Abstract
The olive fruit, a symbol of Mediterranean diets, is a rich source of antioxidants and oleic acid (55–83%). Olive genetic resources, including cultivated olives (cultivars), wild olives as well as related subspecies, are distributed widely across the Mediterranean region and other countries. Certain cultivars have a high commercial demand and economical value due to the differentiating organoleptic characteristics. This might result in economically motivated fraudulent practices and adulteration. Hence, tools to ensure the authenticity of constituent olive cultivars are crucial, and this can be achieved accurately through DNA-based methods. The present review outlines the applications of microsatellite markers, one of the most extensively used types of molecular markers in olive species, particularly referring to the use of these DNA-based markers in cataloging the vast olive germplasm, leading to identification and authentication of the cultivars. Emphasis has been given on the need to adopt a uniform platform where global molecular information pertaining to the details of available markers, cultivar-specific genotyping profiles (their synonyms or homonyms) and the comparative profiles of oil and reference leaf samples is accessible to researchers. The challenges of working with microsatellite markers and efforts underway, mainly advancements in genotyping methods which can be effectively incorporated in olive oil varietal testing, are also provided. Such efforts will pave the way for the development of more robust microsatellite marker-based olive agri-food authentication platforms.
1. Introduction
The olive tree has been cultivated for approximately 6000 years in Mediterranean countries, where 95% of olive germplasm is located. Its habitat is determined by the Mediterranean climate, and it stands as the most highly cultivated fruit crop among temperate crops in the world. According to data published by International Olive Council (IOC) (www.international.oliveoil.org (accessed on 10 February 2021)), in the last 25 years, olive oil production and consumption has increased by 1 million tons. The olive crop is mainly located in the Mediterranean Basin (the leading producers being Spain, Italy and Greece). Moreover, the olive is also a crop under increasing cultivation in non-traditional countries such as Argentina, Australia, Chile, China, Japan and the United States.
Both olive oil and fruits have been found to be a rich source of antioxidants and various other secondary metabolites (phenolics, carotenoids, tocopherols, anthocyanins and oleosides). Olive oil in particular has an unique lipid fatty acid composition and health benefits such as defense against chronic degenerative diseases, and reduced cardiovascular risks are attributed to the consumption of olive oils and table olives [1,2,3]. The increased demand of nutritionally superior olive oil such as extra virgin olive oil (EVOO) and virgin olive oil (VOO) and table olives has also led to increased adulteration of premium quality oils and fruits. Hence, regulations and certifications such as protected designation of origin (PDO) and protected geographical indication (PGI) (EC Regulation no. 510/2006) have been laid out to check product authenticity and traceability.
The exchange of germplasm in ancient times and increased commerce among olive growing nations has established complex genetic relationships among different olive gene pools [4]. The cultivation of cultivars in new climatic conditions and the adoption of local names for new introduced material have led to confusion in the denominations of varieties [5,6]. More than 1200 cultivars of olive spread across the Mediterranean region, with around 600 olive cultivars under cultivation in Italy itself, have been described in the olive germplasm database [7]. The characterization and recognition of many other cultivars and ancient and wild forms is still an ongoing process, and several studies have been undertaken in this direction using morphological as well as molecular tools [8]. Germplasm banks have been established to ensure ex situ conservation of olive genetic resources, and emphasis is being given to the use of microsatellite markers or simple sequence repeats (SSRs) as tools to better inventory these valuable repositories. Molecular characterization or genotypic profiling of available germplasm will not only provide unique identification keys but also help in the development of molecular authentication platforms, wherein these cultivars, wild forms or related species can easily and accurately be identified. Ever since being developed, SSRs or microsatellites are among the most frequently used molecular markers in olives. This is also evident from the large number of publications available pertaining to the use of SSRs in olive research. The characteristic features such as the multiallelic nature, wide genomic distribution, codominant inheritance, locus specificity, high mutation rates, utility as functional markers (present in transcribed regions), cross-transferability, amenability to automation, easy in silico mining and primer design have established SSRs as the markers of choice in most species [9,10]. Detailed reviews are already available, explaining the development, uses and advantages of SSR markers in plants [11,12,13], and these can be consulted for more elaborate information.
In olives, microsatellite markers have been used in various applications such as cultivar identification, characterization of autochthonous olives (ancient olive trees and oleasters), the management of olive germplasm banks, phylogenetics, diversity analysis and mapping. Moreover, these have also been widely utilized in the authentication and traceability of cultivars in olive agri-food products. Most of the studies involved the use of nuclear genomic SSRs, and recently expressed sequence tag (EST)-based SSRs or the EST-SSRs are also being exploited in several olive genetic studies. Olive SSRs have also been used in combination with other marker systems such as amplified fragment length polymorphisms (AFLPs), inter simple sequence repeats (ISSRs), single nucleotide polymorphisms (SNPs) and random amplified polymorphic DNA (RAPD) in various studies related to mapping, cultivar discrimination and genetic relationships [14,15,16,17,18,19,20,21,22,23,24]. Microsatellites, being so extensively applied in olive germplasm cataloging, authentication and traceability studies, need to be reviewed in detail, and therefore, the present review aims to elaborate on the development of SSRs in olives and specifically targets their use in olive cultivar identification, cataloging of germplasm and the traceability of oils and table olives. Information generated through such studies has been thoroughly compiled and presented in this review through extensive literature searching, mainly using Google (www.google.com accessed on 26 July 2021) and Google Scholar (scholar.google.com, accessed on 26 July 2021). Research articles and reviews covering a wide timeframe and encompassing information about olive distribution, the development of SSR markers and databases on olives and their vast applications were referred. Since the aim of the review is to mainly highlight the utility of SSR markers in the characterization of germplasm banks and local, wild and centennial olive germplasm, thereby leading to proper cultivar identification and cataloging and utilization of such information in olive agri-food authentication and traceability, articles pertaining to these fields were mainly included in this review. The review should be useful to researchers working in the above-mentioned areas. Key factors that affect the applicability and usefulness of microsatellites in olive varietal identification are also emphasized and discussed in the manuscript.
2. The Olive Germplasm
The olive (Olea europaea L.) belongs to the family Oleaceae, which comprises around 30 genera and over 600 species. The genus Olea has some 35 species, including both O. europaea subsp. europaea var. sativa (cultivated olive) and Olea europaea subsp. europaea var. sylvestris (wild olive or oleasters). In addition, the wild olive includes feral forms which are seedlings of the cultivated olives or the result of hybridizations between the oleasters and cultivars [25,26]. Additionally, five subspecies, namely laperrinei (Saharan massifs), cuspidata (Afro-Asiatic), guanchica (Canary Islands), maroccana (Morocco) and cerasiformis (Madeira), comprise the Olea europaea complex.
The olive was probably domesticated in the Middle East about 6000 years ago [27]. Afterward, commercial shipping spread this crop westward across the Mediterranean Basin, leading to complex genetic relationships among cultivars [4]. The empiric selection of outstanding individuals within wild olives, crosses between the previous selected or introduced cultivars and other local cultivars or wild olives in all growing areas have yielded a huge number of local cultivars. The easy vegetative propagation of the olive cultivars has allowed for maintaining the characteristics by which they were selected, such as greater productivity, fruit size, oil production and environmental adaptation. It is estimated that there are more than 2000 olive varieties worldwide [28]. The denomination of olive cultivars is usually a process synchronous to their diffusion. Initially, olive cultivars were named using generic criteria, like their outstanding morphological traits, utility of production or the locality of origin of the propagated material, or based on other characteristics [8]. Consequently, in olives, the existence of synonymy (different names for the same cultivar) and homonymy (same name for different cultivars) among and within olive-growing countries is very frequent [5,6,7,8].
Germplasm banks are facilities that permit us to ensure “ex situ” conservation of genetic resources. Clonally propagated fruit crops such as olives are typically conserved in “live collections”, which are suitable selected field plantations where the crop can fulfill its normal biological cycle [29]. Prospecting surveys of olive cultivars in many countries and the exchanges of cultivars between countries have contributed to the high number of conserved accessions in “ex situ” collections. Bartolini et al. [7,30] reviewed for the FAO the accessions conserved in approximately 100 regional and national collections in 54 countries, which include more than 4000 accessions supposedly belonging to 1250 cultivars [31]. Most of these cultivars come from major producer countries like Italy (538 cultivars), Spain (183), France (88) and Greece (52) [32]. Since 1994, the IOC has been promoting a network of banks to preserve the heritage of olive varieties grown in countries around the world. The network presently includes a total of 23 germplasm banks, housing over 1700 varieties andis composed of 3 international banks—Cordoba (Spain), Marrakech (Morocco) and Izmir (Turkey)—and 20 national banks (Albania, Algeria, Argentina, Croatia, Cyprus, Egypt, France, Greece, Iran, Israel, Italy, Jordan, Lebanon, Libya, Montenegro, State of Palestine, Portugal, Slovenia, Tunisia and Uruguay) (https://www.internationaloliveoil.org/the-ioc-network-of-germplasm-banks/, accessed on 5 February 2021). The Olive World Olive Germplasm Bank of Cordoba (Spain) (WOGBC) was established in 1970, and it is one of the largest with more than 1000 accessions from 29 countries [33,34]. The second international bank (WOGBM) was established in 2003 in Marrakech (Morocco) and contains around 560 accessions from 14 countries (mainly from the Mediterranean region) [35]. The third international bank was recently established (2017) in Izmir (Turkey), including 274 accessions [36]. The national olive banks preserve the local as well as important international cultivars.
Despite these efforts, the exploration and conservation of the genetic patrimony of olives is still incomplete. In recent years, numerous initiatives have been promoted to explore, preserve and exploit unknown material, including minority local varieties, centenary trees and wild olive populations (see Section 4.1). It is indeed very clear from the above information that a vast collection of olive cultivars is presently available, but challenges related to correct denominations, geographical origin and proper cataloging of these germplasm still persist, and molecular tools such as SSR markers can be a preferred choice for addressing these aspects, contributing to the proper authentication of agro-food products.
3. Microsatellites in Olives
3.1. Development and Available SSRs
The earliest reports of the development of microsatellites in olives are from the year 2000 by two independent groups. Rallo et al. [37] developed 13 SSR loci (prefixed as IAS-oli) by sequencing 43 clones screened as positive on a GA-enriched olive genomic library of the cultivar “Arbequina”. Among these, only five were found to be polymorphic when analyzed for polymorphism in 46 olive cultivars. The occurrence of repeats, other than the enriched “GA” repeats, was found in the form of compound microsatellites and presumed to be common in the olive genome. Sefc et al. [38] screened a size-selected olive genomic library for GA and CA repeats and designed primers (prefixed as ssrOeUA-DCA or DCA) for 28 microsatellite loci. Among these 15 loci, amplified specific products were polymorphic across a set of 47 olive trees from Iberian Peninsula and Italy. In the year 2002, other groups simultaneously reported the genomic library-based development of microsatellites in olives. Carriero et al. [39] screened a (GA/CT)n-enriched genomic library and characterized 20 SSR primer pairs (prefixed as GAPU) in 6 olive cultivars and finally reported 10 polymorphic SSR loci after testing on a set of 20 olive accessions. An average of 5.7 alleles per SSR loci was obtained with these markers. Although enriched for dinucleotide repeats, clones in the library also possessed “CCT” and “TTC” trinucleotide repeat motifs. Cipriani et al. [40] also reported the selection and sequencing of 52 SSRs from (AC/GT) and (AG/CT) repeat-enriched genomic libraries of the olive cultivar “Frantoio”. Out of these, a set of 30 SSR primers (prefixed as UDO99) were screened for polymorphism in 13 olive Italian cultivars. GA and CA repeat-enriched libraries were also developed by De La Rosa et al. [41] from the cultivar “Picual” and designed 13 primer pairs (EMO prefixed), out of which only 6 were found to be polymorphic in a set of 23 olive cultivars and were also tested for cross-species transferability.
