Collaborative Ring Trial of the Applicability of a Reference Plasmid DNA Calibrant in the Quantitative Analysis of GM Maize Event MON810
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and DNA Extraction
2.2. Oligonucleotide Primers and TaqMan Probes
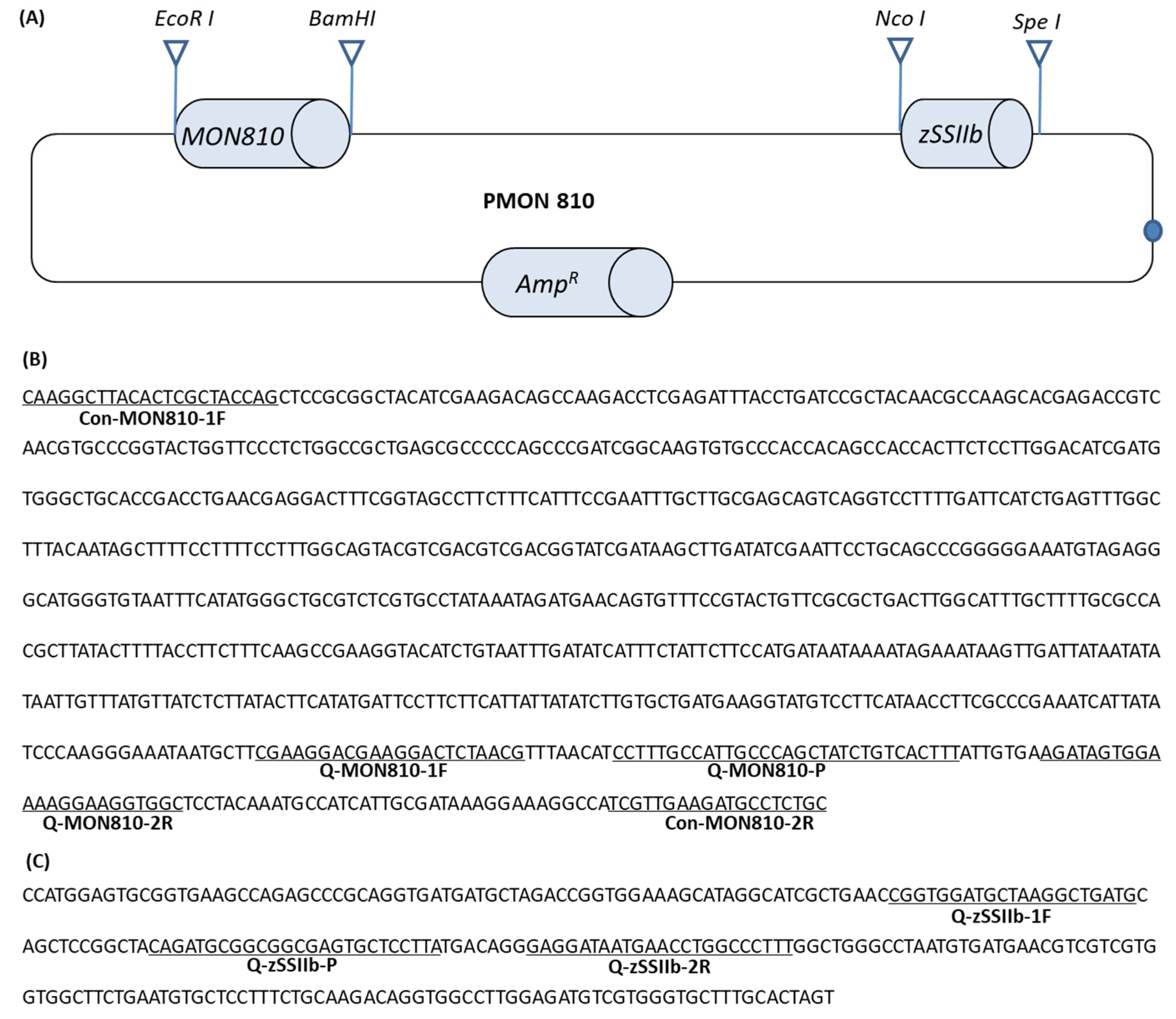
2.3. Construction of pMON810
2.4. Real-Time PCR
2.5. Collaborative Ring Trial
2.6. Statistical Analysis
