- Safe Minimum Cooking Temperatures Chart. Available online: https://www.foodsafety.gov/food-safety-charts/safe-minimum-cooking-temperature (accessed on 20 February 2022).
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef]
- Hokajärvi, A.M.; Rytkönen, A.; Tiwari, A.; Kauppinen, A.; Oikarinen, S.; Lehto, K.M.; Kankaanpää, A.; Gunnar, T.; Al-Hello, H.; Blomqvist, S.; et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021, 770, 145274. [Google Scholar] [CrossRef]
- Yekta, R.; Vahid-Dastjerdi, L.; Norouzbeigi, S.; Mortazavian, A.M. Food products as potential carriers of SARS-CoV-2. Food Control 2021, 123, 107754. [Google Scholar] [CrossRef]
- Venugopal, V.; Doke, S.N.; Thomas, P. Radiation processing to improve the quality of fishery products. Crit. Rev. Food Sci. 1999, 39, 391–440. [Google Scholar] [CrossRef]
- Yang, Z.; Mammel, M.; Papafragkou, E.; Hida, K.; Elkins, C.A.; Kulka, M. Application of next generation sequencing toward sensitive detection of enteric viruses isolated from celery samples as an example of produce. Int. J. Food Microbiol. 2017, 261, 73–81. [Google Scholar] [CrossRef]
- Zhang, L.; Xue, L.; Gao, J.; Cai, W.; Jiang, Y.; Zuo, Y.; Liao, Y.; Qin, Z.; Wu, H.; Cheng, T.; et al. Development of a high-efficient concentrated pretreatment method for noroviruses detection in independent oysters: An extension of the ISO/TS 15216-2:2013 standard method. Food Control 2020, 111, 107032. [Google Scholar] [CrossRef]
- Zhang, W.; He, H.; Zhu, L.; Liu, G.; Wu, L. Food Safety in Post-COVID-19 Pandemic: Challenges and Countermeasures. Biosensors 2021, 11, 71. [Google Scholar] [CrossRef]
- Rizou, M.; Galanakis, I.M.; Aldawoud, T.M.S.; Galanakis, C.M. Safety of foods, food supply chain and environment within the COVID-19 pandemic. Trends Food Sci. Technol. 2020, 102, 293–299. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Liu, P.; Yang, M.; Zhao, X.; Guo, Y.; Wang, L.; Zhang, J.; Lei, W.; Han, W.; Jiang, F.; Liu, W.J.; et al. Cold-chain transportation in the frozen food industry may have caused a recurrence of COVID-19 cases in destination: Successful isolation of SARS-CoV-2 virus from the imported frozen cod package surface. Biosaf. Health 2020, 2, 199–201. [Google Scholar] [CrossRef]
- Mutesa, L.; Ndishimye, P.; Butera, Y.; Souopgui, J.; Uwineza, A.; Rutayisire, R.; Ndoricimpaye, E.L.; Musoni, E.; Rujeni, N.; Nyatanyi, T.; et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature 2021, 589, 276–280. [Google Scholar] [CrossRef]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Fu, Z.; Zhao, S.; Zhang, X.; Polovka, M.; Wang, X. Quality Characteristics Analysis and Remaining Shelf Life Prediction of Fresh Tibetan Tricholoma matsutake under Modified Atmosphere Packaging in Cold Chain. Foods 2019, 8, 136. [Google Scholar] [CrossRef] [PubMed]
- Sarkhosh-Inanlou, R.; Shafiei-Irannejad, V.; Azizi, S.; Jouyban, A.; Ezzati-Nazhad Dolatabadi, J.; Mobed, A.; Adel, B.; Soleymani, J.; Hamblin, M.R. Applications of scaffold-based advanced materials in biomedical sensing. TrAC Trends Anal. Chem. 2021, 143, 116342. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 min. Anal. Chem. 2020, 92, 9454–9458. [Google Scholar] [CrossRef] [PubMed]