To further expand the arsenal, the olive cultivar “Arbequina” was used in genomic library preparation and enrichment for GA, GT and ACT repeats by Diaz et al. [42]. However, inserts with the “ACT” repeat motif were not obtained even after the enrichment step. Specific primers (prefixed as IAS-oli) could be designed from 10 of the sequences containing repeats and an additional 14 sequences available from an earlier report. Gil et al. [43] also employed similar techniques of genomic library enrichment, screening and sequencing with the olive cultivar “Lezzo”, and they reported 12 polymorphic SSR primers (prefixed as ssrOeIGP) when amplified in a set of 33 olive cultivars. All these genomic SSRs have been extensively used in the characterization of olive cultivars and molecular genetic studies in olives, as reviewed in the sections below. Series DCA-, GAPU- and UDO have been very used; nevertheless, others (e.g., the EMO and IAS-oli series) have been scarcely used. Most of these attempts involved dinucleotide repeat-containing sequences for the primer design, and the GA/CT motif was commonly used. An olive genome is presumed to have a relatively frequent occurrence of compound microsatellite motifs, as found in most of the SSR development studies described above. Multiple amplification products were also reported in some genotyping experiments and probably occurred due to, for example, priming at more than one site, ploidy of the species, the presence of compound microsatellites and genome duplication events [37,40].
EST-SSRs have gained interest in recent years, owing to their easy development through user-friendly bioinformatics tools, higher cross-transferability across species and ability to be used as functional markers in marker-assisted breeding [10]. With the beginning of sequencing projects and advanced sequencing technologies, genomic resources in the form of whole genome sequences and transcriptomes have been made available in public databases for olives. These are a rich source for the in silico development of SSR markers in olives. The availability of different transcriptomes has given researchers the opportunity to screen and design primers for microsatellite repeats present in the coding regions of the genome, thus allowing association of marker variability with phenotypic traits in olives. Data from cDNA libraries sequenced as a part of the OLEAGEN project, an olive genomic project in Spain [44] was used to extract sequences with core hexanucleotide repeats, and a set of eight EST SSR primers were designed (prefixed as OLEAGEN-H) which were successfully tested for genotyping as well as paternity testing in olives and were found to be comparable to dinucleotide-based genomic SSRs reported in earlier studies [45].
Adawy et al. [46] identified 8295 SSR repeat motifs after in silico mining of the EST sequences available in the NCBI database and described 1801 EST SSR primers (prefixed as Oe-ESSR) that could be amplified in different genes. Among the set of ESTs, the highest percentage (77.6%) for mononucleotide repeats and lowest for tetranucleotide repeats (0.29%) were reported, with the AAG/CTT repeat dominating among trinucleotide types and AG/CT dominant in the dinucleotide repeats. Twenty-five primers randomly chosen for amplification in a set of 9 cultivars were able to amplify, and 10 of these were found to be polymorphic. Tissue-specific transcriptomes [47,48,49] were utilized for the in silico mining of microsatellite repeats in transcripts in [50]. Trinucleotide and longer repeat motifs containing sequences were BLAST aligned to available olive genome data (oleagenome.org), and after screening for locus redundancy, 80 SSR sequences were targeted for primer design. From a prescreening of 5 olive cultivars for amplifiable loci and expected product size, a set of 26 EST SSRs were finalized (prefixed as OLEST). The authors described a set of the 10 best OLEST SSRs after allele sequencing and validation on a larger set of olive cultivars and related species as potential functional markers in olives. EST SSRs (prefixed as OeUP) were also identified in [51] from a transcriptome of developing fruits of the olive variety “Istrska belica” [52]. Dinucleotide repeats appeared to be abundantly present (36%), with “GA” as a common repeat motif and trinucleotides showing a presence of 33% and “GAA” as a common motif. Out of the 110 EST SSRs chosen for primer designing, 46 showed positive amplification and polymorphism when validated on a set of 8 cultivars and analyzed for diversity among 24 olive varieties. A final set of 27 EST SSRs was recommended on the basis of a low null allele frequency and no deviation from the Hardy–Weinberg equilibrium for diversity and population genetics in olives. Dervishi et al. [53] also performed in silico mining of developing fruit transcriptome of the variety “Istrska belica” for tri- and tetranucleotide repeats and reported 12 primers (prefixed SNB and SiBi) out of 35 EST SSRs for olive genetic studies. Gene annotation for sequences carrying microsatellite repeats was also performed, and genes for disease resistance were reported. Similar to earlier reports, the “AAG” motif was found to be most prevalent among the trinucleotide repeats which were found in 0.18% of the sequences. In the case of tetranucleotides, “AAAT” was most frequent, and the number of repeat units in a sequence ranged from 6 to 21 in the case of trinucleotides and 4–14 for the tetranucleotides. SSRs were also found to exist in compound form in a few of the cases.
More recently, genomic SSRs based on trinucleotide repeats (with at least five core repeats) were retrieved from the whole genome sequence information in olives, and SSR primers were developed (prefixed as BFU), covering most of the chromosomes. Twenty-one SSRs were found to be highly polymorphic and effectively discriminated among a panel of 53 accessions of olives [54]. EST SSRs have also been developed by Gómez-Rodríguez et al. [55], where tetra-, penta- and hexa-nucleotide repeats were retrieved from cDNA sequences, and primers were designed (prefixed as Olea). These newly developed markers could successfully discriminate the cultivars present in the core collection of olives available at the Worldwide Olive Germplasm Bank of Cordoba, Spain. Moreover, both the genomic and EST SSRs in olives have shown transferability across oleasters as well as cultivated olives [41,53,56]. Table 1 depicts the key genetic indices as observed while developing different microsatellite resources in olives. These SSRs are a valuable resource and can be utilized in various studies related to germplasm characterization, cataloging, cultivar identification and authetication in food products as discussed in the sections below.

Table 1.
Key genetic indices as reported for SSR markers developed in olives using enriched genomic libraries and EST sequences.
3.2. SSR Protocols for Cultivar Genotyping
Allele size discrepancies found while comparing the same set of SSRs across different samples and laboratories make the task of fingerprinting cultivars quite challenging, and thus, the utility of SSRs in cultivar authentication or in food traceability is also hampered. SSR protocols for the genotyping of olive cultivars and consensus sets of microsatellites have been proposed by various research groups for uniform data analysis and comparison. With an aim to standardize a set of SSR markers for olive genotyping, Doveri et al. [57] found that among 17 SSR markers, 6 (DCA3, DCA8, DCA11, DCA13, DCA14 and DCA15) showed maximum concordance between data points scored from all partner laboratories. Emphasis was made toward harmonization of SSR profiles for better resolution of the alleles. Baldoni et al. [58] performed an exhaustive exercise across four independent laboratories and proposed a consensus set of 11 SSRs (UDO-043, DCA9, GAPU103A, DCA18, DCA16, GAPU101, DCA3, GAPU71B, DCA5, DCA14 and EMO90) for olive genetic studies. SSRs were ranked according to the peak intensity, stuttering, null alleles, number of amplified loci and allelic error rate, which were calculated to determine the concordance of the SSRs being tested. Allelic ladders were constructed using a set of genotypes which carried true-sized alleles as confirmed by sequencing to identify the corresponding alleles between labs and to reduce the chance of mistyping alleles. The generation of allelic ladders using known profiled cultivars will allow univocal allele binning and assigning correct sizes to the new alleles. The SSRs present in the consensus list have been used in several genotyping and diversity studies of olives since then.
A protocol was also proposed by Trujillo et al. [8] using a nested set of 5, 10 and 17 SSR markers that allowed for quick characterization, authentication and identification of olive cultivars present in the WOGB in Cordoba, Spain and which could be used for management of germplasm resources in any olive gene banks. A molecular key for the identification of cultivars was also proposed by Aksehirli-Pakyurek et al. [59], where a classification binary tree (CBT) was developed and provided sorting of unknown new material that could be originating from any of the cultivars being analyzed. Hence, well-accepted SSR allelic profiles for specific cultivars are absolutely essential in order to avoid any confusion during molecular genotyping by different laboratories. This will also help in adopting a more uniform and application-worthy traceability and authenticity protocol based on SSRs.
3.3. Genotyping Methods
Over the years, genotyping methods used for SSR analysis have advanced to a great extent. When the aim is to specifically use SSRs for food authenticity and traceability, the genotyping methods being used are of the utmost importance, as any discrepancy in allele identification may lead to wrong cultivar identification and hamper the results. Earlier research mainly involved the use of agarose gel electrophoretic separation of SSR amplification products, and the resolution of alleles with 2–4 bp (base pair) differences in size was quite difficult. Denaturing polyacrylamide gels (4–8%) were also used for fragment separation [39,40,60], as these allow for better resolution compared with agarose gels when small base pair differences are to be identified, but these are more cumbersome to prepare, use toxic chemicals like acrylamide and involve silver staining for visualization of the separated bands. Development of more precise separation matrices in the form of high-resolution agarose have been used in amplicon separation in olive SSR analyses to resolve amplicons that differ in size by as little as 2% [37]. With more and more advancement in amplicon resolution and separation methods, matrices such as polyacrylamide and agarose are becoming obsolete and being replaced with automated capillary electrophoresis techniques and sequencing-based instruments which could achieve more sensitive allele separation and base pair calling. These advanced technologies reduced the separation time; hence, results could be obtained faster, and working with a huge sample size became easier. Moreover, integrated data analysis software, multiplexing, better reproducibility and elimination of staining procedures makes automated sequencers quite advantageous over the conventional methods of genotyping. This becomes very important when SSRs are to be used as a potential tool in olive authentication and traceability. Robust allele separation and detection is very crucial in such cases and thus requires high-throughput techniques. One of the major limitations while using microsatellites is the allele calling differences that may emerge due to polymerase slippage, DNA quantity or quality and the use of different instruments and reagents by different laboratories. Additionally, variations in results may arise due to post-PCR handling of samples in the case of gel-based platforms. These factors may cause problems in accurate determination of cultivar-specific SSR profiles and hence need to be taken into consideration while comparing genotyping results across laboratories and identifying correct cultivars [58].
High-resolution melting (HRM) analysis, an advanced method that compares the melting curve profiles of double-stranded DNA products and detects polymorphism, has recently been used as an alternative to gel-based polymorphism detection methods in olives and other species [61,62]. HRM shows greater resolving power compared with conventional melting curves, which are based on only the value of the melting temperature (Tm) and may not give better discrimination between different genotypes [63,64]. More nucleotide variations associated with the flanking regions of repeat sequences, such as single-nucleotide polymorphisms (SNPs), can be detected through this method and hence expand the applicability and potential of SSR marker systems. Refinements in the method are still going on so as to overcome challenges like specificity of the PCR, multilocus markers, and a high number of alleles [64]. Thus, continuous advancements are being made toward achieving more effective and accurate genotyping of the samples. This would help adopt a uniform method for olive genotyping, and hence information could be easily communicated and transferred between laboratories.
