Abstract
Because the robust and rapid determination of spoilage microorganisms is becoming increasingly important in industry, the use of IR microspectroscopy, and the establishment of robust and versatile chemometric models for data processing and classification, is gaining importance. To further improve the chemometric models, bacterial stress responses were induced, to study the effect on the IR spectra and to improve the chemometric model. Thus, in this work, nine important food-relevant microorganisms were subjected to eight stress conditions, besides the regular culturing as a reference. Spectral changes compared to normal growth conditions without stressors were found in the spectral regions of 900–1500 cm−1 and 1500–1700 cm−1. These differences might stem from changes in the protein secondary structure, exopolymer production, and concentration of nucleic acids, lipids, and polysaccharides. As a result, a model for the discrimination of the studied microorganisms at the genus, species and strain level was established, with an accuracy of 96.6%. This was achieved despite the inclusion of various stress conditions and times after incubation of the bacteria. In addition, a model was developed for each individual microorganism, to separate each stress condition or regular treatment with 100% accuracy.
1. Introduction
Because meat and meat products are highly appreciated by consumers, for their nutritional value and taste, the global supply of meat is expected to continue to increase in the coming years [1]. However, meat is highly prone to microbial spoilage and, therefore, rapid and easy identification of contamination is a major concern in food safety [1,2]. This will help to ensure measures to minimize health hazards, and thus prevent foodborne illness and unnecessary food waste along the supply chain [2].
However, as bacteria are subject to constant fluctuations in their growth conditions, both in nature and along the supply chain, they have developed capabilities to constantly adapt to conditions, or even change to a state of viability, but are non-cultivable [3,4,5]. This makes sub-lethally damaged cells difficult to detect with classical laboratory culture techniques [2]. Additionally, standard methods, such as classical microbiology, sensory-mechanical studies, and immunological or genetic techniques, have disadvantages in speed, complexity, and invasiveness [1,6,7,8,9]. However, these viable, but non-culturable, microorganisms can be revived within the supply chain, and thus not only affect the product’s usability, but may also be a health hazard [2,9,10,11].
Infrared (IR) spectroscopy has been successfully used to detect and identify microorganisms [10,12,13,14]. In recent years, many studies dealt with the IR spectroscopic evaluation of specific effects of stress conditions on microorganisms, such as protein misfolding [15], phase behavior of the cell membranes of Escherichia coli (E. coli) during desiccation, rehydration, and growth recovery [16,17], or the sonication injury on Listeria monocytogenes [18]. Moreover, IR spectroscopy was used to study the influence of nanoparticles on E. coli [19,20], and the effects of heavy metals on Brevundimonas sp., Gordonia sp., and Microbacterium oxydans, using the analysis of variance, hierarchical cluster analysis, principal component analysis (PCA), and soft independent modelling of class analogies (SIMCA) [21,22]. Additionally, the influence of heat on Lactococcus lactis, Salmonella enterica, and Listeria monocytogenes was evaluated by the analysis of the IR peak area of amide I and amide II bands, and the extent of injury was predicted by the analysis of the wavenumber area of 900–1300 cm−1 by SIMCA and partial least squares regression analysis (PLSR) [23,24]. Furthermore, the response of E. coli, Campylobacter jejuni, and Pseudomonas aeruginosa that were exposed to cold- [25,26], chemical- [25] and pH-stressors [25,27,28] was studied by DNA microarrays and Fourier-transform (FT) IR analysis, coupled to PCA, discriminant function analysis, and PLSR.
While interesting findings have been reported, IR microspectroscopy, in combination with PCA and canonical discriminant analysis, has not been used so far, to combine different stress conditions on numerous food-related microorganisms at different times after incubation, in one chemometric model.
The food industry is particularly interested in the following most dominant microorganisms that are detected on fresh and chilled meat, and other food products: Pseudomonas spp., especially Pseudomonas fluorescens (Ps. fluor) and Enterobacteriaceae, such as E. coli, Micrococcus luteus (M. luteus), Bacillus thuringiensis israelensis (B. tii), Bacillus coagulans (B. coag), Bacillus subtilis (B. sub), and Brochothrix thermosphacta (B. therm) [29,30,31,32,33,34,35,36].
Therefore, the aim of this study was the development of a rapid and non-destructive analysis method for food-related microorganisms. The influences of numerous stress-conditioned microorganisms, as well as regularly treated microorganisms, over various aging stages on agar plates, were analyzed with IR spectroscopy, to build up an extensive data set. Chemometric models were developed to discriminate the influence of stress, and also to discriminate the selected microorganisms independent of their stress conditions within one model down to the strain level.
2. Materials and Methods
2.1. Bacterial Cultures and Sample Preparation
The following nine microorganisms were cultivated on nutrient agar (10 g/L meat peptone, 10 g/L meat extract, 5 g/L sodium chloride and 18 g/L agar-agar (Merck KGaA, Germany)) and prepared according to our previously published method [12,37,38]: Bacillus subtilis DSM 10, Bacillus coagulans DSM 1, Escherichia coli K12 DSM 498, Escherichia coli TOP10, Micrococcus luteus DSM 20030, Brochothrix thermosphacta DSM 20171, Pseudomonas fluorescens DSM 4358, Pseudomonas fluorescens DSM 50090 and Bacillus thuringiensis israelensis DSM 5724.
As described in our previously published methods [12], the samples were taken by a blotting technique with the sample carrier (stainless steel cylinder) directly from the agar plate without any sampling pre-treatments (e.g., centrifugation, washing, drying). IR spectra were recorded directly (lifetime conditions) after sampling or directly after the stress impact (sampling condition) without any incubation period after sampling. Detailed information can be found in Section 2.2.
The spectral data set of each microorganism, consisting of four independent data sets for each stress condition, were divided into independent training and test data sets. Further information can be found in Section 2.4.
2.2. Sample Treatment
In order to expose the microorganisms to different influences, they were subjected to lifetime conditions (incubation under acidic and alkaline conditions, incubation at different temperatures and incubation under 2-propanol influence) and sampling conditions (cold sampling, heat sampling and desiccation) in a controlled manner, in addition to the regular reference treatment. Incubation was performed for all microorganisms in a Binder BD 240 (BINDER GmbH, Tuttlingen, Germany) incubator.
2.2.1. Reference Samples (Regular Treatment)
All microorganisms were cultivated in accordance to DSMZ (Leibniz Institut DSMZ—German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) guidelines. These samples served as reference samples in this study.
2.2.2. Incubation under Acidic Conditions
To expose microorganisms to acidic pH stress, a hydrochloric acid (HCl) solution (36%, Alfa Aesar, Ward Hill, MA, USA) with pH 1 (verified by means of pH indicator paper, Th. Geyer GmbH & Co. KG, Renningen, Germany) was prepared. The agar plates were completely covered with the hydrochloric acid solution (2 mL) and the inoculation took place onto the hydrochloric acid-covered agar plates. Afterwards the cultivation was performed in accordance to DSMZ guidelines.