- Tayyab, M.; Sami, M.A.; Raji, H.; Mushnoori, S.; Javanmard, M. Potential Microfluidic Devices for COVID-19 Antibody Detection at Point-of-Care (POC): A Review. IEEE Sens. J. 2021, 21, 4007–4017. [Google Scholar] [CrossRef]
- Chi, Y.; Zheng, S.; Liu, C.; Wang, Q. Transmission of SARS-CoV-2 on cold-chain food overpacks: A new challenge. J. Glob. Health 2021, 11, 03071. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Rahbari, R.; Moradi, N.; Abdi, M. rRT-PCR for SARS-CoV-2: Analytical considerations. Clin. Chim. Acta 2021, 516, 1–7. [Google Scholar] [CrossRef]
- Böger, B.; Fachi, M.M.; Vilhena, R.O.; Cobre, A.F.; Tonin, F.S.; Pontarolo, R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control 2021, 49, 21–29. [Google Scholar] [CrossRef]
- Chaimayo, C.; Kaewnaphan, B.; Tanlieng, N.; Athipanyasilp, N.; Sirijatuphat, R.; Chayakulkeeree, M.; Angkasekwinai, N.; Sutthent, R.; Puangpunngam, N.; Tharmviboonsri, T.; et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2022, 17, 177. [Google Scholar] [CrossRef]
- Greninger, A.L.; Zerr, D.M.; Qin, X.; Adler, A.L.; Sampoleo, R.; Kuypers, J.M.; Englund, J.A.; Jerome, K.R. Rapid Metagenomic Next-Generation Sequencing during an Investigation of Hospital-Acquired Human Parainfluenza Virus 3 Infections. J. Clin. Microbiol. 2017, 55, 177–182. [Google Scholar] [CrossRef]
- Chiara, M.; D’Erchia, A.M.; Gissi, C.; Manzari, C.; Parisi, A.; Resta, N.; Zambelli, F.; Picardi, E.; Pavesi, G.; Horner, D.S.; et al. Next generation sequencing of SARS-CoV-2 genomes: Challenges, applications and opportunities. Brief. Bioinform. 2021, 22, 616–630. [Google Scholar] [CrossRef]
- Ji, C.; Xue, S.; Yu, M.; Liu, J.; Zhang, Q.; Zuo, F.; Zheng, Q.; Zhao, L.; Zhang, H.; Cao, J.; et al. Rapid Detection of SARS-CoV-2 Virus Using Dual Reverse Transcriptional Colorimetric Loop-Mediated Isothermal Amplification. ACS Omega 2021, 6, 8837–8849. [Google Scholar] [CrossRef]
- Ding, S.; Chen, G.; Wei, Y.; Dong, J.; Du, F.; Cui, X.; Huang, X.; Tang, Z. Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification. Biosens. Bioelectron. 2021, 178, 113041. [Google Scholar] [CrossRef]
- Day, A.S.; Ulep, T.H.; Safavinia, B.; Hertenstein, T.; Budiman, E.; Dieckhaus, L.; Yoon, J.Y. Emulsion-based isothermal nucleic acid amplification for rapid SARS-CoV-2 detection via angle-dependent light scatter analysis. Biosens. Bioelectron. 2021, 179, 113099. [Google Scholar] [CrossRef]
- Choi, M.H.; Lee, J.; Seo, Y.J. Combined recombinase polymerase amplification/rkDNA-graphene oxide probing system for detection of SARS-CoV-2. Anal. Chim. Acta 2021, 1158, 338390. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Cherkaoui, D.; Huang, D.; Miller, B.S.; Turbé, V.; McKendry, R.A. Harnessing recombinase polymerase amplification for rapid multi-gene detection of SARS-CoV-2 in resource-limited settings. Biosens. Bioelectron. 2021, 189, 113328. [Google Scholar] [CrossRef]
- Shelite, T.R.; Uscanga-Palomeque, A.C.; Castellanos, A.; Melby, P.C.; Travi, B.L. Isothermal recombinase polymerase amplification-lateral flow detection of SARS-CoV-2, the etiological agent of COVID-19. J. Virol. Methods 2021, 296, 114227. [Google Scholar] [CrossRef]
