Antimicrobial Resistance of Non-Typhoid Salmonella in Meat and Meat Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
2.3. Extraction and Data Registry
2.4. Data Analysis
3. Results
3.1. Number of Articles, Countries, and Standard
| Standards Used in the Articles | Antimicrobial Susceptibility Test | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | CLSI, M100-S9 [33] | CLSI, M100-S11 [34] | CLSI, M100-S13 [35] | CLSI, M100-S15 [36] | CLSI, M100-S16 [37] | CLSI, M100-S17 [38] | CLSI, M100-S18 [39] | CLSI, M100-S19 [40] | CLSI, M100-S20 [41] | CLSI, M100 S21 [42] | CLSI, M100-S22 [43] | CLSI, M100-S23 [44] | CLSI, M100 -S24 [45] | CLSI, M100-S25 [46] | CLSI, M100-S28 [47] | CASFM [48] | EUCAST [49] | NARMS [50] | NARMS (It Was Not Cited by Authors); CIPARS [51] | CLSI, M31-S1 [52] | CLSI, M31-A2 [53] | CLSI, M31-A3 [54] | MIC | Disc Diffusion |
| [55] | X | X | ||||||||||||||||||||||
| [56] | X | X | ||||||||||||||||||||||
| [57] | X | X | ||||||||||||||||||||||
| [58] | X | X | X | |||||||||||||||||||||
| [59] | X | X | ||||||||||||||||||||||
| [60] | X | X | ||||||||||||||||||||||
| [61] | X | X | X | |||||||||||||||||||||
| [62] | X | X | X | |||||||||||||||||||||
| [63] | X | X | ||||||||||||||||||||||
| [64] | X | X | ||||||||||||||||||||||
| [65] | X | X | ||||||||||||||||||||||
| [66] | X | X | ||||||||||||||||||||||
| [67] | X | X | ||||||||||||||||||||||
| [68] | X | X | ||||||||||||||||||||||
| [69] | X | X | ||||||||||||||||||||||
| [70] | X | X | ||||||||||||||||||||||
| [71] | X | X | ||||||||||||||||||||||
| [72] | X | X | ||||||||||||||||||||||
| [73] | X | X | ||||||||||||||||||||||
| [74] | X | X | ||||||||||||||||||||||
| [75] | X | X | ||||||||||||||||||||||
| [76] | X | X | ||||||||||||||||||||||
| [21] | X | X | ||||||||||||||||||||||
| [77] | X | X | ||||||||||||||||||||||
| [78] * | X | X | X | |||||||||||||||||||||
| [17] | X | X | ||||||||||||||||||||||
| [79] | X | X | ||||||||||||||||||||||
| [80] | X | X | X | |||||||||||||||||||||
| [18] | X | X | ||||||||||||||||||||||
| [81] | X | X | ||||||||||||||||||||||
| [82] | X | X | X | |||||||||||||||||||||
| [26] | X | X | X | |||||||||||||||||||||
| [83] | X | X | ||||||||||||||||||||||