2.2.3. Incubation under Alkaline Conditions
Complementary to the incubation under acidic conditions, a sodium hydroxide solution (sodium hydroxide pellets, Merck, Darmstadt, Germany) (pH 13 (verified by means of pH indicator paper)) was prepared to expose microorganisms to alkaline pH stress. Afterwards, the cultivation was performed in accordance to DSMZ guidelines.
2.2.4. Incubation at Lower/Higher Temperatures
Microorganisms were incubated at a temperature of 25 °C and 45 °C.
2.2.5. Incubation with 2-Propanol
Complementary to the incubation under acidic and alkaline stress, the microorganisms were stressed with 2-propanol (99.9%, Höfer Chemie GmbH, Kleinblittersdorf, Germany). Afterwards the cultivation was performed in accordance to DSMZ guidelines.
2.2.6. Cold Sampling
Microorganisms were sampled from regular treated samples, covered with liquid nitrogen for 60 s and instantly measured.
2.2.7. Heat-Drying
Microorganisms were sampled from regular treated samples, dried at 50 °C for 60 min and instantly measured.
2.2.8. Desiccation
Microorganisms were sampled from regular treated samples, dried in a desiccator filled with silica gel for 60 min and instantly measured.
2.3. Instrumentation
The samples were examined in reflectance mode, 20 scans per spectrum, 4 cm−1 resolution and 20× magnification (Cassegrain objective (Bruker Ser.910/1022346, numerical aperture: 0.6, working distance: 6 mm) by means of a Hyperion 3000/Vertex 70 Fourier-transform IR microspectrometer (Bruker Optics GmbH, Ettlingen, Germany) with mercury/cadmium/telluride (MCT) detector. Instrument controlling and data acquisition was carried out by the OPUS 7.5 software (Version 7.5, Ettlingen, Germany).
Due to the microscopic component, the morphological properties of heterogeneous samples can be combined with spectral data and samples, with a few hundred microorganisms being determined [39,40]. This often results in no further sample preparation than a transfer of the sample to a sample carrier [41]. In addition, the required analysis time is reduced compared to classical IR spectroscopy [40].
2.4. Data Handling and Visualization
IR spectra were subsequently sum normalized (OriginPro 2019b, OriginLab Corporation, Northampton, MA, USA), data reduced to the range of 915–1750 cm−1 and 2825–3680 cm−1 to exclude the characteristic CO2 region and the lower fingerprint area, the first derivative was built and smoothed with a 13-point Savitzky–Golay filter (LabVIEW 2016; National Instruments, Austin, TX, USA).
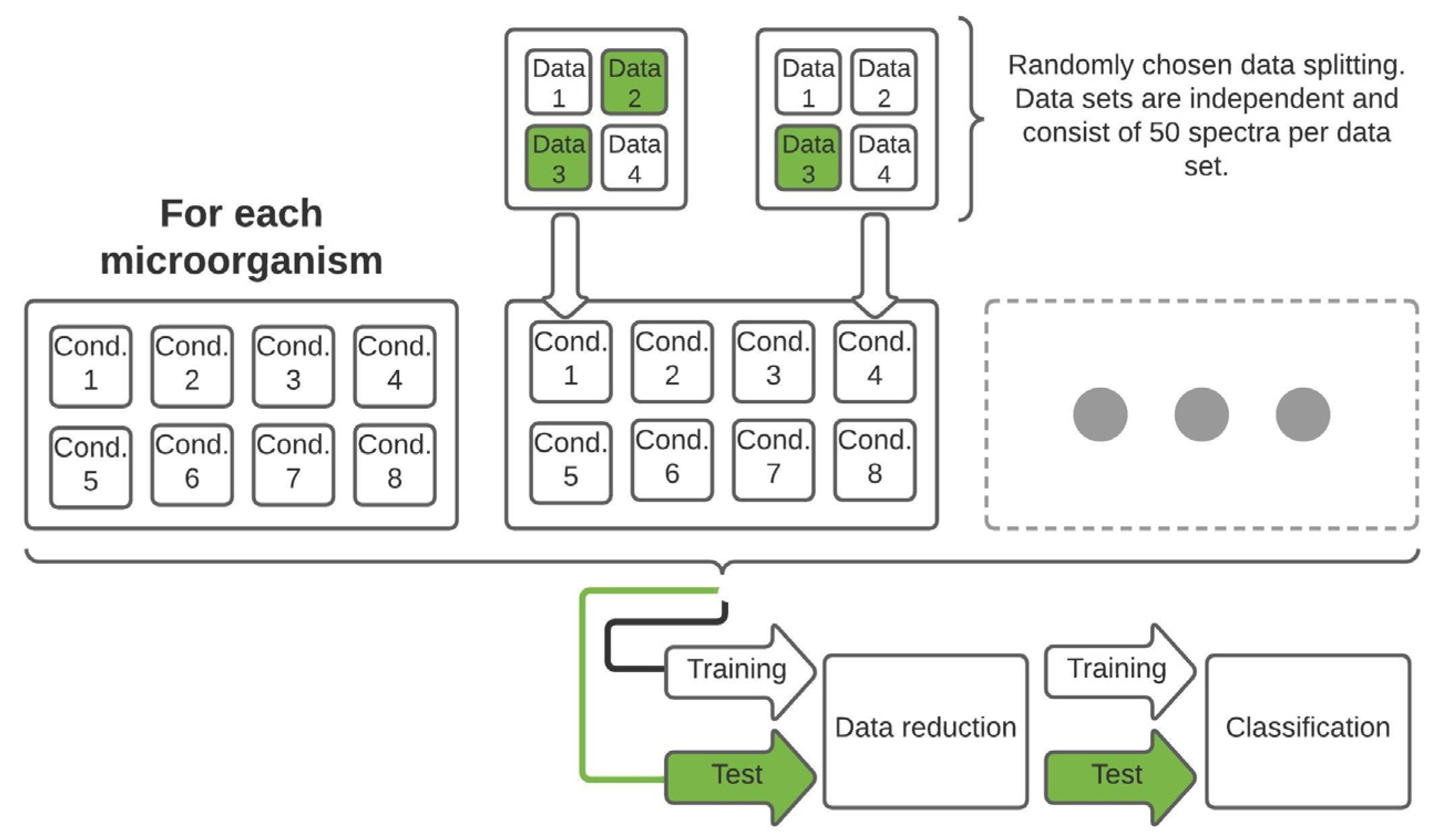
The splitting for training and test data was carried out so that one or two independent data sets, each with 50 spectra for each stress condition and regular treatment, was used as test data (Figure 1).
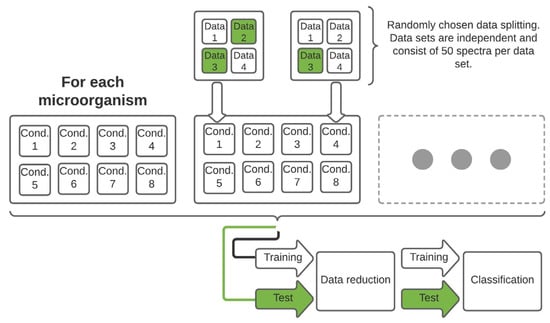
Figure 1.