3.4. SSR Databases
It is indeed very clear that large-scale SSR genotyping projects have generated a vast amount of molecular data for different cultivars across the olive-growing regions of world. Nevertheless, this remains unutilized and inaccessible most of the time. A database is a necessary tool to correctly catalog any germplasm bank and optimize its management. Moreover, the database is the keystone to guarantee that a commercial edible product (oils or table olives) matches the cultivar specified on the label. For these reasons, the data from such independent studies need to be available on uniform platforms for easy access and use of the information. Attempts have been made to develop informative databases for olive trees, such as the Istrian olive database (http://old.iptpo.hr/iod, accessed on 20 January 2021), formed by assembling information about the morphological and molecular profiles of Istrian olive cultivars. This was an outcome of the DNA fingerprinting study of olive varieties of Istria conducted by Poljuha et al. [65]. The OLEA database (http://www.oleadb.it/, accessed on 20 January 2021) was yet another olive molecular database established in 2007 by researchers in Italy, and it comprised SSR marker data of a broad set of olive cultivars. Users could search for cultivars corresponding to a particular data type and variety identity and also look for cultivar information across different olive collection facilities.
With the generation of more and more EST information in public databases and the development of EST SSRs in olives, genetic studies have also been conducted using these SSRs. ReprOlive (http://reprolive.eez.csic.es, accessed on 20 January 2021) is a freely available database that gives access to the reproductive transcriptomes of olive trees, where information can be retrieved about tentative transcripts containing SSR units and suitable primers can be designed [66]. Another comprehensive olive database, the Olive Genetics Diversity Database (OGDD) pertaining to SSR molecular data, was generated by Ben Ayed et al. [67], and it is reported to contain morphological, chemical as well as molecular genetic (SSR) information about several olive varieties and oils. However, it is emphasized that the regular addition of newly generated information, updated software and easy access of these databases are required so that users can access the webpages and information smoothly. Public databases would make comparative studies much easier and more useful in the identification and authentication of cultivars and their products, and the information could be used by breeders, population geneticists and researchers across laboratories.
4. Applications of SSRs: Cataloging of Olive Germplasm, Food Authenticity and Traceability Studies
4.1. Cataloging Olive Germplasm
The varietal cataloging process implies (1) characterization or description of the cultivars at different levels (e.g., morphological, molecular or agronomical); (2) identification, a process that allows us to classify or differentiate one cultivar from the rest; (3) authentication, a process that guarantees that a cultivar corresponds to the original cultivar from its natural area of cultivation or origin; and (4) assigning the correct name to the cultivar once identified and authenticated and defining its synonyms and homonyms [68]. Therefore, the cataloging of any bank should be an essential requisite before using plant material for conservation, propagation and breeding purposes. Varietal information is also a key identifier in quality control for high-value virgin olive oils and table olives in the food industry.
In species like the olive, this task becomes particularly challenging. There are several factors that contribute to this, such as the vast number of olive cultivars, the use of generic criteria to name them and the misunderstanding around basic concepts that has led to a confusing scenario. In addition, the heterogeneity of criteria and methodologies applied for cataloging has hampered the completion of varietal cataloging in most traditional olive-growing countries. In this regard, the integration of molecular markers, particularly the microsatellites with the pomological scheme defined by Barranco et al. [69], has allowed for important advancements in the cataloging of olive germplasm [8]. In this work, the challenges of the incorporation of SSR markers both for the cataloging of germplasm and traceability studies in olive oil and table olives are highlighted. Figure 1 summarizes the microsatellites available in olives, the genotyping process and their applications in the cataloging and management of olive germplasm.
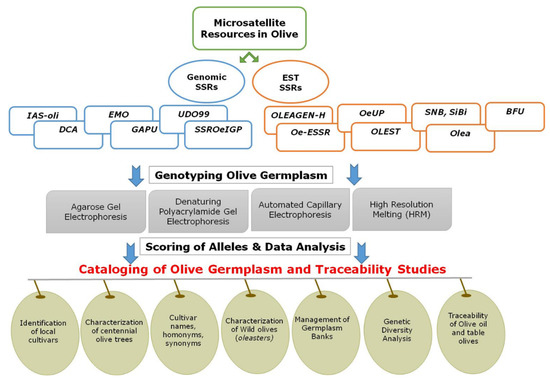
Figure 1.
Microsatellite resources and applications in olive germplasm cataloging, authenticity and traceability.
4.1.1. Cataloging of Germplasm Banks
Collections in a germplasm bank are a proper source of confirming the true identity of the cultivar in question. Hence, proper identification and cataloging of plant material becomes a prerequisite for efficient management of germplasm banks. The cataloging (characterization, identification, authentication and naming of the cultivar) of the accessions of any olive germplasm bank should be compulsory before distribution of any plant material from that bank. Only the diffusion of true-to-type cultivars will avoid worldwide confusion between the denominations and cultivars existing in almost any world germplasm collection [30]. Aside from that, the SSR profiles of correctly identified and authenticated material can be used as a reference when dealing with the authenticity and traceability of olive products. In this direction, Trujillo et al. [8] exhaustively characterized, identified and authenticated the 499 accessions (824 trees in total) present in the WOGBC in Córdoba, Spain, representing samples from 21 countries using both phenotypic characters and molecular profiles generated by 33 available SSR markers. Several cases of synonyms and homonyms were detected and rectified, along with the identification of unique genotypes. The WOGBC has now become one of the most characterized olive germplasm banks and has paved way for other worldwide collections to also be well cataloged.
Trujillo et al. [68] also proposed and presented a guide in the international seminar “The IOC Network of Germplasm Banks and The True Healthy Olive Cultivars Project” held in Cordoba (Spain) in 2019. In this guide, the successive necessary steps and methodologies for accomplishing these goals are described, from the arrival of the vegetal material to the bank to the establishment of the plants in the field collection once identified, authenticated and free of pathogens. The molecular protocol is based on a set of 17 previously selected SSRs. All of them are robust and extremely polymorphic, with almost a limitless capability to catalog olive cultivars [8]. Aside from that, in most of the IOC Network collections, there is a considerable amount of information generated with SSR markers. These exhaustive studies establish the potential of microsatellites as robust markers for the characterization and identification of cultivars in rich olive germplasm. Better management of ex situ collections would in turn facilitate the easy exchange of germplasm material even at international levels, eliminate any mislabeling or misinterpretation of cultivars and ensure a reliable supply of cultivars to research labs, breeders and markets. These are indeed very useful resources in developing olive authentication and traceability studies, where the genotypic profile of any cultivar in question can essentially be matched with its true representative maintained in these worldwide collections.
4.1.2. Local Cultivars and Centennial Trees
In the last 25 years, important socioeconomic changes in many Mediterranean countries have driven significant technological improvements in olive cultivation. These changes are increasing the risk of genetic erosion of olive germplasm because local traditional cultivars are being replaced by a few cultivars that are suitable for new mechanically harvested plantations. Therefore, the identification and conservation of traditional olive cultivars are currently high-priority tasks that are needed to ensure the sustainable use of those cultivars in the future [70]. Microsatellite markers have been proven to be immensely useful in describing olive cultivars cultivated locally in certain regions [71,72,73,74,75,76]. Genotypic data about these local cultivars are useful information when authenticating commercial products coming out of these areas and certifying the origins of cultivars.
In Montenegro, when characterized using 10 SSR markers from the consensus set described by Baldoni et al. [58], the genotypic profile of the oldest olive tree, “Stara Maslina”, was found to be quite distinct from other ancient trees and main varieties, including the most diffused “Zutica Bar” variety. In addition, all locally grown and ancient germplasm of Montenegro were grouped together into a separate cluster when analyzed with other foreign cultivars [77]. Similarly, the autochthonous olive germplasm in Crete, Greece, represented by three cultivars (“Koroneiki”, “Mastoidis” and “Throubolia”) were characterized, along with two cultivars from Turkey and some representative wild genotypes from Crete, using seven informative SSR markers (from the DCA, UDO99 and IAS-oli series). The autochthonous cultivars were grouped into separate clusters showing their distinctness, and the cultivar “Throubolia” was found to be close to Turkish cultivars, indicating possible exchange or movement of the germplasm in the past [59]. Such studies supported by SSR-based genotypic information highlight the uniqueness of local germplasm and point toward more targeted genetic evaluation and conservation of such germplasm in olive-growing regions. Additionally, the information thus generated can also be utilized in developing SSR-based cultivar identification keys to be used in any future authentication of agri-food products based on such cultivars.
Since antiquity, olives have been grown and cultivated in the Mediterranean region of the world, and to date, many such centennial olive trees can be found growing in different regions. Microsatellites have been the molecular marker of choice for the characterization and identification of monumental or centennial olives from different olive-growing regions and proved helpful in generating valuable information with respect to the genotypic identities of trees. These studies supported the hypothesis that ancient olive trees might be unknown traditional cultivars that remained uncharacterized. Rotondi et al. [78] reported that most of the 206 ancient olive trees growing across the Emilia-Romagna region in Italy belonged to 10 cultivars that were already characterized, and the origins of the remaining genotypes remained unknown. In yet another study, 4526 ancient olive trees were surveyed in the “Taula del Sénia” (M-TdS) area (northeast Iberian Peninsula), and a subset of 293 trees was molecularly characterized using eight SSR markers, which revealed 43 different genotypic profiles, with 98% of the trees belonging to the local cultivar “Farga” [79]. Erre et al. [80] genotyped 21 wild and 57 cultivated olives in Sardinia using 13 SSR markers, where novel genotypes were identified and cluster analysis grouped the trees into distinct “wild” and “local” gene pools. Hence, valuable information could be deciphered with reference to the cultivar identity and existence of these trees using molecular as well as phenotypic tools. This would also be helpful in devising strategies for the cataloging, conservation and protection of such a rich ancient resource. The molecular information in the form of SSR profiles generated through such genetic studies can be very useful in cases where local cultivars are blended with commercial ones or when any high-value local cultivars are being adulterated.
4.1.3. Characterization of Wild Olive Populations
Wild germplasm in olives, also known as oleasters, can be exploited in breeding and genetic improvement programs as a rich source of variation in the development of varieties with improved traits, such as biotic and abiotic resistance and increased growth and yields. Microsatellite marker-based diversity studies and the estimation of genetic relationships within wild olive populations and between cultivated and wild forms were reported [80,81,82,83,84,85,86,87,88,89,90]. This would give better insights into the history of the domestication of olives, the regional distribution of genetic diversity and any gene flow between oleasters, feral forms and cultivated types. This topic has been recently reviewed by Belaj et al. [91]. Therefore, to obtain more detailed information, the reader is referred to this review.
In summary, Table 2 provides a list of studies highlighting the various applications of microsatellites in the characterization of olive genetic resources. These studies actually provide useful information about the various microsatellite markers used, and the different genetic indices thus generated can help in the selection of the most appropriate set of SSRs for any future work related to characterization or cultivar authentication. High genetic variability can be utilized in selecting superior genotypes and cultivars for future breeding programs and cultivation. Broad genetic diversity in olive germplasm is also reflected by high heterozygosity levels (both expected and observed heterozygosity) obtained through SSR analysis. For the most part, the expected heterozygosity (He) values were lower than the observed heterozygosity (Ho) in olives, as represented in Table 2. Another informative genetic index for SSR usefulness is the polymorphic information content value (PIC value), which in the case of olive SSRs was >0.5 in most of the cases for different SSR loci and reported to be as high as 0.95 by Omrani-Sabbaghi et al. [92].