- Wang, D.; He, S.; Wang, X.; Yan, Y.; Liu, J.; Wu, S.; Liu, S.; Lei, Y.; Chen, M.; Li, L.; et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1150–1158. [Google Scholar] [CrossRef]
- Fan, Z.; Yao, B.; Ding, Y.; Zhao, J.; Xie, M.; Zhang, K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021, 178, 113015. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, R.; Lv, B.; Yu, X.; Liu, Y.; Yang, J.; Li, W.; Wang, Y.; Zhang, H.; Yan, G.; et al. Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction for detection of SARS-CoV-2 and HPV. Talanta 2021, 227, 122154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, J.; Li, Y.; Tan, G.; Sun, M.; Shan, Y.; Zhang, Y.; Wang, X.; Song, K.; Shi, R.; et al. SARS-CoV-2 detection using quantum dot fluorescence immunochromatography combined with isothermal amplification and CRISPR/Cas13a. Biosens. Bioelectron. 2022, 202, 113978. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Akashi, Y.; Kato, D.; Kuwahara, M.; Muramatsu, S.; Ueda, A.; Notake, S.; Nakamura, K.; Ishikawa, H.; Suzuki, H. The evaluation of a newly developed antigen test (QuickNavi™-COVID19 Ag) for SARS-CoV-2: A prospective observational study in Japan. J. Infect. Chemother. 2021, 27, 890–894. [Google Scholar] [CrossRef]
- Li, Y.; Ma, P.; Tao, Q.; Krause, H.J.; Yang, S.; Ding, G.; Dong, H.; Xie, X. Magnetic graphene quantum dots facilitate closed-tube one-step detection of SARS-CoV-2 with ultra-low field NMR relaxometry. Sens. Actuators B Chem. 2021, 337, 129786. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef]
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. SERS based lateral flow immunoassay for point-of-care detection of SARS-CoV-2 in clinical samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995. [Google Scholar] [CrossRef]
- Elledge, S.K.; Zhou, X.X.; Byrnes, J.R.; Martinko, A.J.; Lui, I.; Pance, K.; Lim, S.A.; Glasgow, J.E.; Glasgow, A.A.; Turcios, K.; et al. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2020, 39, 928–935. [Google Scholar] [CrossRef]
- Wang, Q.; Du, Q.; Guo, B.; Mu, D.; Lu, X.; Ma, Q.; Guo, Y.; Fang, L.; Zhang, B.; Zhang, G. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. J. Clin. Microbiol. 2020, 58, e00375–e00420. [Google Scholar] [CrossRef]
- Padoan, A.; Cosma, C.; Sciacovelli, L.; Faggian, D.; Plebani, M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab. Med. 2020, 58, 1081–1088. [Google Scholar] [CrossRef]
- Niedbala, R.S.; Feindt, H.; Kardos, K.; Vail, T.; Burton, J.; Bielska, B.; Li, S.; Milunic, D.; Bourdelle, P.; Vallejo, R. Detection of analytes by immunoassay using up-converting phosphor technology. Anal. Biochem. 2001, 293, 22–30. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Wei, X.; Zheng, S.W.; Lenhart, B.J.; Xu, P.; Li, J.; Pan, J.; Albrecht, H.; Liu, C. Multiplex quantitative detection of SARS-CoV-2 specific IgG and IgM antibodies based on DNA-assisted nanopore sensing. Biosens. Bioelectron. 2021, 181, 113134. [Google Scholar] [CrossRef]
- Bayin, Q.; Huang, L.; Ren, C.; Fu, Y.; Ma, X.; Guo, J. Anti-SARS-CoV-2 IgG and IgM detection with a GMR based LFIA system. Talanta 2021, 227, 122207. [Google Scholar] [CrossRef]