Schematic representation of the data set structure as well as the splitting process with subsequent data reduction and classification. The training and test data set contains the corresponding information of each measured microorganism and thus of each growth and sampling condition.
The splitting process of training and test data is depicted in Table 1. Detailed information about the exact splitting pattern, the time after incubation of the microorganisms at the time of measurement and thus how long the microorganisms were exposed to the lifetime stress conditions can be found in the Table S1 (Supplementary Material).

Table 1.
Training and test data set sizes and data splitting ratio for the trained and tested microorganisms.
Because balanced training data sets are important, not only for data reduction by PCA, but also for robust, reliable and unweighted model development [42,43], the data sets were split into training and test data in different ways for the reason that not all stress conditions could be measured for all bacteria (see Section 3).
For the following data evaluation, principal component analysis (PCA) was applied to the training data; the test data were converted into the vector space of the training data and the data were classified by a canonical discriminant analysis (CDA) by means of LabVIEW 2016 and OriginPro 2019b.
3. Results and Discussion
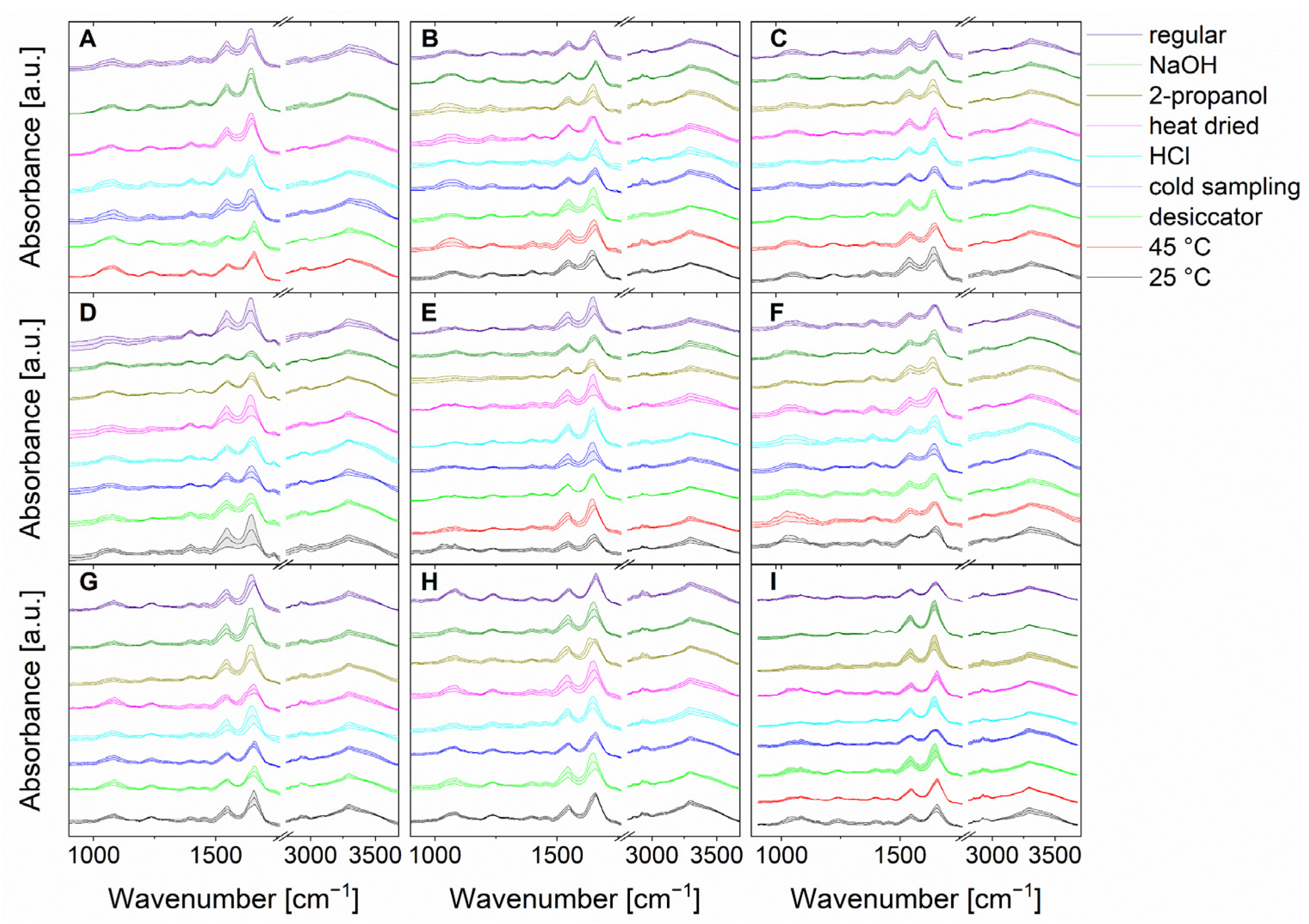
First, the microorganisms were exposed to the different bacterial stress conditions mentioned above, and IR microspectroscopic data were carefully acquired. Figure 2 shows the mean IR spectra, including their standard deviations of bacteria under normal culture conditions versus bacteria under stress conditions. It is important to note that for B. coag no bacterial growth was detected at 25 °C, and when 2-propanol was used. Also, B. tii and Ps. fluor did not grow at 45 °C. This suggests that these stress conditions lead a non-culturable state. As a result, these conditions are not shown in Figure 2, and cannot be used for further evaluation.
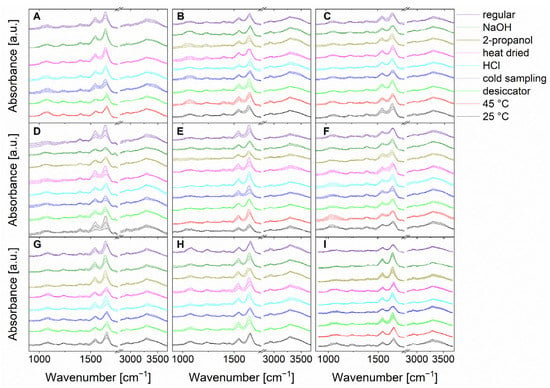
Figure 2.
Stacked mean IR spectra of the normalized data of in total 15,200 spectra (200 spectra of each stress condition and regular treatment) of B. coag (A), B. sub (B), B. therm (C), B. tii (D), E. coli K12 (E), M. luteus (F), Ps. fluor 4358 (G), Ps. fluor 50090 (H) and E. coli TOP10 (I). Standard deviations are indicated by color-coded bands around the mean value.
Because spectral differences between individual microorganisms and individual stress conditions are based on different compositions in proteins, nucleic acids, lipopolysaccharides, or lipids of the cell, visual discrimination of 15,200 spectra in total (Figure 2) is almost impossible [10]. The fine spectral differences are in the range of the P=O vibrations of phospholipids (1085 cm−1 and 1240 cm−1) and the C–O–C vibrations in polysaccharides (900–1200 cm−1) [10,40]. In addition, differences can be noted in the C=O, C–H, and C–O–H vibrations of fatty acids and proteins [10,40]. In the area of proteins, strong bands of amide I and amide II vibrations (1550–1675 cm−1) can also be observed [10,40]. Furthermore, various C–H and N–H stretching vibrations from fatty acids and proteins can be identified in the range of 2850 cm−1 [10,40]. Here, chemometric approaches can aid in the classification and were carefully optimized, as discussed below.