3. Results and Discussion
3.1. Quantification of the Event-Specific Sequence of MON810 and Maize Endogenous zSSIIb Gene in pMON810 using qPCR
3.2. Inter-Laboratory Ring Trial of pMON810 as a Calibrant in MON810 Maize Quantification
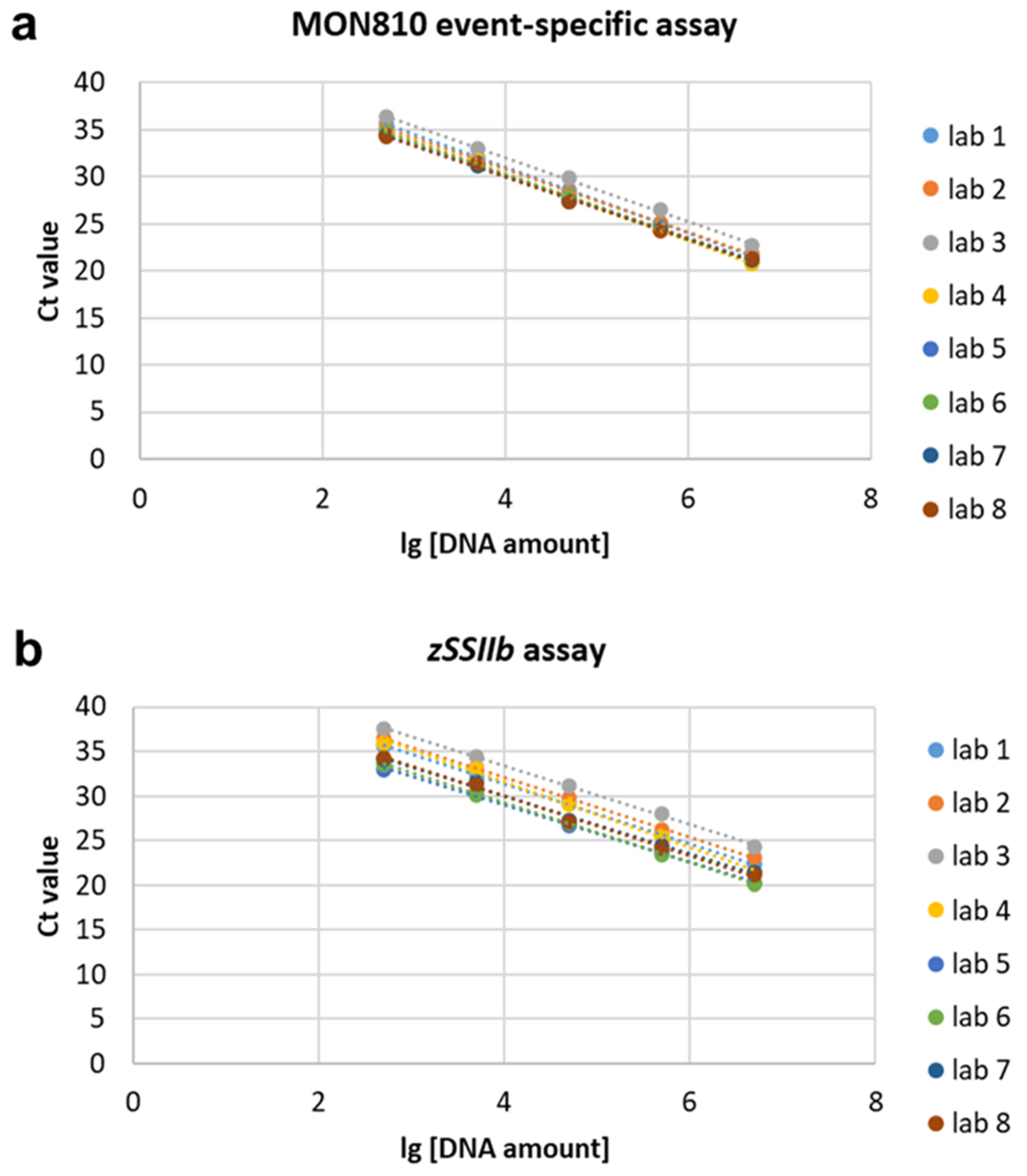
3.3. qPCR Efficiency and Linearity
3.4. qPCR Repeatability and Reproducibility Using pMON810 Plasmid DNA