| [84] | X | X | ||||||||||||||||||||||
| [85] | X | X | ||||||||||||||||||||||
| [86] | X | X | ||||||||||||||||||||||
| [87] | X | X | ||||||||||||||||||||||
| [19] | X | X | ||||||||||||||||||||||
| [88] | X | X | X | |||||||||||||||||||||
| [89] | X | X | ||||||||||||||||||||||
| [90] | X | X | ||||||||||||||||||||||
| [91] | X | X | ||||||||||||||||||||||
| [92] ** | X | X | ||||||||||||||||||||||
| [93] | X | X | ||||||||||||||||||||||
| [94] | X | X | ||||||||||||||||||||||
| [95] | X | X | ||||||||||||||||||||||
| [96] | X | X | ||||||||||||||||||||||
| [97] | X | X | ||||||||||||||||||||||
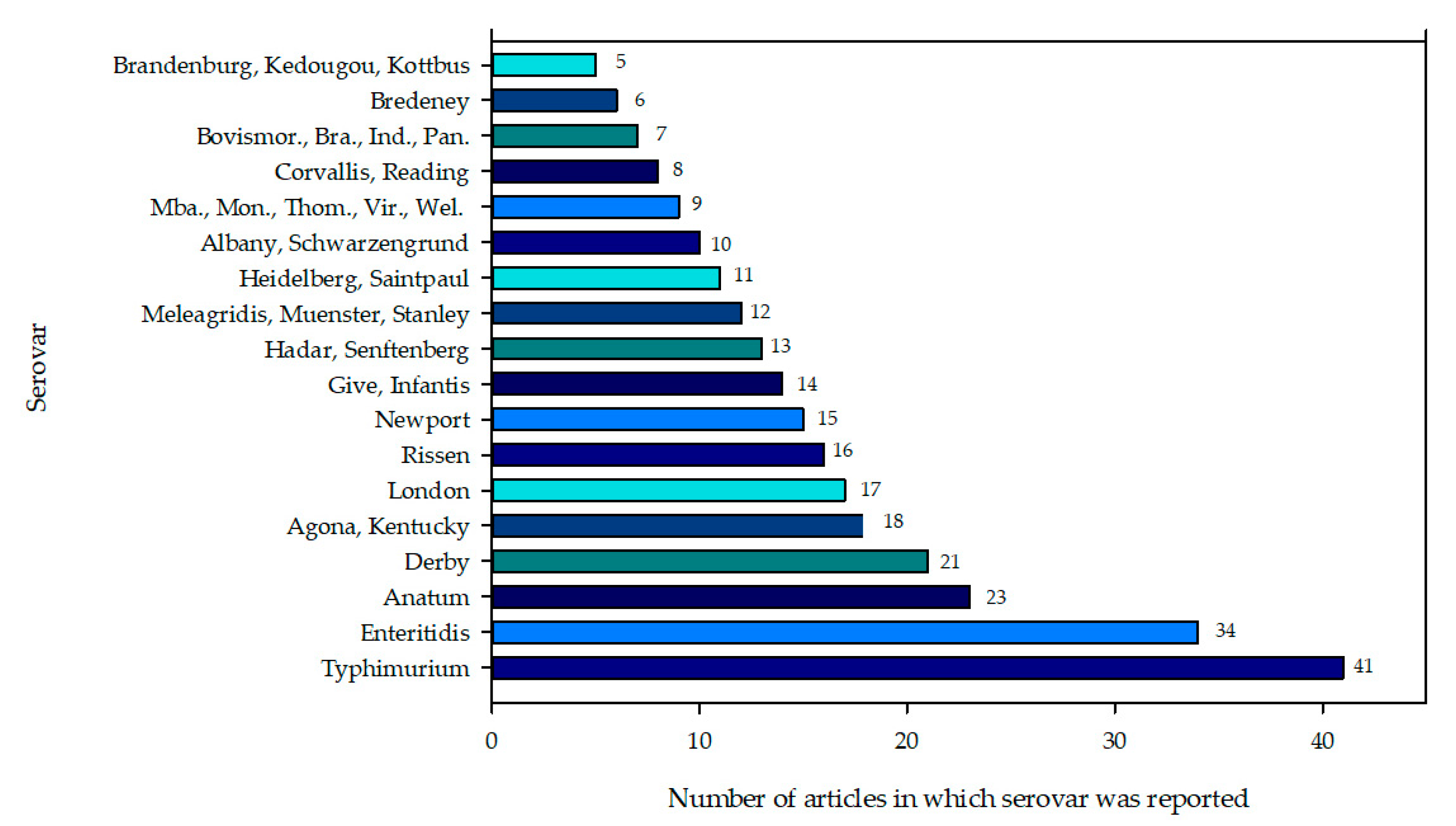
3.2. Reported Salmonella Enterica Serotypes
3.3. Analysis of Antibiotic Concentration Used According to Each Method
3.4. Breakpoints and Interpretative Criteria for Antimicrobial Susceptibility Testing
3.5. Resistance to Salmonella Serotypes Assayed by the MIC Method
| Antibiotic Agent | Prevalence Analysis | Serovar (References) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Typhimurium * [64,66,80] | Enteritidis * [66,80] | Agona [80] | Heidelberg [66] | Indiana [80] | Infantis [66] | Kentucky [64] | Mbandaka [64] | Thompson [80] | ||
| AMP | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 97 | 185 | 22 | 16 | 105 | 6 | 5 | 1 | 66 | |
| Reported frequency (%) | 47.8 | 100 | 92.1 | 31.6 | ND | ND | 76.7 | |||
| Calculated frequency (%) | 65.5 | 69.0 | 47.8 | 100 | 92.1 | 31.6 | 20.8 | 33.3 | 120 | |
| AMC | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 58 | 111 | 16 | ND | 91 | ND | 5 | 1 | 56 | |
| Reported frequency (%) | 34.8 | 0 | 79.8 | ND | ND | ND | 65.1 | |||
| Calculated frequency (%) | 39.2 | 41.4 | 34.8 | 0 | 79.8 | 20.8 | 33.3 | 101.8 | ||
| CRO | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 12 | 45 | 6 | 12 | 73 | 0 | ND | ND | 29 | |
| Reported frequency (%) | 15.4 | 75 | 64 | ND | ND | ND | 34.5 | |||
| Calculated frequency (%) | 8.1 | 16.8 | 13.0 | 75 | 64 | 52.7 | ||||
| FOX | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 22 | 71 | 8 | 4 | 42 | 0 | 5 | 1 | 28 | |
| Reported frequency (%) | 17.4 | 25 | 37 | ND | ND | 33 | 32.6 | |||
| Calculated frequency (%) | 14.9 | 26.5 | 17.4 | 25 | 37.0 | 20.8 | 33.0 | 50.9 | ||