3.1. Bacteria Prediction Model
To ensure optimal model development and to avoid overfitting (performance plot: Figure S1 (Supplementary Material), the first 20 PCs (Figures S2 and S3) were used for model building using discriminant analysis.
As the covariance matrices of the training data classes had no significant equality, a quadratic, instead of a linear discriminant function, was chosen [44,45,46].
The error for classification and cross-validation of the training data was 0.01%; one spectrum of E. coli K12 was assigned to E. coli TOP10. In order to test the developed model for the classification of food-related microorganisms for robustness, accuracy, and reproducibility, independent test data sets of the trained classes were added to the model.
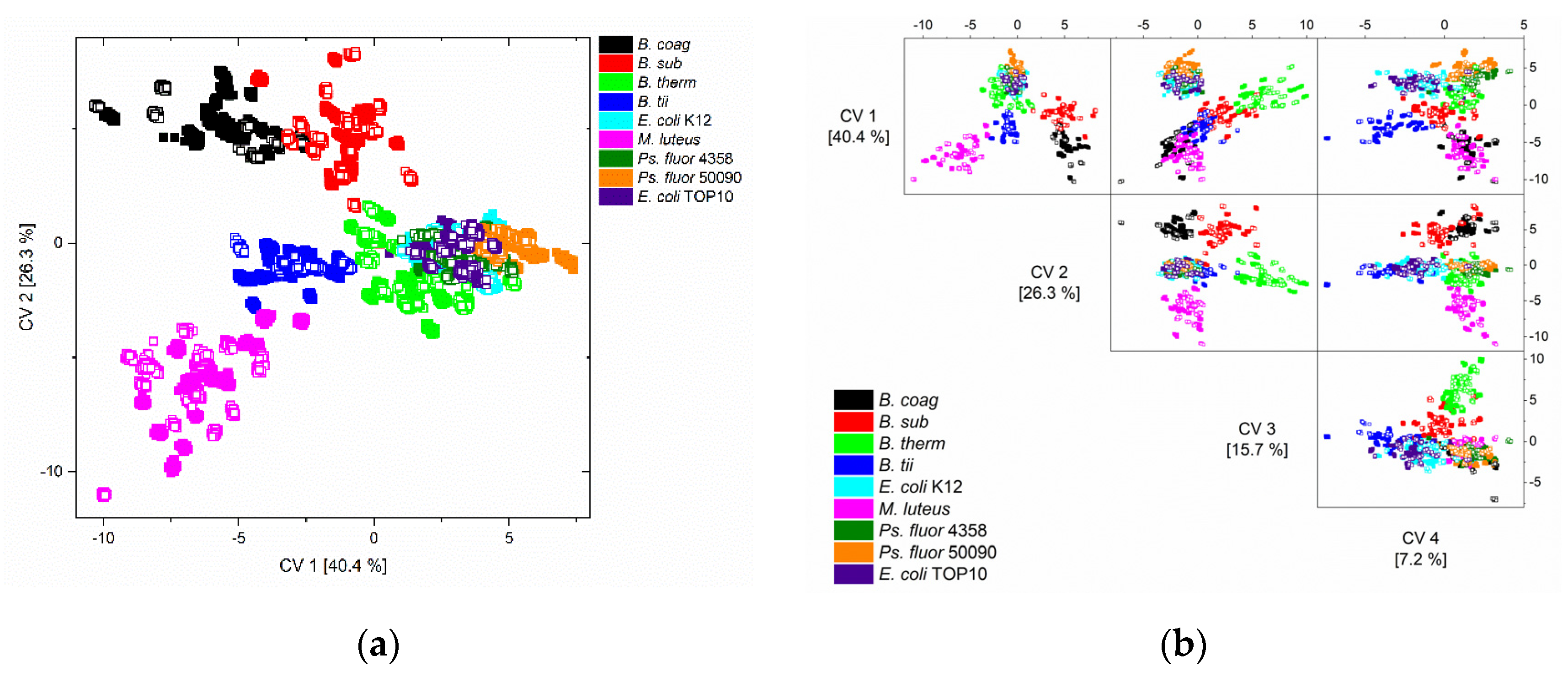
The first two (a) and the first four (b) canonical variables (CV) of the quadratic discriminant analysis (QDA) of the training data (solid squares) and test data (unfilled squares) are depicted in Figure 3a. On closer examination, it is noticeable that the test data are located exactly in the space of the training data.
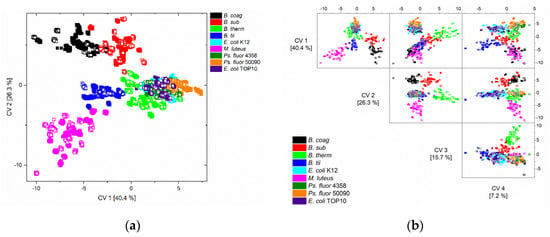
Figure 3.
Scatter (a) and scatter matrix (b) plot of canonical variable one and two, and one to four of the QDA of the training data (solid squares) and the independent test data (unfilled squares) of all nine microorganisms and all nine conditions.
M. luteus and all Bacillus spp. (B. coag, B. sub, B. therm, B. tii) could be separated from each other by the first two canonical variables, and Pseudomonas fluorescens from E. coli could be separated by the first four canonical variables (Figure 3b).
In conclusion, the data of the classification of the independent test data are presented in a confusion matrix (Table 2), which gives the number of spectra that were classified to the correct (diagonal) or wrong predicted class.

Table 2.
Confusion matrix for the independent test data set. All nine conditions were pooled as one class per microorganism. The rows show the observed groups and the columns show the predicted groups. The values in the diagonal of the table reflect the correct classifications of observations into groups.
It can be observed that the classification of the test data, and thus the model development of a robust and meaningful model, was successful. The error rate of the classification of the independent test data was only 3.4%, and can be found in the classes of B. coag, E. coli K12 and TOP10 and Ps. fluorescens.
Because the complete dataset consists of a large number of sub-datasets per microorganism, a detailed analysis of the classification errors on the sub-datasets is given in Table 3.

Table 3.
Detailed analysis of the classification errors of the independent test data set on the sub-dataset level. The numbers are the total numbers of misclassified spectra in the specific sub-dataset.
The detailed analysis of the error rate shows that a major part of the error was due to the misclassification of E. coli K12 to TOP10, and vice versa, and the assignment of Ps. fluorescens 4358 to Ps. fluorescens 50090. In addition, another part of the misclassification was due to the assignment of E. coli K12 to Ps. fluorescens.
These misclassifications were also indicated by the graphical representations (Figure 3) of the classification results, where it is at least visually apparent that the distinction between the respective E. coli and Ps. fluorescens strains, as well as the separation between E. coli and Ps. Fluorescens, appears difficult. However, the detailed error analysis shows that the separation between the Ps. fluorescens strains was feasible, but the separation between the E. coli strains was more difficult.