Table 2.
List of studies highlighting applications of microsatellites in the characterization of olive genetic resources.
4.2. Agri-Food Traceability: Olive Oil and Table Olives
Two of the essential components of Mediterranean cuisine, table olives and olive oil, are among the most valuable agri-food products, especially in European markets. Their rich nutritional value and antioxidant properties have also attracted customers from non-olive oil producing areas such as the U.S. and Asia. Increasing demands and market value have tempted certain groups toward fraud and adulteration of high-quality extra virgin olive oils as well as table olives, with the mixing of cheaper low-quality oils such as other vegetable oils [105] and mislabeling of products produced from high-value cultivars or olive-growing regions being among the identified adulterations [106]. To prevent such fraudulent practices, the European Union (EU) has enacted regulations and introduced certifications (European Council Regulation EEC/2081/1992) in the form of “protected designation of origin” (PDO) and “protected geographical indication” (PGI) and launched a consortium-led project called “OLIV-TRACK” to work on olive oil traceability. Additionally, recent projects such as the OLEUM project (http://www.oleumproject.eu/, accessed 20 June 2021) and the Food Integrity Project (https://secure.fera.defra.gov.uk/foodintegrity/index.cfm?sectionid=21, accessed 20 June 2021) have also presented strategies to tackle olive oil fraud. Apart from the geographical origin of the cultivar and processing methods, the cultivar genotype is one of the key determinants in defining these designations. Therefore, methods are needed that can ascertain the authenticity of cultivars present in a particular batch of olive oil. The concept of cultivar authentication has primarily been used in the context of modern food technology to guarantee that the commercial edible product matches the cultivar specified on the label [107,108,109]. The authenticity of olive oil and table olives has been assessed through conventional methods, including chemical analyses where the presence of the key metabolites responsible for a peculiar flavor and taste is examined. These mainly include the HPLC-based identification of distinct metabolites [110,111]. Several other techniques such as lipid profiling, stable isotope analysis and chromatography-based metabolite analysis have also been used in olive product authentication and traceability [112,113]. Although these methods allow the identification of cultivars and their origins, complex multivariate analyses and statistical procedures are generally needed, which makes these challenging tasks. Additionally, many of these methods are affected by the environment and physiological conditions during the growth of plants, and hence variations in compositions may be seen. Therefore, DNA-based detection methods have gained interest in recent years, as DNA is unaffected by environmental conditions, and thus more specific, accurate and sensitive results could be obtained regarding the origin and identity of a cultivar. Various DNA-based molecular markers have been used in the authentication of olive trees and oil [114,115]. As already detailed in this review, SSRs possess a high power of discrimination and are among the most widely employed molecular marker systems in olives. Difference in SSR profiles between olive oil-producing cultivars can be used to identify their presence in monovarietal oils as well as mixtures of olive oils. The isolation of DNA in adequate amounts and quality from difficult matrices like olive oil is a challenging task, and the success and reproducibility of PCR amplification and marker analysis largely depends on this. Over the last decade, different isolation protocols and kits have been tested and modified for better DNA extraction from fruits and oils, and these studies highlight the importance of DNA quality and its impact on molecular marker-based tests [116,117,118,119,120]. Recently, Piarulli et al. [121] compared four DNA isolation methods referenced in the literature and came up with a modified method based on the work of Consolandi et al. [122] for the extraction of DNA from extra virgin olive oil in a much smaller time frame (4 h as compared with the 30 h reported) and involving low-cost options. A washable and reusable miniaturized device has been developed as well and tested for highly efficient DNA purification from olive oil, providing an increased surface-area-to-volume ratio when compared with other approaches, allowing highly efficient DNA purification and concentration from samples with minute DNA contents [123]. Molecular markers that amplify shorter fragments are supposed to work efficiently with low-quality or fragmented DNA isolated from oil, and SSRs and SNPs are the favored choice in such cases. Here, key achievements in the field of olive oil and table olive traceability using microsatellite or SSR markers are reviewed and summarized in Table 3 with details of the SSR markers and sample types used.

Table 3.
Applications of microsatellites in olive oil and table olive traceability.
Breton et al. [116] used magnetic beads for DNA purification and amplified SSR alleles from leaves as well oil DNA. The SSR patterns were verified in virgin oil samples of known origins, either in separate cultivars or in mixtures, as well as in commercial virgin oil samples available from markets. Virgin olive oil originating from 10 different olive cultivars were also identified by Pasqualone et al. [124], and a set of three primers (DCA4, DCA17 and GAPU89) was used to describe an identification key for olive cultivars and oil traceability. Testolin and Lain [117] reported DNA extraction from olive oil, comparing different protocols and commercial kits and utilizing conventional and nested SSR-PCR to identify specific cultivar DNA in oil. Similarly, Muzzalupo et al. [118] performed SSR-based authentication of virgin olive oil from “Ogliarola salentina” and Pasqualone et al. [125] identified a PDO-designated extra virgin olive oil (Collina di Brindisi) which contained aminimum of 70% oil from the cultivar “Ogliarola salentina”.These studies established the utility of microsatellites in authenticating a cultivar in a mixture of oils as well.
The use of principal component analysis (PCA) was emphasized in a study with 23 olive oil samples of Portuguese origin (11 monovarietal and 12 commercial oils), which were fingerprinted using 4 SSR loci in combination with 2 RAPD and 4 ISSR markers. No correlation among the common denominations was revealed and commercial samples from the same olive oil brand as well as the samples from the PDO olive oil Tras-os-Montes were found to be distributed in different PCA quadrants. The use of a larger set of markers was therefore required in order to cluster the cultivars and identify each denomination. The study found PCA analysis to be useful in the categorization of samples according to the regions of origin [126]. While dealing with oil traceability through genetic markers, the presence of alleles from pollinators also needs to be distinguished in order to infer denominations correctly, as observed by Ben-Ayed et al. [119] and Alba et al. [127], where parental contributions are assessed while comparing the microsatellite profiles generated from the DNA of the leaves and oil of certain cultivars. The utility of microsatellites in the genetic traceability of oil in agro-food chains was also established when DNA isolated from the drupes or leaves samples of three olive oil cultivars, namely “Pisciottana”, “Frantoio” and “Leccino”, were genotyped using six SSRs, and similar genetic profiles were obtained with their monovarietal oils. A 1:1 DNA mixture from two extra virgin monovarietal oils was also tested and could detect the expected alleles in the mixture [99].
Microsatellites have also been used in traceability analyses for PDO table olives. Three Italian PDO olives could be reliably identified among a set of 10 olive cultivars using 16 SSR primer pairs. A power of discrimination as high as 0.9 was obtained in the microsatellite set used for analysis [130]. A combination of genetic and biochemical tools in olive oil traceability studies can add to the accuracy of the experiments. Correlation between the SSR genetic data from cultivars and chemical and sensory profiles of nine monovarietal oils was observed by Rotondi et al. [137]. However, no correlation was obtained between genetic and pleasant flavor profiles. A bunch of parameters could play a role in the success of a traceability system based on genetic markers like microsatellites. An evaluation of such parameters was conducted by Vietina et al. [128] through the genotyping of 21 monovarietal oils obtained from 16 cultivars using 11 microsatellite markers. Each marker was assessed for its amplification ability over different oil DNA, reproducibility across a set of replicates in an experiment and correspondence of alleles in oil as well as leaf DNA. Significant correlation was found between the amplification ability and DNA yield, indicating the role of the extraction method. SSR marker GAPU89 gave a total correspondence and amplification ability value of 49.32%, and marker DCA5 was found to have the highest reproducibility, being 71.43 ± 21.82%. The high standard deviation values were attributed to variations within the samples caused by DNA extraction. Microsatellites were also successfully used by Ben-Ayed et al. [129] in the authenticity and traceability of virgin olive oils, and they also reported the non-correspondence of SSR profiles between oil and leaf DNA in some cases, thereby further strengthening the importance of distinguishing the pollinator and maternal alleles. Figure 2 summarizes the process and the main factors that may potentially affect the molecular traceability of olive oils and table olives when using SSR markers. As depicted in the figure, during DNA isolation, DNA that is too fragmented and very low yields may not always provide sufficient target templates and hence do not amplify the correct alleles. Similarly, the presence of inhibitor compounds from DNA extracts may lead to poor PCR amplification. Amplificability of the markers is also required to be checked for different SSRs in DNA isolated by different methods. Only those markers which give a consistent result in one or two methods should be used further. For the genotyping methods, the resolution of alleles needs to be highly precise for using SSRs in traceability and authenticity testing. Methods like capillary electrophoresis and high-resolution melting have proven to be useful. The correspondence of alleles is yet another important factor, where any microsatellite that generate similar profiles in a target oil and corresponding leaf sample of the cultivar in question can be used as a traceability marker. Ideally, the allelic pattern should be similar, but knowledge of the pollinating behavior of the cultivar is beneficial for result interpretation. As for reproducibility, an ideal SSR used for traceability should be highly reproducible irrespective of the laboratories, instruments and reagents used.
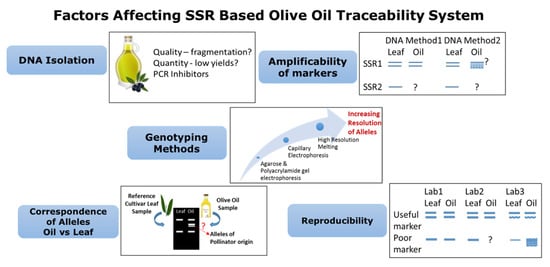
Figure 2.
Process and main factors influencing the applicability of a microsatellite marker in the authentication and traceability of olive oils and table olives.
Concerns with respect to the presence of traces of pollinator DNA in extractions made out of oil matrices leading to differences in the allelic profiling of oil and leaf samples also attracted researchers toward the applicability of plastid-based markers. However, chloroplast DNA (cpDNA) among cultivars has shown low levels of variation, which has limited its use in authenticity testing or traceability analyses.
Pérez-Jiménez et al. [131] utilized nine cpDNA loci that consisted of microsatellites and small insertion–deletions (indels) to identify the olive cultivar in leaves and corresponding oil DNA. Six haplotypes could be fingerprinted, and a rare haplotype was identified in genotypes producing regionally high-valued commercial oil. The available olive plastid genome can therefore be analyzed for the presence of more such microsatellite regions. In order to overcome the challenges of DNA isolation from oil matrices, Muzzalupo et al. [132] reported a direct DNA amplification method which avoided the routine extraction step and instead used KAPA3G plant DNA polymerase (an engineered DNA polymerase which could tolerate plant PCR inhibitors) for SSR amplification of membrane-filtered DNA molecules. DNA isolated from this method was used to check the traceability of three distinct types of virgin olive oil. The diagnostics power of microsatellite markers was further proven in the analysis of processed olives by Crawford et al. [138], where a panel of 5 SSRs was selected out of the 15 tested to authenticate California-style olive cultivars, widely marketed as packed forms. Based on the differences in allele combinations generated through these markers, any two samples could be differentiated. While comparing the genotyping method based on SSR alongside fatty acid analysis, phenolic content and nuclear magnetic resonance (NMR) analysis, Crawford et al. [139] found NMR to be able to discriminate all four tested cultivars in their processed forms. However, the five SSR markers could still detect genetic similarity between Sevillano and Gordal cultivars and indicated possible synonymy between the two.