- Yao, Z.; Drecun, L.; Aboualizadeh, F.; Kim, S.J.; Li, Z.; Wood, H.; Valcourt, E.J.; Manguiat, K.; Plenderleith, S.; Yip, L.; et al. A homogeneous split-luciferase assay for rapid and sensitive detection of anti-SARS CoV-2 antibodies. Nat. Commun. 2021, 12, 1086. [Google Scholar] [CrossRef]
- Bundschuh, C.; Egger, M.; Wiesinger, K.; Gabriel, C.; Clodi, M.; Mueller, T.; Dieplinger, B. Evaluation of the EDI enzyme linked immunosorbent assays for the detection of SARS-CoV-2 IgM and IgG antibodies in human plasma. Clin. Chim. Acta 2020, 509, 79–82. [Google Scholar] [CrossRef]
- Al-Jighefee, H.T.; Yassine, H.M.; Nasrallah, G.K. Evaluation of antibody response in symptomatic and asymptomatic COVID-19 patients and diagnostic assessment of new IgM/IgG ELISA kits. Pathogens 2021, 10, 161. [Google Scholar] [CrossRef]
- Skalnikova, H.K.; Kepkova, K.V.; Vodicka, P. Luminex xMAP Assay to Quantify Cytokines in Cancer Patient Serum. In Immune Mediators in Cancer; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Torretta, S.; Zuccotti, G.; Cristofaro, V.; Ettori, J.; Solimeno, L.; Battilocchi, L.; D’Onghia, A.; Bonsembiante, A.; Pignataro, L.; Marchisio, P.; et al. Diagnosis of SARS-CoV-2 by RT-PCR Using Different Sample Sources: Review of the Literature. Ear Nose Throat J. 2021, 100, 131s–138s. [Google Scholar] [CrossRef]
- Sule, W.F.; Oluwayelu, D.O. Real-time RT-PCR for COVID-19 diagnosis: Challenges and prospects. Pan Afr. Med. J. 2020, 35, 121. [Google Scholar] [CrossRef]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test to Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438–e01520. [Google Scholar] [CrossRef] [PubMed]
- Yelin, I.; Aharony, N.; Tamar, E.S.; Argoetti, A.; Messer, E.; Berenbaum, D.; Shafran, E.; Kuzli, A.; Gandali, N.; Shkedi, O.; et al. Evaluation of COVID-19 RT-qPCR Test in Multi sample Pools. Clin. Infect. Dis. 2020, 71, 2073–2078. [Google Scholar] [CrossRef] [PubMed]
- Meza-Robles, C.; Barajas-Saucedo, C.E.; Tiburcio-Jimenez, D.; Mokay-Ramírez, K.A.; Melnikov, V.; Rodriguez-Sanchez, I.P.; Martinez-Fierro, M.L.; Garza-Veloz, I.; Zaizar-Fregoso, S.A.; Guzman-Esquivel, J.; et al. One-step nested RT-PCR for COVID-19 detection: A flexible, locally developed test for SARS-CoV2 nucleic acid detection. J. Infect. Dev. Ctries. 2020, 14, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Anderson, E.M.; Maldarelli, F. Quantification of HIV DNA Using Droplet Digital PCR Techniques. Curr. Protoc. Microbiol. 2018, 51, e62. [Google Scholar] [CrossRef]
- Yin, J.; Hu, J.; Sun, J.; Wang, B.; Mu, Y. A fast nucleic acid extraction system for point-of-care and integration of digital PCR. Analyst 2019, 144, 7032–7040. [Google Scholar] [CrossRef]
- Carraturo, F.; Del Giudice, C.; Morelli, M.; Cerullo, V.; Libralato, G.; Galdiero, E.; Guida, M. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020, 265, 115010. [Google Scholar] [CrossRef]
- Rödel, J.; Egerer, R.; Suleyman, A.; Sommer-Schmid, B.; Baier, M.; Henke, A.; Edel, B.; Löffler, B. Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J. Clin. Virol. 2020, 132, 104616. [Google Scholar] [CrossRef]