In contrast to other approaches, the presented results for the general discrimination of food-related bacteria were carried out with 20 scans per sample, as comparatively short measurement times [15,19,21,22,23,41,47] and in the absence of further sample preparation steps [15,18,21,22,25,26,47]. Nonetheless, the results demonstrate a non-inferior classification, even compared to macroscopic and microscopic studies, in which standardized work was performed in other approaches [12,23,26,41,48,49,50]. The novel aspect of this model, in comparison to the literature, is the inclusion of numerous stress conditions on microorganisms and the consideration of these stress conditions on numerous bacteria in one model, because often only the influence of a few stress conditions on single or a few selected microorganisms were investigated [19,21,22,23,26].
3.2. Stress Condition Prediction Model
Because clusters within a class (microorganisms) were visible, but do not seem to significantly influence the model for the discrimination of food-related microorganisms, it was reasonable to investigate what general influence the stress conditions have on the bacteria, or on the model. Therefore, we tested whether this type of model was also able to separate different stress conditions from each other.
All data were preprocessed, as already described for the discrimination model. The model building for the discrimination of different stress conditions per microorganism was executed as in the previously described model. For this purpose, the fourth of the independent data sets for each stress condition was used as the test data, and the first three independent data sets were used as the training data.
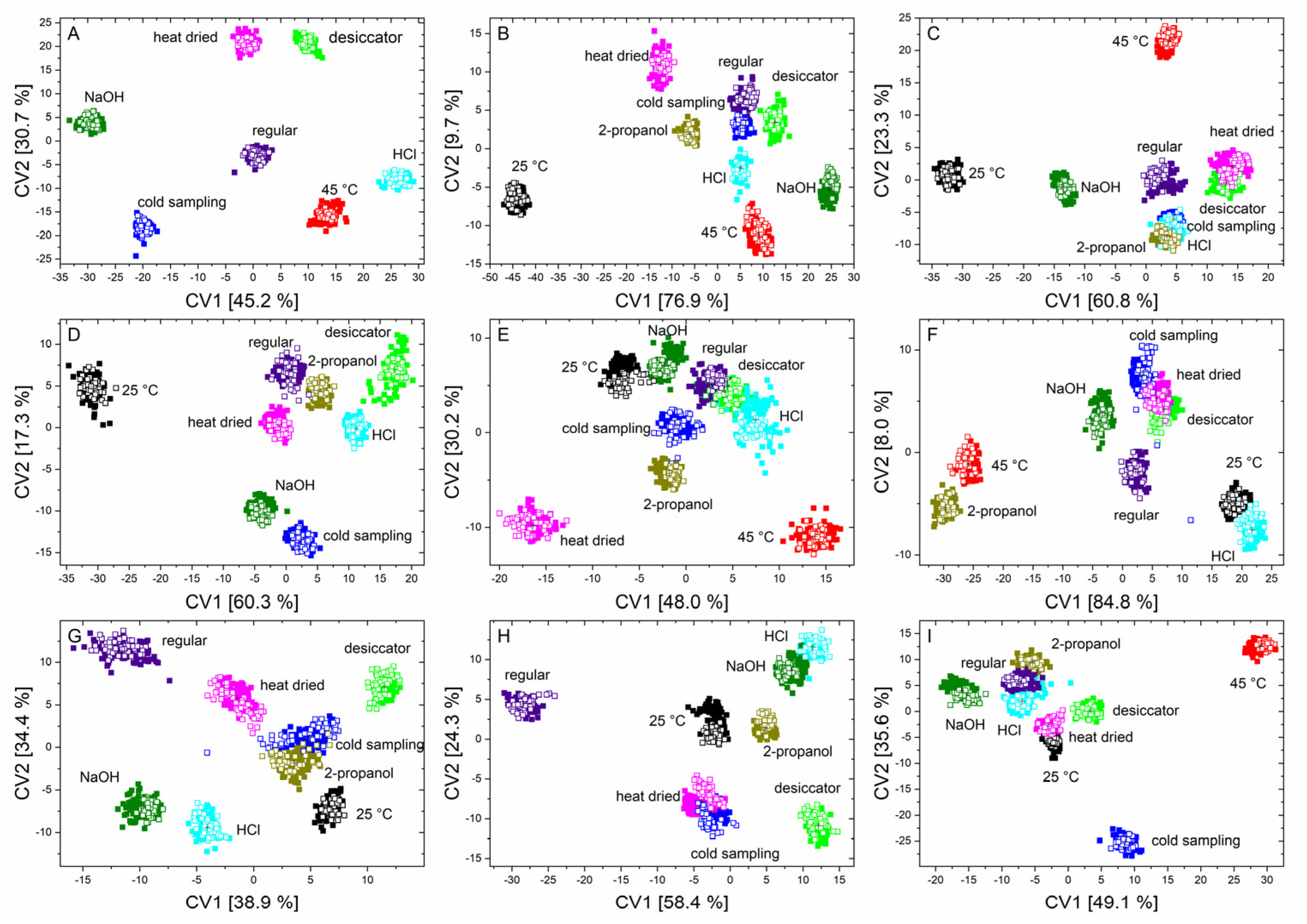
The summary of the quadratic discriminant analyses of each microorganism is depicted in Figure 4.
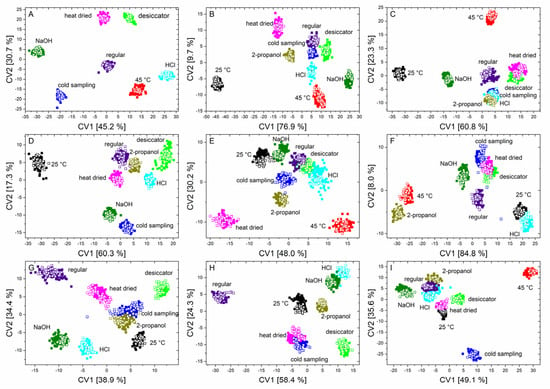
Figure 4.
Scatter plots of canonical variable one and two of the QDA of the training data (solid squares) and the independent test data (unfilled squares) of B. coag (A), B. sub (B), B. therm (C), B. tii (D), E. coli K12 (E), M. luteus (F), Ps. fluor 4358 (G), Ps. fluor 50090 (H) and E. coli TOP10 (I) to discriminate all nine conditions.
The first two canonical variables clearly separate almost all the stress conditions for each microorganism. Furthermore, it was also evident that the test data can again be found exactly in the data clouds of the training data.
The corresponding confusion matrices (Tables S2–S10 in the Supplementary Material) show that, for each microorganism, an error-free classification of the test data of each stress condition was possible, and all stress conditions were located in mostly isolated data clouds.