More recently, techniques like high-resolution melting (HRM) have been reported to be coupled to SSR genotyping for the identification of target cultivars in commercial olive oil samples. HRM gives an additional advantage of closed-tube analysis post-PCR and is a sensitive and cost-effective method. Montemurro et al. [133] identified the constituent cultivars of PDO, designated “Terra di Bari” extra virgin olive oil, using HRM curve analysis of the SSR marker DCA18, and Gomes et al. [135] also applied this method for varietal identification in monovarietal PDO as well as blended olive oils using three SSRs from the UDO99 series (UDO99-011, UDO99-039 and UDO99-024) and one SSR from the DCA series (ssrOeUA-DCA16). In addition, Pasqualone et al. [134] evaluated the effect of talc addition during olive oil processing on DNA by comparing the SSR-HRM profiles of treated as well as control samples. Similarly, Pasqualone et al. [136] carried out varietal authentication in samples from crude olive pomace oil and corresponding virgin olive oil. Chedid et al. [140] performed both SSR-HRM and SNP-HRM for authentication and trace adulteration in olive oils and found that the discrimination power of SSRs was greater in the case of monovarietal olive oils, while SNPs were the marker of choice when the oils were blended together or adulterated.
Overall, microsatellites present a desirable system for formulating olive oil and table olive traceability studies, and key parameters like DNA extraction efficiency, reproducibility of the SSR profiles, knowledge about the breeding and pollinating behavior of the cultivars in question and correspondence levels between the oil and reference leaf SSR profiles should be focused on in order to utilize the method as a successful detection tool.
5. Concluding Remarks
A vast amount of genetic information about olive populations, wild relatives, local cultivars and germplasm banks around the world is now available to researchers, which can be utilized for developing cultivar breeding programs and better management of global olive genetic resources. However, organizing this valuable information in the form of easy-to-access and routinely updated databases is essential for the smooth transfer and sharing of scientific knowledge to the olive research community and control laboratories for the olive industry. Olives and olive oil have been an essential part of the diets for many populations, especially the Mediterranean region, with a notable presence nowadays in the non-olive growing nations of the world as well. Therefore, genetic characterization of the available unexplored germplasm is an important step for the introduction of new and improved cultivars. There are challenges associated with use of SSRs as tools to identify olive cultivars and obtain reproducible DNA profiles extracted from its oils.
One of the main limitations in implementing a traceability system based on microsatellites or any of the marker systems is the reproducibility of genotypic profiles across different laboratories. There can be variations due to the quality of the DNA extracted and the genotyping method used, and therefore, results need to be carefully interpreted while using the same set of cultivars and markers under different conditions. Additionally, identifying pollinator origin alleles while comparing olive oil and corresponding leaf DNA is crucial for correct result interpretation. A set of reference cultivars and their respective SSR profiles should be defined globally, and this can be used as a set of controls during experiments by all the laboratories working in cultivar identification and traceability of oil and table olives in order to maintain the authenticity of the data. Olive oil and table olive quality and authenticity is a topic of concern nowadays, and continuous efforts are being made to develop traceability tools based on chemical as well as molecular methods. The available literature indicates that microsatellites are a potential marker system with excellent utility in cultivar identification and coupling with high-throughput platforms, like automated sequencers, and high-resolution melting provides much faster and more sensitive and accurate results. As developments are being made in sophisticated techniques of genotyping, the problems associated with microsatellite profiling, such as mis-scoring of alleles or poor resolution of the electrophoresis gels, are being overcome, allowing users to obtain robust and reliable molecular profiles from samples of commercial olive oil and table olives.
With the use of next-generation sequencing (NGS) technologies in olive trees, more and more genomic information is being added and can be used as a rich source for the development of new sets of long core repeats containing microsatellite markers to overcome limitations while using dinucleotide repeat-rich SSRs. The increasing number of available genomic as well as EST SSRs will not only escalate the existing molecular arsenal but also pave the way for their application in the development of functional markers and linkage, as well as association mapping, map-based cloning and marker-assisted selection in the future, in addition to variety identification in high-quality food products such as table olives and olive oil. The use of techniques like HRM has opened new ways of analyzing microsatellites and exploring their potential beyond length polymorphisms. The development and applications of SNP markers in olives have also gained attention in recent years, but SSRs still remain a marker of choice to initiate preliminary genetic studies in a collection of cultivars, especially in resource-limited laboratories.
Author Contributions
S.Y. and M.P. conceptualized the manuscript design and contents; S.Y. and J.C. wrote the manuscript and carried out the detailed literature search and compilation; M.P. and I.T. contributed to meticulous inputs for refinement and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NANOEATERS: Valorization and transfer of NANOtechnologies to EArly adopTERS of the Euroregion Galicia-Norte Portugal (200005902—NANOEATERS—Aceite de oliva), supported by INTERREG V-A España-Portugal (POCTEP) 2014–2020, and by the project Nanotechnology-Based Functional Solutions (NORTE-01-0145-FEDER-000019), supported by the Norte Portugal Regional Operational Programme (NORTE2020) under the PORTUGAL 2020 Partnership Agreement through the European Regional Development Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The author (S.Y.) acknowledges the Indian Council of Forestry Research and Education (ICFRE) in Dehradun, India for administrative support to work as a post-doctoral visiting researcher at the Food Quality and Safety Research Group of the International Iberian Nanotechnology Laboratory (INL) in Braga, Portugal.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pérez-Jiménez, F.; Ruano, J.; Perez-Martinez, P.; Lopez-Segura, F.; Lopez-Miranda, J. The influence of olive oil on human health: Not a question of fat alone. Mol. Nutr. Food Res. 2007, 51, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Cicerale, S.; Lucas, L.; Keast, R. Biological activities of phenolic compounds present in virgin olive oil. Int. J. Mol. Sci. 2010, 11, 458–479. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Hu, F.B.; Martínez-González, M.A.; Fitó, M.; Bulló, M.; Estruch, R.; Ros, E.; Corella, D.; Recondo, J.; Gómez-Gracia, E.; et al. Olive oil intake and risk of cardiovascular disease and mortality in the PREDIMED Study. BMC Med. 2014, 12, 78. [Google Scholar] [CrossRef]
- Besnard, G.; Bakkali, A.E.; Haouane, H.; Baali-Cherif, D.; Moukhli, A.; Khadari, B. Population genetics of Mediterranean and Saharan olives: Geographic patterns of differentiation and evidence for early generations of admixture. Ann. Bot. 2013, 112, 1293–1302. [Google Scholar] [CrossRef]
- Barranco, D.; Trujillo, I.; Rallo, P. Are “Oblonga” and “Frantoio” olives the same cultivar? HortScience 2000, 35, 1323–1325. [Google Scholar] [CrossRef]
- Corrado, G.; La Mura, M.; Ambrosino, O.; Pugliano, G.; Varricchio, P.; Rao, R. Relationships of Campanian olive cultivars: Comparative analysis of molecular and phenotypic data. Genome 2009, 52, 692–700. [Google Scholar] [CrossRef]
- Bartolini, G.; Cerreti, S. Olive Germplasm (Olea europaea L.) Cultivars, Synonyms, Cultivation Area, Collections, Descriptors. Available online: www.oleadb.it/olivodb.html (accessed on 10 February 2021).
- Trujillo, I.; Ojeda, M.A.; Urdiroz, N.M.; Potter, D.; Barranco, D.; Rallo, L.; Diez, C.M. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genet. Genomes 2014, 10, 141–155. [Google Scholar] [CrossRef]
- Gupta, P.K.; Varshney, R.K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 2000, 113, 163–185. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef]
- Morgante, M.; Olivieri, A.M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C. de F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Belaj, A.; Satovic, Z.; Cipriani, G.; Baldoni, L.; Testolin, R.; Rallo, L.; Trujillo, I. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor. Appl. Genet. 2003, 107, 736–744. [Google Scholar] [CrossRef]
- De la Rosa, R.; Angiolillo, A.; Guerrero, C.; Pellegrini, M.; Rallo, L.; Besnard, G.; Bervillé, A.; Martin, A.; Baldoni, L. A first linkage map of olive (Olea europaea L.) cultivars using RAPD, AFLP, RFLP and SSR markers. Theor. Appl. Genet. 2003, 106, 1273–1282. [Google Scholar] [CrossRef]
- Bandelj, D.; Jakše, J.; Javornik, B. Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica 2004, 136, 93–102. [Google Scholar] [CrossRef]
- Wu, S.; Collins, G.; Sedgley, M. A molecular linkage map of olive (Olea europaea L.) based on RAPD, microsatellite, and SCAR markers. Genome 2004, 47, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, C.; Simeone, R.; Pasqualone, A.; Ferrara, E.; Blanco, A. Genetic relationships and cultivar identification among 112 olive accessions using AFLP and SSR markers. J. Hortic. Sci. Biotechnol. 2005, 80, 105–110. [Google Scholar] [CrossRef]