- Schermer, B.; Fabretti, F.; Damagnez, M.; Di Cristanziano, V.; Heger, E.; Arjune, S.; Tanner, N.A.; Imhof, T.; Koch, M.; Ladha, A.; et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS ONE 2020, 15, e0238612. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef]
- Rabe, B.A.; Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA 2020, 117, 24450–24458. [Google Scholar] [CrossRef]
- Xiang, X.; Qian, K.; Zhang, Z.; Lin, F.; Xie, Y.; Liu, Y.; Yang, Z. CRISPR-cas systems based molecular diagnostic tool for infectious diseases and emerging 2019 novel coronavirus (COVID-19) pneumonia. J. Drug Target. 2020, 28, 727–731. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef]
- Palaz, F.; Kalkan, A.K.; Tozluyurt, A.; Ozsoz, M. CRISPR-based tools: Alternative methods for the diagnosis of COVID-19. Clin. Biochem. 2021, 89, 1–13. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Lepej, S.Z.; Poljak, M. Portable molecular diagnostic instruments in microbiology: Current status. Clin. Microbiol. Infect. 2020, 26, 411–420. [Google Scholar] [CrossRef]
- Batule, B.S.; Seok, Y.; Kim, M.G. Paper-based nucleic acid testing system for simple and early diagnosis of mosquito-borne RNA viruses from human serum. Biosens. Bioelectron. 2020, 151, 111998. [Google Scholar] [CrossRef]
- Mitchell, S.L.; George, K.S. Evaluation of the COVID19 ID NOW EUA assay. J. Clin. Virol. 2020, 128, 104429. [Google Scholar] [CrossRef]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses-Current challenges and future perspectives. Biosens. Bioelectron. 2020, 163, 112291. [Google Scholar] [CrossRef]
- Van Tan, L.; Hong, N.T.T.; Ngoc, N.M.; Thanh, T.T.; Lam, V.T.; Nguyet, L.A.; Nhu, L.N.T.; Ny, N.T.H.; Minh, N.N.Q.; Man, D.N.H.; et al. SARS-CoV-2 and co-infections detection in nasopharyngeal throat swabs of COVID-19 patients by metagenomics. J. Infect. 2020, 81, e175–e177. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). bioRxiv 2021, 118, e2114828118. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Jiao, H.; Liu, Q.; Zhang, Z.; Xiong, Q.; Wang, B.-J.; Wang, X.; Guo, M.; Wang, L.-F.; Lan, K.; et al. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat. Ecol. Evol. 2021, 5, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.E.; Yount, B.L.; Graham, R.L.; Leist, S.R.; Hou, Y.J.; Dinnon, K.H.; Sims, A.C.; Swanstrom, J.; Gully, K.; Scobey, T.D.; et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA 2020, 117, 26915–26925. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.; Smith, G.; Pinette, M.; Embury-Hyatt, C.; Moffat, E.; Marszal, P.; Lewis, C.E. Susceptibility of domestic swine to experimental infection with SARS-CoV-2. bioRxiv 2020, 27, 104–112. [Google Scholar] [CrossRef]
- Santini, J.M.; Edwards, S.J. Host range of SARS-CoV-2 and implications for public health. Lancet Microbe 2020, 1, e141–e142. [Google Scholar] [CrossRef]
- Wu, J.L.; Tseng, W.P.; Lin, C.H.; Lee, T.F.; Chung, M.Y.; Huang, C.H.; Chen, S.Y.; Hsueh, P.R.; Chen, S.C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. [Google Scholar] [CrossRef]
- Van Elslande, J.; Houben, E.; Depypere, M.; Brackenier, A.; Desmet, S.; André, E.; Van Ranst, M.; Lagrou, K.; Vermeersch, P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1082–1087. [Google Scholar] [CrossRef]