It can be observed that stressed microorganisms, in comparison to regularly treated microorganisms, show altered signals in the range of nucleic acids, polysaccharides, lipids, and in the region of –CH2/–CH3 stretching vibrations. Additionally, in most cases, a shift in the peak position was noticeable in the spectral region that was assigned to proteins (amide I and amide II vibrations) (Figure S4 (Supplementary Material)). Particularly notable were features such as those in Figure 4, where incubation at 25 °C for B. sub (B) causes this point cloud to be far removed from all other influencing conditions. This is because, in this case, the above-mentioned peaks have an increased intensity compared to regularly treated bacteria. It was also noticeable that, in the region of the amide I vibration, there is a shift to smaller wavenumbers. Observations of this nature were also found during stress reactions of the other microorganisms. For example, the data from B. therm (C), stressed with 2-propanol and HCl, cluster together as a result of a negative shift of the amide I band in both cases. Furthermore, a reduction in the bands in the fatty acid region was generally detected in B. tii, and desiccation of E. coli K12 only results in marginal changes in the spectrum (Figure S3 (Supplementary Material)). In addition, the heat drying of E. coli TOP10 results in a change in the ratio of nucleic acids, phospholipids, and polysaccharides. M. luteus shows significant changes in the ratio of lipids, nucleic acids, and proteins at incorrect incubation temperatures, and under NaOH and 2-propanol influence. Additionally, the two Pseudomonas species behave largely similarly under stress, but tend to undergo an opposite shift, Ps. fluor 4358 to higher and Ps. fluor 50090 to lower wavenumbers, in the amide I band (Figure S3 (Supplementary Material)).
For the range of 2800–3000 cm−1, our observations were consistent with the findings of Saulou et al. and Loffhagen et al., who presented that the spectra of viable microorganisms did not shift to lower wavenumbers, thus the microorganisms did not alter their membrane fluidity, but continued to show the presence of unsaturated bonds in lipids [19,51]. Furthermore, our results confirm the findings that stressed microorganisms show changes in the region of amide bands as part of their stress response mechanisms, indicating the alteration of the proteins secondary structure [19,23]. In addition, with the changes in the range of nucleic acids, polysaccharides, and lipids, resulting from the denaturation of nucleic acids, the production of exopolymer and effects on polysaccharides of the cell wall, in the range of 900–1300 cm−1, could be confirmed [21,23,26,27].
In summary, the spectral changes of sub-lethally stressed microorganisms, such as the change in the ranges 900–1500 cm−1 and 1500–1700 cm−1, which indicate an altered concentration of nucleic acids, lipids, polysaccharides, as well as the shift of the amide bands, indicated by a change in the secondary structure of the proteins, are reproducible and extensively described in the literature mentioned before, for living or stressed microorganisms. Thus, it can be stated that a rapid, robust and meaningful model for the discrimination of food-related microorganisms down to the strain level, irrespective of sample age, lifetime stress conditions, and sampling stress conditions, could be established.
4. Conclusions
The response of food-related bacteria to stress gives rise to changes in their spectral features in FT-IR. Specifically, a method using simple sample preparation, fast measurement by IR microspectroscopy, and chemometrics, was carefully developed for the rapid and non-destructive analysis of food-relevant bacteria, independent of their time after incubation, cultivation conditions, and sampling condition. Classification, using canonical discriminant analysis, showed that a robust and meaningful model was developed to discriminate nine different microorganisms at the genus, species, and strain levels, with 96.6% accuracy. Furthermore, it was demonstrated that sub-lethally stressed microorganisms, irrespective of the lifetime or sampling condition, showed changes in the spectral range associated with nucleic acids, polysaccharides, lipids, –CH2/–CH3 stretching vibrations, and especially in the range of proteins (amide I and amide II vibrations), compared to reference microorganisms that were grown under well-established guidelines. These spectral changes were discussed and could indicate, for example, changes in the secondary structure of proteins and the production of the exopolymer.
The results obtained not only confirm the potential of IR microspectroscopy for the rapid differentiation of microorganisms and elucidation of the stress response of bacteria, but also show that the existing highly standardized databases should be expanded to include stress conditions, and reconsidered in terms of sample preparation and spectra quality. Continuing this approach, these models should be progressively supplemented by, for example, food samples, in order to take into account the influence of food matrices to the models.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods10081850/s1, Table S1: Data splitting scheme for the trained and tested microorganisms. The time period after incubation is given in days for each data sets. Each data set consists of 50 spectra. Stress conditions are divided into lifetime conditions, in which the influence is already applied at inoculation and thus active over a life cycle, and sampling condition, in which the influence is a short major stress during sampling; Figure S1: Check for overfitting for the general discrimination of food-related microorganisms; Figure S2: Loadings (PC1–PC9) of the PCA of the training data set for the bacteria discrimination model. For a better spectral comparison in the graphs for PC1–PC3 the average spectrum of B. coag is given in gray; Figure S3: Loadings (PC10–PC20) of the PCA of the training data set for the bacteria discrimination model. For a better spectral comparison in the graphs for PC10–PC12 the average spectrum of B. coag is given in gray; Figure S4: Raw IR spectra of each stress condition vs. regular treated of B. coag (A), B. sub (B), B. therm (C), B. tii (D), E. coli K12 (E), M. luteus (F), Ps. fluor 4358 (G), Ps. fluor 50090 (H) and E. coli TOP10 (I); Table S2: Confusion matrix for the independent test data set for the classification of all stress condition for B. coag. The values of the table reflect the correct classifications of observations; Table S3: Confusion matrix for the independent test data set for the classification of all stress condition for B. sub. The values of the table reflect the correct classifications of observations; Table S4: Confusion matrix for the independent test data set for the classification of all stress condition for B. therm. The values of the table reflect the correct classifications of observations; Table S5: Confusion matrix for the independent test data set for the classification of all stress condition for B. tii. The values of the table reflect the correct classifications of observations; Table S6: Confusion matrix for the independent test data set for the classification of all stress condition for E. coli K12. The values of the table reflect the correct classifications of observations; Table S7: Confusion matrix for the independent test data set for the classification of all stress condition for M. luteus. The values of the table reflect the correct classifications of observations; Table S8: Confusion matrix for the independent test data set for the classification of all stress condition for Ps. fluor 4358. The values of the table reflect the correct classifications of observations; Table S9: Confusion matrix for the independent test data set for the classification of all stress condition for Ps. fluor 50090. The values of the table reflect the correct classifications of observations; Table S10: Confusion matrix for the independent test data set for the classification of all stress condition for E. coli TOP10. The values of the table reflect the correct classifications of observations.