- Ganino, T.; Beghè, D.; Valenti, S.; Nisi, R.; Fabbri, A. RAPD and SSR markers for characterization and identification of ancient cultivars of Olea europaea L. in the Emilia region, Northern Italy. Genet. Resour. Crop Evol. 2007, 54, 1531–1540. [Google Scholar] [CrossRef]
- Gomes, S.; Martins-Lopes, P.; Lima-Brito, J.; Meirinhos, J.; Lopes, J.; Martins, A.; Guedes-Pinto, H. Evidence for clonal variation in “Verdeal-Transmontana” olive using RAPD, ISSR and SSR markers. J. Hortic. Sci. Biotechnol. 2008, 83, 395–400. [Google Scholar] [CrossRef]
- Khadari, B.; El Aabidine, A.Z.; Grout, C.; Ben Sadok, I.; Doligez, A.; Moutier, N.; Santoni, S.; Costes, E. A Genetic Linkage Map of Olive Based on Amplified Fragment Length Polymorphism, Intersimple Sequence Repeat and Simple Sequence Repeat Markers. J. Am. Soc. Hortic. Sci. 2010, 135, 548–555. [Google Scholar] [CrossRef]
- Linos, A.; Nikoloudakis, N.; Katsiotis, A.; Hagidimitriou, M. Genetic structure of the Greek olive germplasm revealed by RAPD, ISSR and SSR markers. Sci. Hortic. 2014, 175, 33–43. [Google Scholar] [CrossRef]
- Kaya, H.B.; Cetin, O.; Kaya, H.; Sahin, M.; Sefer, F.; Kahraman, A.; Tanyolac, B. SNP Discovery by Illumina-Based Transcriptome Sequencing of the Olive and the Genetic Characterization of Turkish Olive Genotypes Revealed by AFLP, SSR and SNP Markers. PLoS ONE 2013, 8, e93146. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.B.; Cetin, O.; Kaya, H.S.; Sahin, M.; Sefer, F.; Tanyolac, B. Association Mapping in Turkish Olive Cultivars Revealed Significant Markers Related to Some Important Agronomic Traits. Biochem. Genet. 2016, 54, 506–533. [Google Scholar] [CrossRef]
- Angiolillo, A.; Mencuccini, M.; Baldoni, L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor. Appl. Genet. 1999, 98, 411–421. [Google Scholar] [CrossRef]
- Lumaret, R.; Ouazzani, N.; Michaud, H.; Vivier, G.; Deguilloux, M.-F.; Di Giusto, F. Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity 2004, 92, 343–351. [Google Scholar] [CrossRef]
- Zohary, D.; Spiegel-Roy, P. Beginnings of Fruit Growing in the Old World. Science 1975, 187, 319–327. [Google Scholar] [CrossRef]
- Lavee, S. ¿Por qué la necesidad de nuevas variedades de olivos? Fruticultura 1994, 62, 29–37. [Google Scholar]
- Van Hintum, T.J.L.; Brown, A.H.D.; Spillane, C.; Hodgkin, T. Core Collections of Plant Genetic Resources; IPGRI Technical Bulletin No. 3; International Plant Genetic Resources Institute: Rome, Italy, 2000; ISBN 92-9043-454-6. [Google Scholar]
- Bartolini, G.; Prevost, G.; Messeri, C.; Carignani, G. Olive Germplasm: Cultivars and World-Wide Collections; FAO: Rome, Italy, 1998. [Google Scholar]
- Rallo, L.; Barranco, D.; Díez, C.M.; Rallo, P.; Suárez, M.P.; Trapero, C.; Pliego-Alfaro, F. Strategies for Olive (Olea europaea L.) Breeding: Cultivated Genetic Resources and Crossbreeding. In Advances in Plant Breeding Strategies: Fruits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 3, pp. 535–600. ISBN 978-3-319-91944-7. [Google Scholar]
- Baldoni, L.; Belaj, A. Olive. In Oil Crops; Vollmann, J., Rajcan, I., Eds.; Springer: New York, NY, USA, 2009; pp. 397–421. ISBN 978-0-387-77594-4. [Google Scholar]
- Belaj, A. Germplasm bank: WOGBC-IFAPA (ESP046). In Proceedings of the International Olive Council (IOC) Network of Germplasm Banks and Phytosanitary Management II Workshop True Healthy Olive Cultivars, Videoconference, 9–11 December 2020. [Google Scholar]
- Morello, P. Germplasm Bank WOGBC-UCO Spain. In Proceedings of the International Olive Council (IOC) Network of Germplasm Banks and Phytosanitary Management II Workshop True Healthy Olive Cultivars, Videoconference, 9–11 December 2020. [Google Scholar]
- Haouane, H.; El Bakkali, A.; Moukhli, A.; Tollon, C.; Santoni, S.; Oukabli, A.; El Modafar, C.; Khadari, B. Genetic structure and core collection of the World Olive Germplasm Bank of Marrakech: Towards the optimised management and use of Mediterranean olive genetic resources. Genetica 2011, 139, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Veral, G.M. Germplasm bank: Izmir World Olive and Collection Turkish National Olive Collection. In Proceedings of the International Olive Council (IOC) Network of Germplasm Banks and Phytosanitary Management II Workshop True Healthy Olive Cultivars, Videoconference, 9–11 December 2020. [Google Scholar]
- Rallo, P.; Dorado, G.; Martín, A. Development of simple sequence repeats (SSRs) in olive tree (Olea europaea L.). Theor. Appl. Genet. 2000, 101, 984–989. [Google Scholar] [CrossRef]
- Sefc, K.M.; Lopes, M.S.; Mendonça, D.; Dos Santos, M.R.; Da Câmara Machado, M.L.; Da Câmara Machado, A. Identification of microsatellite loci in olive (Olea europaea) and their characaterization in Italian and Iberian olive trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). TAG Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef]
- Cipriani, G.; Marrazzo, M.T.; Marconi, R.; Cimato, A.; Testolin, R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor. Appl. Genet. 2002, 104, 223–228. [Google Scholar] [CrossRef]
- De La Rosa, R.; James, C.M.; Tobutt, K.R. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol. Ecol. Notes 2002, 2, 265–267. [Google Scholar] [CrossRef]
- Díaz, A.; De La Rosa, R.; Martín, A.; Rallo, P. Development, characterization and inheritance of new microsatellites in olive (Olea europaea L.) and evaluation of their usefulness in cultivar identification and genetic relationship studies. Tree Genet. Genomes 2006, 2, 165–175. [Google Scholar] [CrossRef]
- Gil, F.S.; Busconi, M.; Da Câmara Machado, A.; Fogher, C. Development and characterization of microsatellite loci from Olea europaea. Mol. Ecol. Notes 2006, 6, 1275–1277. [Google Scholar] [CrossRef]
- Munoz-Merida, A.; Gonzalez-Plaza, J.J.; Canada, A.; Blanco, A.M.; Garcia-Lopez, M.d.C.; Rodriguez, J.M.; Pedrola, L.; Sicardo, M.D.; Hernandez, M.L.; De la Rosa, R.; et al. De Novo Assembly and Functional Annotation of the Olive (Olea europaea) Transcriptome. DNA Res. 2013, 20, 93–108. [Google Scholar] [CrossRef]
- De la Rosa, R.; Belaj, A.; Munoz-Merida, A.; Trelles, O.; Ortiz-Martin, I.; Gonzalez-Plaza, J.J.; Valpuesta, V.; Beuzon, C.R. Development of EST-derived SSR Markers with Long-core Repeat in Olive and Their Use for Paternity Testing. J. Am. Soc. Hort. Sci. 2013, 138, 290–296. [Google Scholar] [CrossRef]
- Adawy, S.S.; Mokhtar, M.M.; Alsamman, A.M.; Sakr, M.M. Development of Annotated EST-SSR Database in Olive (Olea europaea). Int. J. Sci. Res. 2015, 14, 1063–1073. [Google Scholar]
- Alagna, F.; D’Agostino, N.; Torchia, L.; Servili, M.; Rao, R.; Pietrella, M.; Giuliano, G.; Chiusano, M.L.; Baldoni, L.; Perrotta, G. Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development. BMC Genom. 2009. [Google Scholar] [CrossRef]
- Alagna, F.; Cirilli, M.; Galla, G.; Carbone, F.; Daddiego, L.; Facella, P.; Lopez, L.; Colao, C.; Mariotti, R.; Cultrera, N.; et al. Transcript analysis and regulative events during flower development in olive (Olea europaea L.). PLoS ONE 2016. [Google Scholar] [CrossRef]
- Corrado, G.; Alagna, F.; Rocco, M.; Renzone, G.; Varricchio, P.; Coppola, V.; Coppola, M.; Garonna, A.; Baldoni, L.; Scaloni, A.; et al. Molecular interactions between the olive and the fruit fly Bactrocera oleae. BMC Plant Biol. 2012, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, R.; Cultrera, N.G.M.; Mousavi, S.; Baglivo, F.; Rossi, M.; Albertini, E.; Alagna, F.; Carbone, F.; Perrotta, G.; Baldoni, L. Development, evaluation, and validation of new EST-SSR markers in olive (Olea europaea L.). Tree Genet. Genomes 2016, 12, 120. [Google Scholar] [CrossRef]
- Arbeiter, A.B.; Hladnik, M.; Jakše, J.; Bandelj, D.; Arbeiter, A.B.; Hladnik, M.; Jakše, J.; Bandelj, D. Identification and validation of novel EST-SSR markers in olives. Sci. Agric. 2017, 74, 215–225. [Google Scholar] [CrossRef]
- Resetic, T.; Stajner, N.; Bandelj, D.; Javornik, B.; Jakse, J. Validation of candidate reference genes in RT-qPCR studies of developing olive fruit and expression analysis of four genes involved in fatty acids metabolism. Mol. Breed. 2013, 32, 211–222. [Google Scholar] [CrossRef]
- Dervishi, A.; Jakše, J.; Ismaili, H.; Javornik, B.; Štajner, N. Comparative assessment of genetic diversity in Albanian olive (Olea europaea L.) using SSRs from anonymous and transcribed genomic regions. Tree Genet. Genomes 2018, 14, 53. [Google Scholar] [CrossRef]
- Li, D.; Long, C.; Pang, X.; Ning, D.; Wu, T.; Dong, M.; Han, X.; Guo, H. The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions. PeerJ 2020, 8, e8573. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Rodríguez, M.V.; Beuzon, C.; González-Plaza, J.J.; Fernández-Ocaña, A.M. Identification of an olive (Olea europaea L.) core collection with a new set of SSR markers. Genet. Resour. Crop Evol. 2021, 68, 117–133. [Google Scholar] [CrossRef]
- Rallo, P.; Tenzer, I.; Gessler, C.; Baldoni, L.; Dorado, G.; Martín, A. Transferability of olive microsatellite loci across the genus Olea. Theor. Appl. Genet. 2003, 107, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Doveri, S.; Sabino Gil, F.; Díaz, A.; Reale, S.; Busconi, M.; da Câmara Machado, A.; Martín, A.; Fogher, C.; Donini, P.; Lee, D. Standardization of a set of microsatellite markers for use in cultivar identification studies in olive (Olea europaea L.). Sci. Hortic. 2008, 116, 367–373. [Google Scholar] [CrossRef]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojeda, M.A.; et al. A consensus list of microsatellite markers for olive genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Aksehirli-Pakyurek, M.; Koubouris, G.C.; Petrakis, P.V.; Hepaksoy, S.; Metzidakis, I.T.; Yalcinkaya, E.; Doulis, A.G. Cultivated and Wild Olives in Crete, Greece—Genetic Diversity and Relationships with Major Turkish Cultivars Revealed by SSR Markers. Plant Mol. Biol. Report. 2017, 35, 575–585. [Google Scholar] [CrossRef]
- Bandelj, D.; Jakse, J.; Javornik, B. DNA fingerprinting of olive varieties by microsatellite markers. Food Technol. Biotechnol. 2002, 40, 185–190. [Google Scholar]