- Roy, V.; Fischinger, S.; Atyeo, C.; Slein, M.; Loos, C.; Balazs, A.; Luedemann, C.; Astudillo, M.G.; Yang, D.; Wesemann, D.R.; et al. SARS-CoV-2-specific ELISA development. J. Immunol. Methods 2020, 484–485, 112832. [Google Scholar] [CrossRef]
- Fang, W.H.; Yang, J.R.; Lin, C.Y.; Hsiao, P.J.; Tu, M.Y.; Chen, C.F.; Tsai, D.J.; Su, W.; Huang, G.S.; Chang, H.; et al. Accuracy augmentation of body composition measurement by bioelectrical impedance analyzer in elderly population. Medicine 2020, 99, e19103. [Google Scholar] [CrossRef]
- Mak, G.C.; Cheng, P.K.; Lau, S.S.; Wong, K.K.; Lau, C.S.; Lam, E.T.; Chan, R.C.; Tsang, D.N.C. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020, 129, 104500. [Google Scholar] [CrossRef]
- Goode, J.A.; Rushworth, J.V.; Millner, P.A. Biosensor Regeneration: A Review of Common Techniques and Outcomes. Langmuir 2015, 31, 6267–6276. [Google Scholar] [CrossRef]
- Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in rapid detection of COVID-19. Biotech 2020, 10, 385. [Google Scholar] [CrossRef]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef]
- Xu, L.; Li, D.; Ramadan, S.; Li, Y.; Klein, N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020, 170, 112673. [Google Scholar] [CrossRef]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Thijsen, S.; Heron, M.; Gremmels, H.; van der Kieft, R.; Reusken, C.; Kremer, K.; Limonard, G.; Bossink, A. Elevated nucleoprotein-induced interferon-γ release in COVID-19 patients detected in a SARS-CoV-2 enzyme-linked immunosorbent spot assay. J. Infect. 2020, 81, 452–482. [Google Scholar] [CrossRef]
- Mauro, G.; Scavone, C.; Rafaniello, C.; Rossi, F.; Capuano, A. SARS-Cov-2 infection: Response of human immune system and possible implications for the rapid test and treatment. Int. Immunopharmacol. 2020, 84, 106519. [Google Scholar] [CrossRef]
| Methods | Category | Subcategory | LOD | Specificity | Sensitivity | Cost | Time | Description | References |
|---|---|---|---|---|---|---|---|---|---|
| Nucleic acid test | RT-PCR | 1–10 copies | 97.06–99.69% | 91.06–99.96% | USD 25–200 | 4-6 h | The gold standard for SARS-CoV-2 diagnosis is suitable for the large-scale test but needs specialized laboratory equipment and trained technicians | [51,52] | |
| Whole-genome sequencing | ND | ND | 98.33–99.83% | USD 2000 | 48-72 h | The first complete genomic sequences of SARS-CoV-2 were obtained through metatranscriptomics approaches | [53,54] | ||
| Isothermal amplification technology | Transcriptional colorimetric loop-mediated isothermal amplification | 100 copies/μL | 100% | 85% | ND | 21 h | Effectively reduce the false positive rate and improve the detection efficiency | [55] | |
| Proofreading enzyme-mediated isothermal amplification | 100 copies | Effectively distinguish SARS-CoV-2 from SARS-CoV | Effectively detect as few as 100 copies of gene N RNA in 1 h | ND | 50 min | Show similar analytical performance with the conventional RT-PCR | [56] | ||
| Emulsion loop-mediated isothermal amplification | 10, 103, and 105 copies/μL | ND | ND | ND | 5–10 min | Limit of detection of 1 copy per microliter sample and portable device using a miniature spectrometer or a smartphone | [57] | ||