Author Contributions
Conceptualization, D.K.; methodology, D.K.; software, D.K. and R.B.; validation, D.K. and R.B.; formal analysis, D.K. and J.R.; investigation, D.K. and J.R.; resources, P.K. and C.E.; data curation, D.K.; writing—original draft preparation, D.K.; writing—review and editing, D.K., R.B., J.R., C.E. and P.K.; visualization, D.K.; supervision, C.E. and P.K.; project administration, P.K.; funding acquisition, P.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Research Foundation (DFG) as part of the research training group GRK 1564 ‘Imaging New Modalities’. Additionally, this work was supported and financed by the Safety and Security Research Institute as well as the Graduate Institute of the Bonn-Rhein-Sieg University of Applied Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Candoğan, K.; Altuntas, E.G.; İğci, N. Authentication and Quality Assessment of Meat Products by Fourier-Transform Infrared (FTIR) Spectroscopy. Food Eng. Rev. 2020, 13, 66–91. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Mouwen, D.J.M.; López, M.; Prieto, M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J. Microbiol. Methods 2011, 84, 369–378. [Google Scholar] [CrossRef]
- Oliver, J.D. The viable but nonculturable state in bacteria. J. Microbiol. 2005, 43, 93–100. [Google Scholar]
- Price, P.B.; Sowers, T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. USA 2004, 101, 4631–4636. [Google Scholar] [CrossRef]
- Marles-Wright, J.; Lewis, R.J. Stress responses of bacteria. Curr. Opin. Struct. Biol. 2007, 17, 755–760. [Google Scholar] [CrossRef]
- Ajaykumar, V.J.; Mandal, P.K. Modern concept and detection of spoilage in meat and meat products. In Meat Quality Analysis; Elsevier: Amsterdam, The Netherlands, 2020; pp. 335–349. ISBN 9780128192337. [Google Scholar]
- Gurbanov, R.; Gozen, A.G.; Severcan, F. Rapid classification of heavy metal-exposed freshwater bacteria by infrared spectroscopy coupled with chemometrics using supervised method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 189, 282–290. [Google Scholar] [CrossRef]
- Teng, L.; Wang, X.; Wang, X.; Gou, H.; Ren, L.; Wang, T.; Wang, Y.; Ji, Y.; Huang, W.E.; Xu, J. Label-free, rapid and quantitative phenotyping of stress response in E. coli via ramanome. Sci. Rep. 2016, 6, 34359. [Google Scholar] [CrossRef]
- Lu, X.; Al-qadiri, H.M.; Lin, M.; Rasco, B.A. Application of Mid-infrared and Raman Spectroscopy to the Study of Bacteria. Food Bioprocess Technol. 2011, 4, 919–935. [Google Scholar] [CrossRef]
- Davis, R.; Mauer, L. Fourier transform infrared (FT-IR) spectroscopy: A rapid tool for detection and analysis of foodborne pathogenic bacteria. Curr. Res. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 2010, 2, 1582–1594. [Google Scholar]
- Bozoglu, F.; Alpas, H.; Kaletunc, G. Injury recovery of foodborne pathogens in high hydrostatic pressure treated milk during storage. FEMS Immunol. Med. Microbiol. 2004, 40, 243–247. [Google Scholar] [CrossRef]
- Klein, D.; Breuch, R.; Reinmüller, J.; Engelhard, C.; Kaul, P. Rapid detection and discrimination of food-related bacteria using IR-microspectroscopy in combination with multivariate statistical analysis. Talanta 2021, 232, 122424. [Google Scholar] [CrossRef]
- Helm, D.; Labischinski, H.; Naumann, D. Elaboration of a procedure for identification of bacteria using Fourier-Transform IR spectral libraries: A stepwise correlation approach. J. Microbiol. Methods 1991, 14, 127–142. [Google Scholar] [CrossRef]
- Breuch, R.; Klein, D.; Siefke, E.; Hebel, M.; Herbert, U.; Wickleder, C.; Kaul, P. Differentiation of meat-related microorganisms using paper-based surface-enhanced Raman spectroscopy combined with multivariate statistical analysis. Talanta 2020, 219, 121315. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A.; Schultz, T.; Gatti-Lafranconi, P.; Lotti, M.; Doglia, S.M.; de Marco, A. Effects of recombinant protein misfolding and aggregation on bacterial membranes. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 263–269. [Google Scholar] [CrossRef]
- Scherber, C.M.; Schottel, J.L.; Aksan, A. Membrane phase behavior of Escherichia coli during desiccation, rehydration, and growth recovery. Biochim. Biophys. Acta Biomembr. 2009, 1788, 2427–2435. [Google Scholar] [CrossRef]
- Beney, L.; Mille, Y.; Gervais, P. Death of Escherichia coli during rapid and severe dehydration is related to lipid phase transition. Appl. Microbiol. Biotechnol. 2004, 65, 457–464. [Google Scholar] [CrossRef]
- Lin, M.; Al-Holy, M.; Al-Qadiri, H.; Kang, D.-H.; Cavinato, A.G.; Huang, Y.; Rasco, B.A. Discrimination of Intact and Injured Listeria monocytogenes by Fourier Transform Infrared Spectroscopy and Principal Component Analysis. J. Agric. Food Chem. 2004, 52, 5769–5772. [Google Scholar] [CrossRef]
- Saulou, C.; Jamme, F.; Girbal, L.; Maranges, C.; Fourquaux, I.; Cocaign-Bousquet, M.; Dumas, P.; Mercier-Bonin, M. Synchrotron FTIR microspectroscopy of Escherichia coli at single-cell scale under silver-induced stress conditions. Anal. Bioanal. Chem. 2013, 405, 2685–2697. [Google Scholar] [CrossRef]
- Liu, Y.; He, L.; Mustapha, A.; Li, H.; Hu, Z.Q.; Lin, M. Antibacterial activities of zinc oxide nanoparticles against Escherichia coli O157:H7. J. Appl. Microbiol. 2009, 107, 1193–1201. [Google Scholar] [CrossRef]
- Kepenek, E.S.; Severcan, M.; Gozen, A.G.; Severcan, F. Discrimination of heavy metal acclimated environmental strains by chemometric analysis of FTIR spectra. Ecotoxicol. Environ. Saf. 2020, 202, 110953. [Google Scholar] [CrossRef] [PubMed]
- Kepenek, E.S.; Gozen, A.G.; Severcan, F. Molecular characterization of acutely and gradually heavy metal acclimated aquatic bacteria by FTIR spectroscopy. J. Biophotonics 2019, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Al-Qadiri, H.M.; Lin, M.; Al-Holy, M.A.; Cavinato, A.G.; Rasco, B.A. Detection of Sublethal Thermal Injury in Salmonella enterica Serotype Typhimurium and Listeria monocytogenes Using Fourier Transform Infrared (FT-IR) Spectroscopy (4000 to 600 cm−1). J. Food Sci. 2008, 73, M54–M61. [Google Scholar] [CrossRef]
- Kilimann, K.V.; Doster, W.; Vogel, R.F.; Hartmann, C.; Gänzle, M.G. Protection by sucrose against heat-induced lethal and sublethal injury of Lactococcus lactis: An FT-IR study. Biochim. Biophys. Acta Proteins Proteom. 2006, 1764, 1188–1197. [Google Scholar] [CrossRef]