3.5. Measurement of the Conversion Factors (Cfs)
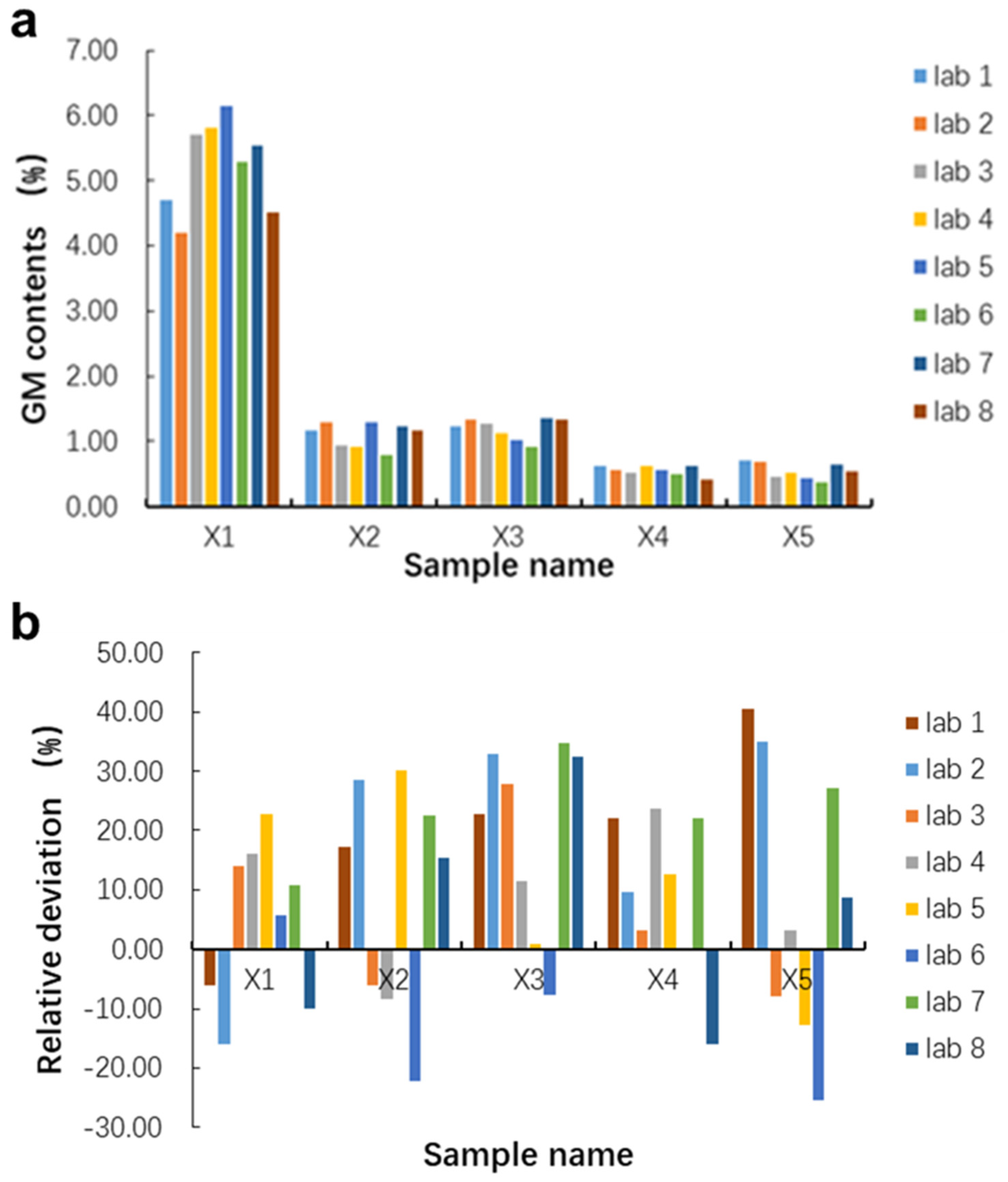
3.6. Blind Sample Quantification
3.7. Two-Factor ANOVA Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ISAAA. Global Status of Commercialized Biotech/GM Crops; Brief. No. 55; ISAAA: Ithaca, NY, USA, 2020; Available online: https://www.isaaa.org/resources/publications/briefs/55/default.asp (accessed on 1 April 2022).