| Recombinase polymerase amplification (RPA) | Combined RPA with rkDNA-graphene oxide probing system | 6.0 aM | ND | ND | ND | 1.6 h | Exhibit high selectivity and sensitivity for the diagnosis of COVID-19 | [58] | |
| Recombinase polymerase amplification | 7.659 copies/μL | 100% | 98% | USD 4.3 | 5–20 min | High specificity | [59,60] | ||
| Isothermal RPA-lateral flow detection | 0.25–2.5 copies/μL | 100% | 94% | ND | 5 min | The detection limit of RPA-LF for SARS-CoV-2 was 35.4 nucleocapsid (N) gene copies/L; the sensitivity was similar to that of qualitative real-time PCR | [61] | ||
| Hybrid capture immunofluorescence assay | Hybrid capture immunofluorescence assay | 500 copies per mL | 99% | 100% | ND | 45 min | The detection sensitivity is consistent with similar products on the market; however, this technique can only give qualitative results | [62] | |
| Entropy-driven amplified electrochemiluminescence | 2.67 fM | ND | ND | ND | 10–20 h | High selectivity and stability | [63] | ||
| CRISPR-based test | Cas12a | 10 copies per μL reaction | 100% | 95% | USD 6 | 40–60 min | Enables rapid, ultrasensitive (few copies), and highly specific nucleic acid detections | [43] | |
| Cas13a | 10–100 copies per μL | 100% | 96% | USD 3.5 | 40–57 min | Rapid, sensitive, and with low instrument requirement | [64] | ||
| Pyrococcus furiosus Argonaute coupled with modified ligase chain reaction | 10 aM | ND | ND | Cheaper than CRISPR | ~70 min | High sensitivity, high specificity, and multiplexing detection; without the use of RNA as guidance | [65] | ||
| Immunological test | Antigen immunological test | Quantum dot immunochromatographic assay | 4.9 pg/mL | 100% | 75 pg/mL | USD 1.5 | 3 min | One single test that can cover hs-CRP and routine-range CRP with a detection range from 1 to 200 μg mL−1 | [66] |
| QuickNavi™-COVID-19 Ag immunochromatographic test | ND | 100% | 86.7% | Cheaper than nucleic acid amplification tests | 5 min | The overall sensitivity was 86.7%, and the positive detection rate in patients with CT < 30 was comparable to that of RT-PCR | [67] | ||
| Magnetic graphene quantum dots | 248 Particles mL−1 | Related to SARS-CoV-2 antigen protein | No response to MERS-CoV | USD 1.25 | 2 min | Sensitive detection without sample pretreatment in one step with a LOD of 248 Particles mL−1 | [68] | ||
| Binax-CoV2 | 1.6 × 104–4.3 × 104 viral RNA copies | 99.9% | 93.3% | USD 5 | 15 min | The sensitivity of Binax-CoV2 was 93.3% and the specificity was 99.9% | [69] | ||
| SERS biosensor | 80 copies mL−1 | Related to the sensing environment | Suffers from non-specific binding | More expensive than ELISA | 5 min | The low detection limit (LOD) can be reduced to 80 parts mL−1 | [70] | ||
| Interdigitated microelectrode chip | 2.29 × 10−6 ng/mL | 4.27 × 10−4 ng/mL | 234:1 | USD 1 | 20 s | The linear range is 10−5–10−1 ng/mL; the strategy is real-time, sensitive, selective, and large-scale in cold-chain food quarantine | [25] | ||
| Serum antibody immunological test | Split luciferase antibody biosensors | ND | > 99% | > 98% | ∼15 ¢ | 5 min | The sensitivity to detect anti-S protein antibodies was 89% and anti-N protein antibodies were 98%, and the specificity of both was more than 99% | [71] | |