- Moen, B.; Janbu, A.O.; Langsrud, S.; Langsrud, Ø.; Hobman, J.L.; Constantinidou, C.; Kohler, A.; Rudi, K. Global responses of Escherichia coli to adverse conditions determined by microarrays and FT-IR spectroscopy. Can. J. Microbiol. 2009, 55, 714–728. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, Q.; Wu, D.; Al-Qadiri, H.M.; Al-Alami, N.I.; Kang, D.-H.; Shin, J.-H.; Tang, J.; Jabal, J.M.F.; Aston, E.D.; et al. Using of infrared spectroscopy to study the survival and injury of Escherichia coli O157:H7, Campylobacter jejuni and Pseudomonas aeruginosa under cold stress in low nutrient media. Food Microbiol. 2011, 28, 537–546. [Google Scholar] [CrossRef]
- Hlaing, M.M.; Wood, B.R.; McNaughton, D.; Rood, J.I.; Fox, E.M.; Augustin, M.A. Vibrational spectroscopy combined with transcriptomic analysis for investigation of bacterial responses towards acid stress. Appl. Microbiol. Biotechnol. 2018, 102, 333–343. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Boutou, E.; Zoumpopoulou, G.; Tarantilis, P.A.; Polissiou, M.; Vorgias, C.E.; Tsakalidou, E. RNA Arbitrarily Primed PCR and Fourier Transform Infrared Spectroscopy Reveal Plasticity in the Acid Tolerance Response of Streptococcus macedonicus. Appl. Environ. Microbiol. 2008, 74, 6068–6076. [Google Scholar] [CrossRef] [PubMed]
- Stanborough, T.; Fegan, N.; Powell, S.M.; Tamplin, M.; Chandry, P.S. Insight into the Genome of Brochothrix thermosphacta, a Problematic Meat Spoilage Bacterium. Appl. Environ. Microbiol. 2017, 83, 1–20. [Google Scholar] [CrossRef]
- Mallidis, C.G.; Frantzeskakis, P.; Balatsouras, G.; Katsaboxakis, C. Thermal treatment of aseptically processed tomato paste. Int. J. Food Sci. Technol. 2007, 25, 442–448. [Google Scholar] [CrossRef]
- Lucas, R.; Grande, M.J.; Abriouel, H.; Maqueda, M.; Ben Omar, N.; Valdivia, E.; Martínez-Cañamero, M.; Gálvez, A. Application of the broad-spectrum bacteriocin enterocin AS-48 to inhibit Bacillus coagulans in canned fruit and vegetable foods. Food Chem. Toxicol. 2006, 44, 1774–1781. [Google Scholar] [CrossRef]
- Apetroaie-Constantin, C.; Mikkola, R.; Andersson, M.A.; Teplova, V.; Suominen, I.; Johansson, T.; Salkinoja-Salonen, M. Bacillus subtilis and B. mojavensis strains connected to food poisoning produce the heat stable toxin amylosin. J. Appl. Microbiol. 2009, 106, 1976–1985. [Google Scholar] [CrossRef]
- Vilar, I.; Garcia Fontan, M.C.; Prieto, B.; Tornadijo, M.E.; Carballo, J. A survey on the microbiological changes during the manufacture of dry-cured lacon, a Spanish traditional meat product. J. Appl. Microbiol. 2000, 89, 1018–1026. [Google Scholar] [CrossRef]
- Rosenquist, H.; Smidt, L.; Andersen, S.R.; Jensen, G.B.; Wilcks, A. Occurrence and significance of Bacillus cereus and Bacillus thuringiensis in ready-to-eat food. FEMS Microbiol. Lett. 2005, 250, 129–136. [Google Scholar] [CrossRef]
- Gospavic, R.; Kreyenschmidt, J.; Bruckner, S.; Popov, V.; Haque, N. Mathematical modelling for predicting the growth of Pseudomonas spp. in poultry under variable temperature conditions. Int. J. Food Microbiol. 2008, 127, 290–297. [Google Scholar] [CrossRef]
- Herbert, U. Assessment of Different Packaging Atmospheres for the Poultry Meat Industry Based on an Overall Quality Index. Ph.D. Thesis, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn, Germany, 2014. [Google Scholar]
- Leibniz Institut, D. Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Catalogue of Microorganisms. Available online: https://www.dsmz.de/catalogues/catalogue-microorganisms.html (accessed on 28 September 2020).
- Klein, D.; Breuch, R.; von der Mark, S.; Wickleder, C.; Kaul, P. Detection of spoilage associated bacteria using Raman-microspectroscopy combined with multivariate statistical analysis. Talanta 2019, 196, 325–328. [Google Scholar] [CrossRef]
- Sabbatini, S.; Conti, C.; Orilisi, G.; Giorgini, E. Infrared spectroscopy as a new tool for studying single living cells: Is there a niche? Biomed. Spectrosc. Imaging 2017, 6, 85–99. [Google Scholar] [CrossRef]
- Naumann, D. Infrared Spectroscopy in Microbiology. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2006; pp. 1–29. ISBN 9780470027318. [Google Scholar]
- Janbu, A.O.; Møretrø, T.; Bertrand, D.; Kohler, A. FT-IR microspectroscopy: A promising method for the rapid identification of Listeria species. FEMS Microbiol. Lett. 2008, 278, 164–170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, J.; Qiu, Z. The effect of imbalanced data sets on LDA: A theoretical and empirical analysis. Pattern Recognit. 2007, 40, 557–562. [Google Scholar] [CrossRef]
- Burgos-Paz, W.; Ramos-Onsins, S.E.; Pérez-Enciso, M.; Ferretti, L. Correcting For Unequal Sampling in Principal Component Analysis of Genetic Data. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Cananda, 19 August 2014; pp. 1–3. [Google Scholar]
- Moore, D.H. Combining linear and quadratic discriminants. Comput. Biomed. Res. 1973, 6, 422–429. [Google Scholar] [CrossRef]
- Fulcomer, M.C.; Schönemann, P.H.; Molnar, G. Classification by linear and quadratic discriminant scores. Behav. Res. Methods Instrum. 1974, 6, 443–445. [Google Scholar] [CrossRef]
- Wu, W.; Mallet, Y.; Walczak, B.; Penninckx, W.; Massart, D.L.; Heuerding, S.; Erni, F. Comparison of regularized discriminant analysis linear discriminant analysis and quadratic discriminant analysis applied to NIR data. Anal. Chim. Acta 1996, 329, 257–265. [Google Scholar] [CrossRef]
- Grewal, M.K.; Jaiswal, P.; Jha, S.N. Detection of poultry meat specific bacteria using FTIR spectroscopy and chemometrics. J. Food Sci. Technol. 2015, 52, 3859–3869. [Google Scholar] [CrossRef]
- Rebuffo-Scheer, C.A.; Dietrich, J.; Wenning, M.; Scherer, S. Identification of five Listeria species based on infrared spectra (FTIR) using macrosamples is superior to a microsample approach. Anal. Bioanal. Chem. 2008, 390, 1629–1635. [Google Scholar] [CrossRef]
- Campos, J.; Sousa, C.; Mourão, J.; Lopes, J.; Antunes, P.; Peixe, L. Discrimination of non-typhoid Salmonella serogroups and serotypes by Fourier Transform Infrared Spectroscopy: A comprehensive analysis. Int. J. Food Microbiol. 2018, 285, 34–41. [Google Scholar] [CrossRef]
- Wenning, M.; Büchl, N.R.; Scherer, S. Species and strain identification of lactic acid bacteria using FTIR spectroscopy and artificial neural networks. J. Biophotonics 2010, 3, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Loffhagen, N.; Härtig, C.; Geyer, W.; Voyevoda, M.; Harms, H. Competition betweencis, trans and Cyclopropane Fatty Acid Formation and its Impact on Membrane Fluidity. Eng. Life Sci. 2007, 7, 67–74. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).