- Dong, W.; Yang, L.; Shen, K.; Kim, B.; Kleter, G.A.; Marvin, H.J.; Guo, R.; Liang, W.; Zhang, D. GMDD: A database of GMO detection methods. BMC Bioinform. 2008, 9, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamse, P.; Dagand, E.; Bohmert-Tatarev, K.; Wahler, D.; Bendiek, J. GMO Genetic Elements Thesaurus (GMO-GET): A controlled vocabulary for the consensus designation of introduced or modified genetic elements in genetically modified organisms. BMC Bioinform. 2021, 22, 48. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.M.; Shan, G.; Layton, D.T.; Bell, T.A.; Whipkey, S.; Shillito, R.D. Application of DNA- and Protein-Based Detection Methods in Agricultural Biotechnology. J. Agric. Food Chem. 2019, 67, 1019–1028. [Google Scholar] [CrossRef]
- Yang, L.; Chen, Y.; Li, R.; Xu, W.; Zhang, X. Universal LNA Probe-Mediated Multiplex Droplet Digital Polymerase Chain Reaction for Ultrasensitive and Accurate Quantitative Analysis of Genetically Modified Organisms. J. Agric. Food Chem. 2021, 69, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Salisu, I.B.; Shahid, A.A.; Amina, Y.; Qurban, A.; Bajwa, K.S.; Rao, A.Q.; Tayyab, H. Molecular approaches for high throughput detection and quantification of genetically modified crops: A Review. Front. Plant Sci. 2017, 8, 1670–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurel, J.; Schbath, S.; Bougeard, S.; Rolland, M.; Touzain, F. DUGMO: Tool for the detection of unknown genetically modified organisms with high-throughput sequencing data for pure bacterial samples. BMC Bioinform. 2020, 21, 284. [Google Scholar] [CrossRef]
- Sánchez-Paniagua López, M.; Manzanares-Palenzuela, C.L.; López-Ruiz, B. Biosensors for GMO Testing: Nearly 25 Years of Research. Crit. Rev. Anal. Chem. 2018, 48, 391–405. [Google Scholar] [CrossRef]
- Anklam, E.; Gadani, F.; Heinze, P.; Pijnenburg, H.; Eede, G. Analytical methods for detection and determination of genetically modified organisms in agricultural crops and plant-derived food products. Eur. Food Res. Technol. 2002, 214, 3–26. [Google Scholar] [CrossRef]
- Li, R.; Shi, J.; Liu, B.; Wang, C.; Zhang, D.; Zhao, X.; Yang, L. Inter-laboratory Validation of Visual Loop-mediated Isothermal Amplification Assays for GM Contents Screening. Food Chem. 2018, 274, 659–663. [Google Scholar] [CrossRef]
- Yang, Y.; Li, L.; Yang, H.; Li, X.; Zhang, X.; Xu, J.; Zhang, D.; Jin, W.; Yang, L. Development of Certified Matrix-Based Reference Material as a Calibrator for Genetically Modified Rice G6H1 Analysis. J. Agric. Food Chem. 2018, 66, 3708–3715. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Li, X.; Zhai, S.; Gao, H.; Li, Y.; Zhang, X.; Wu, G. Development and strategy of reference materials for the DNA-based detection of genetically modified organisms. Anal. Bioanal. Chem. 2019, 411, 1729–1744. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, H.; Quan, S.; Liu, Y.; Shen, P.; Yang, L. Development of certified matrix-based reference material of genetically modified rice event TT51-1 for real-time PCR quantification. Anal. Bioanal. Chem. 2015, 407, 6731–6739. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Li, L.; Li, X.; Zhang, X.; Zhai, S.; Gao, H.; Li, Y.; Wu, G.; Wu, Y. Development of Genomic DNA Certified Reference Materials for Genetically Modified Rice Kefeng 6. ACS Omega 2020, 5, 21602–21609. [Google Scholar] [CrossRef]