| GEN | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 40 | 81 | 6 | 0 | 93 | 5 | ND | ND | 44 | |
| Reported frequency (%) | 13.0 | 0 | 81.6 | 26.3 | ND | ND | 51.2 | |||
| Calculated frequency (%) | 27.0 | 30.2 | 13.0 | 0 | 81.6 | 26.3 | 80.0 | |||
| STR | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 78 | 155 | 14 | 16 | 83 | 18 | 12 | 1 | 18 | |
| Reported frequency (%) | 30.4 | 100 | 73.5 | 94.7 | ND | 33 | 20.9 | |||
| Calculated frequency (%) | 52.7 | 57.8 | 30.4 | 100 | 72.8 | 94.7 | 50.0 | 33 | 32.7 | |
| CIP | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 22 | 32 | 10 | 0 | 93 | 0 | ND | ND | 8 | |
| Reported frequency (%) | 21.7 | 0 | 81.6 | ND | 0 | ND | 9.3 | |||
| Calculated frequency (%) | 14.9 | 11.9 | 21.7 | 0 | 81.6 | 14.5 | ||||
| NAL | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 81 | 275 | 23 | 2 | 106 | 3 | ND | ND | 39 | |
| Reported frequency (%) | 50.0 | 12.5 | 93 | 15.8 | ND | ND | 45.4 | |||
| Calculated frequency (%) | 54.7 | 102.6 | 50.0 | 12.5 | 93.0 | 15.8 | 70.9 | |||
| SXT | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 70 | 123 | 23 | 0 | 105 | 2 | ND | ND | 60 | |
| Reported frequency (%) | 50.0 | 0 | 92.9 | 10.5 | 0 | ND | 69.8 | |||
| Calculated frequency (%) | 47.3 | 45.9 | 50.0 | 0 | 92.1 | 10.5 | 109.1 | |||
| TET | N° isolates | 148 | 268 | 46 | 16 | 114 | 19 | 24 | 3 | 55 |
| N° resistant isolates | 123 | 191 | 30 | 0 | 104 | 0 | 15 | 0 | 70 | |
| Reported frequency (%) | 65.2 | 0 | 91.2 | ND | ND | ND | 81.4 | |||
| Calculated frequency (%) | 83.1 | 71.3 | 65.2 | 0 | 91.2 | 62.5 | 127.3 | |||
3.6. Salmonella Serotype Resistance Assayed by the Disc Diffusion Method
| Serovar | Number of Isolates | Calculated Frequency of Resistant Serovar (%) | Ref. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | GEN | AMK | KAN | STR | CIP | OFX | NAL | SXT | TMP | SUL | CHL | TET | |||
| Agona | 71 | 22.5 | 28.2 | 28.2 | 23.9 | 39.4 | [79] | ||||||||
| 1 | 100 | 100 | 100 | 100 | [87] | ||||||||||
| 56 | 16.1 | 1.8 | 16.1 | 1.8 | 91.1 | 19.6 | 17.9 | 41.1 | 89.3 | 48.2 | [92] | ||||
| Anatum | 5 | 100 | 20.0 | 20.0 | 60.0 | 40.0 | 40.0 | 60.0 | [17] | ||||||
| 7 | 100 | 57.1 | 85.7 | 57.1 | 100 | [87] | |||||||||