- Mackay, J.F.; Wright, C.D.; Bonfiglioli, R.G. A new approach to varietal identification in plants by microsatellite high resolution melting analysis: Application to the verification of grapevine and olive cultivars. Plant Methods 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulou, A.; Ganopoulos, I.; Koubouris, G.; Tsaftaris, A.; Sergendani, C.; Kalivas, A.; Madesis, P. Microsatellite high-resolution melting (SSR-HRM) analysis for genotyping and molecular characterization of an Olea europaea germplasm collection. Plant Genet. Resour. 2014, 12, 273–277. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 2011, 22, 532–541. [Google Scholar] [CrossRef]
- Distefano, G.; Caruso, M.; La Malfa, S.; Gentile, A.; Wu, S.B. High Resolution Melting Analysis Is a More Sensitive and Effective Alternative to Gel-Based Platforms in Analysis of SSR—An Example in Citrus. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Poljuha, D.; Sladonja, B.; Šetić, E.; Milotić, A.; Bandelj, D.; Jakše, J.; Javornik, B. DNA fingerprinting of olive varieties in Istria (Croatia) by microsatellite markers. Sci. Hortic. 2008, 115, 223–230. [Google Scholar] [CrossRef]
- Carmona, R.; Zafra, A.; Seoane, P.; Castro, A.J.; Guerrero-Fernández, D.; Castillo-Castillo, T.; Medina-García, A.; Cánovas, F.M.; Aldana-Montes, J.F.; Navas-Delgado, I.; et al. ReprOlive: A database with linked data for the olive tree (Olea europaea L.) reproductive transcriptome. Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Ben Hassen, H.; Ennouri, K.; Ben Marzoug, R.; Rebai, A. OGDD (Olive Genetic Diversity Database): A microsatellite markers’ genotypes database of worldwide olive trees for cultivar identification and virgin olive oil traceability. Database 2016, 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, I.; Barranco, D.; Cabello, A.; Gordon, P.; Morello, C.; Díez, M.; Rallo, L. Proposal of a Guide for the cataloging, sanitation and management of the Germplasm Banks of the IOC Network (UCOLIVO). In Proceedings of the International Seminar. The IOC Network of Germplasm Banks and the True Healthy Olive Cultivars Project, Córdoba, Spain, 21–24 October 2019. [Google Scholar]
- Barranco, D.; Cimato, A.; Fiorino, P.; Rallo, L.; Touzani, A.; Castañeda, C.; Serafini, F.; Trujillo, I. World Catalogue of Olive Cultivars; International Olive Oil Council: Madrid, Spain, 2000. [Google Scholar]
- Rallo, L.; Barranco, D.; Castro-García, S.; Connor, D.J.; Gómez del Campo, M.; Rallo, P. High-Density Olive Plantations. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; Volume 41, pp. 303–384. [Google Scholar]
- Fernández i Martí, A.; Font i Forcada, C.; Socias i Company, R.; Rubio-Cabetas, M.J. Genetic relationships and population structure of local olive tree accessions from Northeastern Spain revealed by SSR markers. Acta Physiol. Plant. 2015, 37. [Google Scholar] [CrossRef]
- Sakar, E.; Unver, H.; Bakir, M.; Ulas, M.; Sakar, Z.M. Genetic Relationships Among Olive (Olea europaea L.) Cultivars Native to Turkey. Biochem. Genet. 2016, 54, 348–359. [Google Scholar] [CrossRef]
- Sakar, E.; Unver, H.; Ulas, M.; Lazovic, B.; Ercisli, S. Genetic Diversity and Relationships among Local Olive (Olea europeaea L.) Genotypes from Gaziantep Province and Notable Cultivars in Turkey, Based on SSR Markers. Not. Bot. Horti Agrobot. Cluj-Napoca 2016, 44, 557–562. [Google Scholar] [CrossRef]
- Unver, H.; Sakar, E.; Ulas, M.; Ercisli, S.; Ak, B. Molecular characterization of indigenous olive genotypes based on SSR analysis. Genetika 2016, 48, 1017–1025. [Google Scholar] [CrossRef]
- Hmmam, I.; Mariotti, R.; Ruperti, B.; Cultrera, N.; Baldoni, L.; Barcaccia, G. Venetian olive (Olea europaea) germplasm: Disclosing the genetic identity of locally grown cultivars suited for typical extra virgin oil productions. Genet. Resour. Crop Evol. 2018, 65, 1733–1750. [Google Scholar] [CrossRef]
- Reboredo-Rodríguez, P.; González-Barreiro, C.; Cancho-Grande, B.; Simal-Gándara, J.; Trujillo, I. Genotypic and phenotypic identification of olive cultivars from north-western Spain and characterization of their extra virgin olive oils in terms of fatty acid composition and minor compounds. Sci. Hortic. 2018, 232, 269–279. [Google Scholar] [CrossRef]
- Lazović, B.; Adakalić, M.; Pucci, C.; Perović, T.; Bandelj, D.; Belaj, A.; Mariotti, R.; Baldoni, L. Characterizing ancient and local olive germplasm from Montenegro. Sci. Hortic. 2016, 209, 117–123. [Google Scholar] [CrossRef]
- Rotondi, A.; Ganino, T.; Beghè, D.; Di Virgilio, N.; Morrone, L.; Fabbri, A.; Neri, L. Genetic and landscape characterization of ancient autochthonous olive trees in northern Italy. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2018, 152, 1067–1074. [Google Scholar] [CrossRef]
- Ninot, A.; Howad, W.; Aranzana, M.J.; Senar, R.; Romero, A.; Mariotti, R.; Baldoni, L.; Belaj, A. Survey of over 4, 500 monumental olive trees preserved on-farm in the northeast Iberian Peninsula, their genotyping and characterization. Sci. Hortic. 2018, 231, 253–264. [Google Scholar] [CrossRef]
- Erre, P.; Chessa, I.; Muñoz-Diez, C.; Belaj, A.; Rallo, L.; Trujillo, I. Genetic diversity and relationships between wild and cultivated olives (Olea europaea L.) in Sardinia as assessed by SSR markers. Genet. Resour. Crop Evol. 2010, 57, 41–54. [Google Scholar] [CrossRef]
- Belaj, A.; Muñoz-Diez, C.; Baldoni, L.; Porceddu, A.; Barranco, D.; Satovic, Z. Genetic diversity and population structure of wild olives from the north-western Mediterranean assessed by SSR markers. Ann. Bot. 2007, 100, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Belaj, A.; León, L.; Satovic, Z.; De la Rosa, R. Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by agro-morphological traits and SSR markers. Sci. Hortic. 2011, 129, 561–569. [Google Scholar] [CrossRef]
- Breton, C.; Tersac, M.; Bervillé, A. Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: Several Plio-Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J. Biogeogr. 2006, 33, 1916–1928. [Google Scholar] [CrossRef]
- Hannachi, H.; Breton, C.; Msallem, M.; Ben El Hadj, S.; El Gazzah, M.; Bervillé, A. Are olive cultivars distinguishable from oleaster trees based on morphology of drupes and pits, oil composition and microsatellite polymorphisms? Acta Bot. Gall. 2008, 155, 531–545. [Google Scholar] [CrossRef]
- Hannachi, H.; Breton, C.; Msallem, M.; Ben El Hadj, S.; El Gazzah, M.; Bervillé, A. Differences between native and introduced olive cultivars as revealed by morphology of drupes, oil composition and SSR polymorphisms: A case study in Tunisia. Sci. Hortic. 2008, 116, 280–290. [Google Scholar] [CrossRef]
- Hannachi, H.; Breton, C.; Msallem, M.; Hadj, S.B.E.; Gazzah, M.E.; Berville, A. Genetic Relationships between Cultivated and Wild Olive Trees (Olea europaea L. var. europaea and var. sylvestris) Based on Nuclear and Chloroplast SSR Markers. Nat. Resour. 2010, 1, 95–103. [Google Scholar] [CrossRef]
- Belaj, A.; Muñoz-Diez, C.; Baldoni, L.; Satovic, Z.; Barranco, D. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Sci. Hortic. 2010, 124, 323–330. [Google Scholar] [CrossRef]
- Yoruk, B.; Taskin, V. Genetic diversity and relationships of wild and cultivated olives in Turkey. Plant Syst. Evol. 2014, 300, 1247–1258. [Google Scholar] [CrossRef]
- Boucheffa, S.; Miazzi, M.M.; di Rienzo, V.; Mangini, G.; Fanelli, V.; Tamendjari, A.; Pignone, D.; Montemurro, C. The coexistence of oleaster and traditional varieties affects genetic diversity and population structure in Algerian olive (Olea europaea) germplasm. Genet. Resour. Crop Evol. 2017, 64, 379–390. [Google Scholar] [CrossRef]
- Chiappetta, A.; Muto, A.; Muzzalupo, R.; Muzzalupo, I. New rapid procedure for genetic characterization of Italian wild olive (Olea europaea) and traceability of virgin olive oils by means of SSR markers. Sci. Hortic. 2017, 226, 42–49. [Google Scholar] [CrossRef]
- Belaj, A.; Veral, M.G.; Sikaoui, H.; Moukhli, A.; Khadari, B.; Mariotti, R.; Baldoni, L. Olive Genetic Resources. In The Olive Tree Genome; Rugini, E., Baldoni, L., Muleo, R., Sebastiani, L., Eds.; Springer: Cham, Switzerland, 2016; pp. 27–54. [Google Scholar]
- Omrani-Sabbaghi, A.; Shahriari, M.; Falahati-Anbaran, M.; Mohammadi, S.A.; Nankali, A.; Mardi, M.; Ghareyazie, B. Microsatellite markers based assessment of genetic diversity in Iranian olive (Olea europaea L.) collections. Sci. Hortic. 2007, 112, 439–447. [Google Scholar] [CrossRef]
- Stambuk, S.; Sutlović, D.; Bakarić, P.; Petricević, S.; Andelinović, S. Forensic botany: Potential usefulness of microsatellite-based genotyping of Croatian olive (Olea europaea L.) in forensic casework. Croat. Med. J. 2007, 48, 556–562. [Google Scholar]
- Cantini, C.; Cimato, A.; Autino, A.; Redi, A.; Cresti, M. Assessment of the Tuscan Olive Germplasm by Microsatellite Markers Reveals Genetic Identities and Different Discrimination Capacity among and within Cultivars. J. Am. Soc. Hortic. Sci. 2008, 133, 598–604. [Google Scholar] [CrossRef]
- Ganino, T.; Beghè, D.; Rotondi, A.; Fabbri, A. Genetic resources of Olea europaea L. in the Bologna province (Italy): SSR analysis and identification of local germplasm. Adv. Hortic. Sci. 2008, 22, 149–155. [Google Scholar] [CrossRef]
- Rekik, I.; Salimonti, A.; Grati Kamoun, N.; Muzzalupo, I.; Lepais, O.; Gerber, S.; Perri, E.; Rebai, A. Characterization and Identification of Tunisian Olive Tree Varieties by Microsatellite Markers. HortScience 2008, 43, 1371–1376. [Google Scholar] [CrossRef]
- Ipek, A.; Barut, E.; Gulen, H.; Oz, A.T.; Tangu, N.A.; Ipek, M. SSR analysis demonstrates that olive production in the southern Marmara region in Turkey uses a single genotype. Genet. Mol. Res. 2009, 8, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Noormohammadi, Z.; Hosseini-Mazinani, M.; Trujillo, I.; Belaj, A. Study of intracultivar variation among main Iranian olive cultivars using SSR markers. Acta Biol. Szeged. 2009, 53, 27–32. [Google Scholar]
- Corrado, G.; Imperato, A.; la Mura, M.; Perri, E.; Rao, R. Genetic diversity among olive varieties of southern italy and the traceability of olive oil using SSR markers. J. Hortic. Sci. Biotechnol. 2011, 86, 461–466. [Google Scholar] [CrossRef]