- Yang, L.; Guo, J.; Pan, A.; Zhang, H.; Zhang, K.; Wang, Z.; Zhang, D. Event-specific quantitative detection of nine genetically modified maizes using one novel standard reference molecule. J. Agric. Food Chem. 2007, 55, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Pi, L.; Li, X.; Cao, Y.; Wang, C.; Pan, L.; Yang, L. Development and application of a multi-targeting reference plasmid as calibrator for analysis of five genetically modified soybean events. Anal. Bioanal. Chem. 2015, 407, 2877–2886. [Google Scholar] [CrossRef]
- Meng, Y.; Liu, X.; Wang, S.; Zhang, D.; Yang, L. Applicability of plasmid calibrant pTC1507 in quantification of TC1507 maize: An interlaboratory study. J. Agric. Food Chem. 2012, 60, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, Y.; Wu, G.; Cao, Y.; Lu, C. Correction of the lack of commutability between plasmid DNA and genomic DNA for quantification of genetically modified organisms using pBSTopas as a model. Anal. Bioanal. Chem. 2014, 406, 6385–6397. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Wang, Y.; Li, X.; Li, Y.; Zhu, L.; Li, J.; Wu, G. Development and application of a general plasmid reference material for GMO screening. Plasmid 2016, 87, 28–36. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, S.B.; Roh, H.J.; Park, S.; Shin, M.K.; Moon, G.I.; Hong, J.H.; Kim, H.Y. A simplified and accurate detection of the genetically modified wheat MON71800 with one calibrator plasmid. Food Chem. 2015, 176, 1–6. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Wang, C.; Song, G.; Pi, L.; Zheng, L.; Zhang, D.; Yang, L. One Novel Multiple-Target Plasmid Reference Molecule Targeting Eight Genetically Modified Canola Events for Genetically Modified Canola Detection. J. Agric. Food Chem. 2017, 65, 8489–8500. [Google Scholar] [CrossRef]
- Liu, G.; Yan, L.I.; Li, X.U.; Biometrology, L.O. Plasmid DNA Standard Reference Material for the Specific Detection of Listeria Monocytogenes. Chem. Reag. 2016, 38, 664–668. [Google Scholar] [CrossRef]
- Li, X.U.; Yan, L.I.; Wang, X.; Xiao, Y.; Liu, G. Development of a Plasmid DNA Reference Material for Influenza A (H7N9) Virus detection. Res. Explor. Lab. 2018, 37, 17–21. [Google Scholar]
- Horwitz, W. Protocol for the design, conduct and interpretation of method-performance studies: Revised 1994 (Technical Report). Pure Appl. Chem. 1995, 67, 331–343. [Google Scholar] [CrossRef]
- Wei, J.; Le, H.; Pan, A.; Xu, J.; Li, F.; Li, X.; Quan, S.; Guo, J.; Yang, L. Collaborative trial for the validation of event-specific PCR detection methods of genetically modified papaya Huanong No.1. Food Chem. 2016, 194, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Huang, C.; Zhu, P.; Du, Z.; Wei, S.; Fu, W. Applicability of a General Analytical Approach for Detection of Genetically Modified Organisms: Collaborative Trial. J. Aoac. Int. 2021, 105, 476–482. [Google Scholar] [CrossRef]
- Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