| Colloidal gold immunochromatography assay | 20.00 IU/mL | 96.2% | 71.1% | ND | 10–15 min | The IgM/IgG test assay demonstrated high sensitivity of 71.1% and specificity of 96.2% in 150 suspect COVID-19 cases | [72] | ||
| Chemiluminescence immunoassay | 0.5–1.5 AU/mL | 97.5% | 78.65% | ND | 1 h | The antibody detection rate has high sensitivity, high precision, quantitative detection, and easy automation | [73] | ||
| Upconverting phosphor immunochromatography assay | ND | 99.75% | 89.15% | ND | 10 min | High sensitivity, no interference from the background, and good stability | [74] | ||
| Surface plasmon resonance biosensors (SPRS) | 0.22 pM | ND | ND | ND | ND | The SPR biosensor is feasible in the concentration range of 2 to 1000 ng/mL | [75] | ||
| DNA-assisted nanopore sensing | 50 ng/mL (IgM) 10 ng/mL(IgG) | ND | ND | USD 8 | ND | High sensitivity and specificity compared to laboratory techniques | [76] | ||
| Colorimetric-fluorescent dual-mode lateral flow immunoassay biosensor | 10 ng/mL(IgM) 5 ng/mL(IgG) | 100% | ND | ND | ND | The combined detection sensitivity and specificity of this assay for IgM/IgG is 100%, and it has great potential for rapid and accurate detection | [76] | ||
| The lateral flow immunoassay method | ND | 90.63% | 88.66% | ND | 15 min | The limits of detection for IgM and IgG were 10 ng/mL and 5 ng/mL, respectively | [77] | ||
| Luciferase immunosorbent assay (LISA) | 0.4–75 pg / μl | 100% | 71% | ND | ~60 min | LISA had a sensitivity of 71% in COVID-19 patients and a specificity of 100% in healthy blood donors in the second week after onset | [78] | ||
| Enzyme-linked immunosorbent assays | 0.095 (IgM) 0.083 (IgG) | ND | 98% | ND | 80–120 min | High sensitivity and specificity | [79] | ||
| Enzyme-linked immunosorbent assays | ND | 88.2–99.2% (IgM) 75.6–98.3% (IgG) | 78.2% (IgG) 96.6%(IgM) | ND | 1.5 h | ELISA was used to detect IgG antibodies in confirmed patients with COVID-19, and the sensitivity to detect IgM antibodies was low | [80] | ||
| Enzyme-linked immunosorbent assays (ELISA) | ND | 93–100% | 65–85% | ND | 4 h | The detection precision is similar to ELISA, but the detection range is wider and the sensitivity is higher | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, J.; Li, W.; Hu, J.; Zhao, S.; Yue, T.; Li, Z.; Xia, Y. The Safety of Cold-Chain Food in Post-COVID-19 Pandemic: Precaution and Quarantine. Foods 2022, 11, 1540. https://doi.org/10.3390/foods11111540
Kong J, Li W, Hu J, Zhao S, Yue T, Li Z, Xia Y. The Safety of Cold-Chain Food in Post-COVID-19 Pandemic: Precaution and Quarantine. Foods. 2022; 11(11):1540. https://doi.org/10.3390/foods11111540
Chicago/Turabian StyleKong, Jia, Wenxin Li, Jinyao Hu, Shixuan Zhao, Tianli Yue, Zhonghong Li, and Yinqiang Xia. 2022. "The Safety of Cold-Chain Food in Post-COVID-19 Pandemic: Precaution and Quarantine" Foods 11, no. 11: 1540. https://doi.org/10.3390/foods11111540
APA StyleKong, J., Li, W., Hu, J., Zhao, S., Yue, T., Li, Z., & Xia, Y. (2022). The Safety of Cold-Chain Food in Post-COVID-19 Pandemic: Precaution and Quarantine. Foods, 11(11), 1540. https://doi.org/10.3390/foods11111540