- Ercisli, S.; Ipek, A.; Barut, E. SSR Marker-Based DNA Fingerprinting and Cultivar Identification of Olives (Olea europaea). Biochem. Genet. 2011, 49, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Mailer, R.J.; Belaj, A.; De La Rosa, R.; Raman, H. Microsatellite marker-based identification of mother plants for the reliable propagation of olive (Olea europaea L.) cultivars in Australia. J. Hortic. Sci. Biotechnol. 2012, 87, 647–653. [Google Scholar] [CrossRef]
- El Bakkali, A.; Haouane, H.; Moukhli, A.; Costes, E.; van Damme, P.; Khadari, B. Construction of Core Collections Suitable for Association Mapping to Optimize Use of Mediterranean Olive (Olea europaea L.) Genetic Resources. PLoS ONE 2013, 8, e61265. [Google Scholar] [CrossRef][Green Version]
- Abdessemed, S.; Muzzalupo, I.; Benbouza, H. Assessment of genetic diversity among Algerian olive (Olea europaea L.) cultivars using SSR marker. Sci. Hortic. 2015, 192, 10–20. [Google Scholar] [CrossRef]
- Beghè, D.; Molano, J.F.G.; Fabbri, A.; Ganino, T. Olive biodiversity in Colombia. A molecular study of local germplasm. Sci. Hortic. 2015, 189, 122131. [Google Scholar] [CrossRef]
- Casadei, E.; Valli, E.; Panni, F.; Donarski, J.; Farrús Gubern, J.; Lucci, P.; Conte, L.; Lacoste, F.; Maquet, A.; Brereton, P.; et al. Emerging trends in olive oil fraud and possible countermeasures. Food Control 2021, 124, 107902. [Google Scholar] [CrossRef]
- Kiritsakis, A.; Christie, W.W. Analysis of Edible Oils. In Handbook of Olive Oil; Harwood, J., Aparicio, R., Eds.; Springer US: Boston, MA, USA, 2000; pp. 129–158. ISBN 978-1-4757-5371-4. [Google Scholar]
- Downey, G.; Boussion, J. Authentication of Coffee Bean Variety by Near-infrared Reflectance Spectroscopy of Dried Extract. J. Sci. Food Agric. 1996, 71, 41–49. [Google Scholar] [CrossRef]
- Melchiade, D.; Foroni, I.; Corrado, G.; Santangelo, I.; Rao, R. Authentication of the ‘Annurca’ apple in agro-food chain by ampli-fication of microsatellite loci. Food Biotechnol. 2007, 21, 33–43. [Google Scholar] [CrossRef]
- Mouly, P.P.; Gaydou, E.M.; Faure, R.; Estienne, J.M. Blood Orange Juice Authentication Using Cinnamic Acid Derivatives. Variety Differentiations Associated with Flavanone Glycoside Content. J. Agric. Food Chem. 1997, 45, 373–377. [Google Scholar] [CrossRef]
- Montealegre, C.; Marina Alegre, M.L.; García-Ruiz, C. Traceability Markers to the Botanical Origin in Olive Oils. J. Agric. Food Chem. 2010, 58, 28–38. [Google Scholar] [CrossRef]
- Bajoub, A.; Bendini, A.; Fernández-Gutiérrez, A.; Carrasco-Pancorbo, A. Olive oil authentication: A comparative analysis of regulatory frameworks with especial emphasis on quality and authenticity indices, and recent analytical techniques developed for their assessment. A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 832–857. [Google Scholar] [CrossRef] [PubMed]
- Lukić, I.; Ros, A.D.; Guella, G.; Camin, F.; Masuero, D.; Mulinacci, N.; Vrhovsek, U.; Mattivi, F. Lipid profiling and stable isotopic data analysis for differentiation of extra virgin olive oils based on their origin. Molecules 2020, 25, 4. [Google Scholar] [CrossRef]
- Jiménez-Morillo, N.T.; Palma, V.; Garcia, R.; Dias, C.B.; Cabrita, M.J. Combination of Stable Isotope Analysis and Chemometrics to Discriminate Geoclimatically and Temporally the Virgin Olive Oils from Three Mediterranean Countries. Foods 2020, 9, 1855. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ayed, R.; Kamoun-Grati, N.; Rebai, A. An overview of the authentication of olive tree and oil. Compr. Rev. Food Sci. Food Saf. 2013, 12, 218–227. [Google Scholar] [CrossRef]
- Costa, J.; Mafra, I.; Oliveira, M.B.P.P. Advances in vegetable oil authentication by DNA-based markers. Trends Food Sci. Technol. 2012, 26, 43–55. [Google Scholar] [CrossRef]
- Breton, C.; Claux, D.; Metton, I.; Skorski, G.; Bervillé, A. Comparative Study of Methods for DNA Preparation from Olive Oil Samples to Identify Cultivar SSR Alleles in Commercial Oil Samples: Possible Forensic Applications. J. Agric. Food Chem. 2004, 52, 531–537. [Google Scholar] [CrossRef]
- Testolin, R.; Lain, O. DNA Extraction from Olive Oil and PCR Amplification of Microsatellite Markers. J. Food Sci. 2005, 70, C108–C112. [Google Scholar] [CrossRef]
- Muzzalupo, I.; Pellegrino, M.; Perri, E. Detection of DNA in virgin olive oils extracted from destoned fruits. Eur. Food Res. Technol. 2007, 224, 469–475. [Google Scholar] [CrossRef]
- Ben Ayed, R.; Grati-Kamoun, N.; Moreau, F.; Rebaï, A. Comparative study of microsatellite profiles of DNA from oil and leaves of two Tunisian olive cultivars. Eur. Food Res. Technol. 2009, 229, 757–762. [Google Scholar] [CrossRef]
- Raieta, K.; Muccillo, L.; Colantuoni, V. A novel reliable method of DNA extraction from olive oil suitable for molecular traceability. Food Chem. 2015, 172, 596–602. [Google Scholar] [CrossRef]
- Piarulli, L.; Savoia, M.A.; Taranto, F.; D’Agostino, N.; Sardaro, R.; Girone, S.; Gadaleta, S.; Fucili, V.; De Giovanni, C.; Montemurro, C.; et al. A Robust DNA Isolation Protocol from Filtered Commercial Olive Oil for PCR-Based Fingerprinting. Foods 2019, 8, 462. [Google Scholar] [CrossRef]
- Consolandi, C.; Palmieri, L.; Severgnini, M.; Maestri, E.; Marmiroli, N.; Agrimonti, C.; Baldoni, L.; Donini, P.; De Bellis, G.; Castiglioni, B. A procedure for olive oil traceability and authenticity: DNA extraction, multiplex PCR and LDR-universal array analysis. Eur. Food Res. Technol. 2008, 227, 1429–1438. [Google Scholar] [CrossRef]
- Carvalho, J.; Puertas, G.; Gaspar, J.; Azinheiro, S.; Diéguez, L.; Garrido-Maestu, A.; Vázquez, M.; Barros-Velázquez, J.; Cardoso, S.; Prado, M. Highly efficient DNA extraction and purification from olive oil on a washable and reusable miniaturized device. Anal. Chim. Acta 2018, 1020, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Pasqualone, A.; Montemurro, C.; Caponio, F.; Blanco, A. Identification of Virgin Olive Oil from Different Cultivars by Analysis of DNA Microsatellites. J. Agric. Food Chem. 2004, 52, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Pasqualone, A.; Montemurro, C.; Summo, C.; Sabetta, W.; Caponio, F.; Blanco, A. Effectiveness of Microsatellite DNA Markers in Checking the Identity of Protected Designation of Origin Extra Virgin Olive Oil. J. Agric. Food Chem. 2007, 55, 3857–3862. [Google Scholar] [CrossRef]
- Martins-Lopes, P.; Gomes, S.; Santos, E.; Guedes-Pinto, H. DNA Markers for Portuguese Olive Oil Fingerprinting. J. Agric. Food Chem. 2008, 56, 11786–11791. [Google Scholar] [CrossRef] [PubMed]
- Alba, V.; Sabetta, W.; Blanco, A.; Pasqualone, A.; Montemurro, C. Microsatellite markers to identify specific alleles in DNA extracted from monovarietal virgin olive oils. Eur. Food Res. Technol. 2009, 229, 375–382. [Google Scholar] [CrossRef]
- Vietina, M.; Agrimonti, C.; Marmiroli, M.; Bonas, U.; Marmiroli, N. Applicability of SSR markers to the traceability of monovarietal olive oils. J. Sci. Food Agric. 2011, 91, 1381–1391. [Google Scholar] [CrossRef]
- Ben-Ayed, R.; Grati-Kamoun, N.; Sans-Grout, C.; Moreau, F.; Rebai, A. Characterization and authenticity of virgin olive oil (Olea europaea L.) cultivars by microsatellite markers. Eur. Food Res. Technol. 2012, 234, 263–271. [Google Scholar] [CrossRef]
- Pasqualone, A.; Di Rienzo, V.; Nasti, R.; Blanco, A.; Gomes, T.; Montemurro, C. Traceability of Italian Protected Designation of Origin (PDO) table olives by means of microsatellite molecular markers. J. Agric. Food Chem. 2013, 61, 3068–3073. [Google Scholar] [CrossRef]
- Pérez-Jiménez, M.; Besnard, G.; Dorado, G.; Hernandez, P. Varietal tracing of virgin olive oils based on plastid DNA variation profiling. PLoS ONE 2013, 8, e70507. [Google Scholar] [CrossRef][Green Version]
- Muzzalupo, I.; Pisani, F.; Greco, F.; Chiappetta, A. Direct DNA amplification from virgin olive oil for traceability and authenticity. Eur. Food Res. Technol. 2015, 241, 151–155. [Google Scholar] [CrossRef]
- Montemurro, C.; Miazzi, M.M.; Pasqualone, A.; Fanelli, V.; Sabetta, W.; Di Rienzo, V. Traceability of PDO Olive Oil “Terra di Bari” Using High Resolution Melting. J. Chem. 2015, 2015, 1–7. [Google Scholar] [CrossRef]
- Pasqualone, A.; Rienzo, V.D.; Miazzi, M.M.; Fanelli, V.; Caponio, F.; Montemurro, C. High resolution melting analysis of DNA microsatellites in olive pastes and virgin olive oils obtained by talc addition. Eur. J. Lipid Sci. Technol. 2015, 117, 2044–2048. [Google Scholar] [CrossRef]
- Gomes, S.; Breia, R.; Carvalho, T.; Carnide, V.; Martins-Lopes, P. Microsatellite High-Resolution Melting (SSR-HRM) to Track Olive Genotypes: From Field to Olive Oil. J. Food Sci. 2018, 83, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Pasqualone, A.; Di Rienzo, V.; Sabetta, W.; Fanelli, V.; Summo, C.; Paradiso, V.M.; Montemurro, C.; Caponio, F. Chemical and Molecular Characterization of Crude Oil Obtained by Olive-Pomace Recentrifugation. J. Chem. 2016, 2016, 1–7. [Google Scholar] [CrossRef]
- Rotondi, A.; Beghè, D.; Fabbri, A.; Ganino, T. Olive oil traceability by means of chemical and sensory analyses: A comparison with SSR biomolecular profiles. Food Chem. 2011, 129, 1825–1831. [Google Scholar] [CrossRef]
- Crawford, L.M.; Carrasquilla-Garcia, N.; Cook, D.; Wang, S.C. Analysis of Microsatellites (SSRs) in Processed Olives as a Means of Cultivar Traceability and Authentication. J. Agric. Food Chem. 2020, 68, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Crawford, L.M.; Janovick, J.L.; Carrasquilla-Garcia, N.; Hatzakis, E.; Wang, S.C. Comparison of DNA analysis, targeted metabolite profiling, and non-targeted NMR fingerprinting for differentiating cultivars of processed olives. Food Control 2020, 114, 107264. [Google Scholar] [CrossRef]
- Chedid, E.; Rizou, M.; Kalaitzis, P. Application of high resolution melting combined with DNA-based markers for quantitative analysis of olive oil authenticity and adulteration. Food Chem. X 2020, 6, 100082. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).