| Lab Code | MON810 Event-Specific | zSSIIb | Equipment | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Formula | Efficiency | SD | RSD (%) | R2 | SD | RSD (%) | Formula | Efficiency | SD | RSD (%) | R2 | SD | RSD (%) | ||
| 1 | y = −3.4849x + 45.004 | 0.936 | 0.03 | 3.120 | 0.9997 | 0.001 | 0.093 | y = −3.419x + 44.729 | 0.961 | 0.05 | 5.070 | 0.9995 | 0.002 | 0.216 | Prism ABI 7500 |
| 2 | y = −3.3641x + 44.305 | 0.983 | 0.9996 | y = −3.3643x + 45.581 | 0.983 | 0.9997 | Prism ABI 7500 | ||||||||
| 3 | y = −3.3803x + 45.607 | 0.976 | 0.9991 | y = −3.3085x + 46.641 | 1.006 | 0.9989 | Rotor-Gene Q | ||||||||
| 4 | y = −3.5054x + 44.354 | 0.929 | 0.998 | y = −3.6326x + 46.044 | 0.885 | 0.9971 | CFX96 Touch | ||||||||
| 5 | y = −3.3131x + 43.492 | 1.004 | 0.9992 | y = −3.2092x + 41.843 | 1.049 | 0.9987 | CFX96 Touch | ||||||||
| 6 | y = −3.4103x + 43.977 | 0.964 | 0.9997 | y = −3.3528x + 42.61 | 0.987 | 0.9996 | Prism ABI 7900 | ||||||||
| 7 | y = −3.3181x + 43.275 | 1.002 | 0.9991 | y = −3.244x + 43.019 | 1.034 | 0.9933 | Prism ABI 7900 | ||||||||
| 8 | y = −3.3032x + 43.259 | 1.008 | 0.9971 | y = −3.3571x + 43.372 | 0.986 | 0.9972 | CFX96 Touch | ||||||||
| Lab Code | C1 | C2 | C3 | C4 | C5 | Mean Cf | SD | RSD (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean Ct | Cf | Mean Ct | Cf | Mean Ct | Cf | Mean Ct | Cf | Mean Ct | Cf | |||||||||
| MON810 | zSSIIb | MON810 | zSSIIb | MON810 | zSSIIb | MON810 | zSSIIb | MON810 | zSSIIb | |||||||||
| 1 | 25.53 | 24.87 | 0.60 | 27.34 | 26.87 | 0.70 | 29.86 | 29.26 | 0.66 | 32.30 | 31.68 | 0.67 | 34.74 | 33.90 | 0.60 | 0.65 | 0.04 | 6.78 |
| 2 | 24.24 | 24.82 | 0.62 | 26.53 | 27.39 | 0.75 | 29.13 | 29.65 | 0.60 | 31.84 | 32.66 | 0.73 | 34.39 | 35.28 | 0.77 | 0.69 | 0.08 | 11.48 |
| 3 | 26.31 | 26.68 | 0.47 | 28.17 | 28.97 | 0.66 | 30.69 | 30.94 | 0.47 | 33.10 | 33.17 | 0.43 | 35.50 | 35.82 | 0.52 | 0.51 | 0.09 | 17.78 |
| 4 | 24.43 | 24.50 | 0.56 | 26.64 | 26.94 | 0.62 | 29.17 | 29.46 | 0.58 | 31.75 | 31.95 | 0.52 | 34.35 | 35.04 | 0.66 | 0.59 | 0.06 | 9.37 |
| 5 | 24.97 | 23.21 | 0.61 | 27.26 | 25.40 | 0.60 | 29.79 | 28.05 | 0.69 | 32.34 | 30.36 | 0.62 | 34.55 | 32.65 | 0.69 | 0.64 | 0.04 | 6.92 |
| 6 | 27.07 | 25.05 | 0.53 | 29.57 | 27.37 | 0.48 | 32.06 | 30.16 | 0.60 | 34.48 | 32.78 | 0.71 | 37.01 | 35.16 | 0.66 | 0.60 | 0.10 | 16.15 |
| 7 | 24.58 | 24.23 | 0.70 | 26.93 | 26.58 | 0.72 | 28.91 | 28.41 | 0.67 | 31.43 | 31.04 | 0.75 | 33.88 | 33.15 | 0.61 | 0.69 | 0.05 | 7.73 |
| 8 | 24.97 | 24.25 | 0.69 | 26.90 | 26.21 | 0.69 | 29.36 | 28.62 | 0.65 | 31.70 | 31.01 | 0.66 | 33.64 | 33.10 | 0.71 | 0.68 | 0.02 | 3.63 |
| Lab Code | qPCR Assay | X1 | X2 | X3 | X4 | X5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ct | GM Content (%) | Mean | Ct | GM Content (%) | Mean | Ct | GM Content (%) | Mean | Ct | GM Content (%) | Mean | Ct | GM Content (%) | Mean | ||
| 1 | MON810 | 33.8 | 4.70 | 5.23 | 35.86 | 1.17 | 1.10 | 35.76 | 1.23 | 1.20 | 36.95 | 0.61 | 0.55 | 36.85 | 0.70 | 0.54 |
| zSSIIb | 28.55 | 28.51 | 28.48 | 28.61 | 28.72 | |||||||||||
| 2 | MON810 | 33.05 | 4.20 | 34.86 | 1.29 | 34.72 | 1.33 | 35.91 | 0.55 | 35.57 | 0.68 | |||||
| zSSIIb | 29.16 | 29.24 | 29.15 | 29.05 | 29.01 | |||||||||||
| 3 | MON810 | 33.25 | 5.70 | 35.61 | 0.94 | 35.26 | 1.28 | 36.43 | 0.52 | 36.82 | 0.46 | |||||
| zSSIIb | 29.46 | 29.18 | 29.28 | 29.12 | 29.34 | |||||||||||
| 4 | MON810 | 31.46 | 5.80 | 34.55 | 0.92 | 34.28 | 1.11 | 35.06 | 0.62 | 35.27 | 0.52 | |||||
| zSSIIb | 27.36 | 27.65 | 27.68 | 27.56 | 27.49 | |||||||||||
| 5 | MON810 | 31.65 | 6.14 | 33.49 | 1.30 | 33.68 | 1.01 | 34.28 | 0.56 | 34.72 | 0.44 | |||||
| zSSIIb | 25.86 | 25.48 | 25.31 | 25.08 | 25.15 | |||||||||||
| 6 | MON810 | 32.05 | 5.28 | 34.52 | 0.78 | 34.86 | 0.92 | 35.27 | 0.50 | 35.61 | 0.37 | |||||
| zSSIIb | 25.85 | 25.49 | 25.68 | 25.58 | 25.49 | |||||||||||
| 7 | MON810 | 31.57 | 5.54 | 33.62 | 1.23 | 33.34 | 1.35 | 34.45 | 0.61 | 34.29 | 0.64 | |||||
| zSSIIb | 26.98 | 26.86 | 26.72 | 26.69 | 26.59 | |||||||||||
| 8 | MON810 | 31.54 | 4.50 | 33.68 | 1.15 | 33.59 | 1.33 | 35.05 | 0.42 | 34.82 | 0.54 | |||||
| zSSIIb | 26.38 | 26.57 | 26.68 | 26.49 | 26.63 | |||||||||||
| Unknown Samples GM% | Blind Sample | ||||
|---|---|---|---|---|---|
| X1 | X2 | X3 | X4 | X5 | |
| Laboratories returning results | 8 | 8 | 8 | 8 | 8 |
| Sample per laboratory | 1 | 1 | 1 | 1 | 1 |
| Total data number | 9 | 9 | 9 | 9 | 9 |
| Data excluded | 0 | 0 | 0 | 0 | 0 |
| Reason for exclusion | - | - | - | - | - |
| Mean value (%) | 5.23 | 1.10 | 1.20 | 0.55 | 0.54 |
| True value (%) | 5.00 | 1.00 | 1.00 | 0.50 | 0.50 |
| Repeatability SD | 0.35 | 0.11 | 0.13 | 0.06 | 0.09 |
| Repeatability RSD (%) | 6.69 | 10.03 | 11.14 | 10.94 | 16.58 |
| Reproducibility SD | 0.69 | 0.19 | 0.16 | 0.07 | 0.12 |
| Reproducibility RSD (%) | 13.20 | 17.61 | 13.48 | 12.49 | 21.94 |
| Bias (absolute value) | 0.23% | 0.10% | 0.20% | 0.05% | 0.04% |
| Bias (%) | 4.60 | 10.00 | 20.00 | 10.00 | 8.00 |
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Squares | F | p |
|---|---|---|---|---|---|
| Error | 7246.24 | 28 | 258.80 | ||
| Laboratory | 3802.60 | 7 | 543.23 | 2.10 | 0.08 |
| GM amounts | 957.00 | 4 | 239.25 | 0.92 | 0.46 |
| Total | 12,005.83 | 39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, Y.; Wang, S.; Guo, J.; Yang, L. Collaborative Ring Trial of the Applicability of a Reference Plasmid DNA Calibrant in the Quantitative Analysis of GM Maize Event MON810. Foods 2022, 11, 1538. https://doi.org/10.3390/foods11111538
Meng Y, Wang S, Guo J, Yang L. Collaborative Ring Trial of the Applicability of a Reference Plasmid DNA Calibrant in the Quantitative Analysis of GM Maize Event MON810. Foods. 2022; 11(11):1538. https://doi.org/10.3390/foods11111538
Chicago/Turabian StyleMeng, Yanan, Shu Wang, Jinchao Guo, and Litao Yang. 2022. "Collaborative Ring Trial of the Applicability of a Reference Plasmid DNA Calibrant in the Quantitative Analysis of GM Maize Event MON810" Foods 11, no. 11: 1538. https://doi.org/10.3390/foods11111538
APA StyleMeng, Y., Wang, S., Guo, J., & Yang, L. (2022). Collaborative Ring Trial of the Applicability of a Reference Plasmid DNA Calibrant in the Quantitative Analysis of GM Maize Event MON810. Foods, 11(11), 1538. https://doi.org/10.3390/foods11111538