| 3 | 33.3 | 33.3 | 66.7 | 66.7 | 100 | [97] | |||||||||
| Corvallis | 1 | 100 | [56] | ||||||||||||
| 46 | 17.4 | 17.4 | 17.4 | 87.0 | 43.5 | 17.4 | 73.9 | 52.2 | 80.4 | [92] | |||||
| 3 | 100 | 100 | [97] | ||||||||||||
| Derby | 11 | 9.1 | 45.5 | 18.2 | 81.8 | 18.2 | 100 | [59] | |||||||
| 1 | 100 | 100 | [83] | ||||||||||||
| 1 | 100 | 100 | 100 | [87] | |||||||||||
| 75 | 56.0 | 36.0 | 1.3 | 36.0 | 33.3 | 42.7 | 40.0 | 42.7 | 92.0 | 44.0 | 81.3 | [92] | |||
| 79 | 41.8 | 24.1 | 2.5 | 25.3 | 45.6 | 25.3 | 40.5 | 43.0 | 35.4 | 77.2 | [97] | ||||
| Enteritidis | 6 | 33.3 | 33.3 | 66.7 | 33.3 | 16.7 | 66.7 | [56] | |||||||
| 6 | 16.7 | 66.7 | [61] | ||||||||||||
| 27 | 55.6 | 44.4 | 33.3 | 59.3 | 92.6 | 3.7 | 7.4 | 44.4 | [69] | ||||||
| 30 | 86.7 | 26.7 | 46.7 | 86.7 | 73.3 | 73.3 | 80.0 | [17] | |||||||
| 25 | 8.0 | 44.0 | 56.0 | 88.0 | 52.0 | 68.0 | 48.0 | [81] | |||||||
| 7 | 100 | 14.3 | 14.3 | 14.3 | 71.4 | 14.3 | 100 | 28.6 | 14.3 | 28.6 | [97] | ||||
| Hadar | 6 | 16.7 | 100 | 50.0 | 50.0 | 50.0 | 100 | [56] | |||||||
| 8 | 25.0 | 37.5 | 87.5 | 62.5 | 12.5 | 87.5 | [61] | ||||||||
| 31 | 25.8 | 3.2 | 22.6 | 22.6 | 9.7 | 74.2 | [79] | ||||||||
| 6 | 16.7 | 66.7 | 50.0 | 100 | 83.3 | 66.7 | 16.7 | 100 | [81] | ||||||
| Indiana | 28 | 71.4 | 46.4 | 14.3 | 39.3 | 39.3 | 57.1 | 57.1 | 78.6 | 85.7 | [79] | ||||
| 5 | 100 | 20.0 | 100 | [87] | |||||||||||
| 4 | 100 | 50.0 | 75.0 | 100 | 50.0 | 100 | 100 | 75.0 | 100 | 100 | [97] | ||||
| Kentucky | 27 | 44.4 | 18.5 | 3.7 | 11.1 | 33.3 | 29.6 | 25.9 | 22.2 | [56] | |||||
| 2 | 100 | 100 | 100 | [87] | |||||||||||
| 38 | 21.1 | 7.9 | 2.6 | 13.2 | 28.9 | 65.8 | 28.9 | 68.4 | 92.1 | 78.9 | 86.8 | [92] | |||
| London | 57 | 61.4 | 57.9 | 68.4 | 15.8 | 1.8 | 70.2 | 77.2 | 42.1 | 86.0 | [92] | ||||
| 20 | 50.0 | 25.0 | 5.0 | 45.0 | 50.0 | 35.0 | 50.0 | [97] | |||||||
| Mbandaka | 3 | 33.3 | 100 | [56] | |||||||||||
| 34 | 2.9 | 2.9 | 73.5 | 2.9 | 61.8 | 2.9 | 5.9 | 100 | 97.1 | [92] | |||||
| Muenster | 12 | 50.0 | 33.3 | 75.0 | 75.0 | 75.0 | 33.3 | [56] | |||||||
| 10 | 90.0 | 20.0 | 20.0 | 70.0 | 40.0 | 60.0 | 60.0 | [17] | |||||||
| Panama | 12 | 83.3 | 8.3 | 75.0 | 50.0 | 33.3 | 83.3 | 75.0 | 75.0 | [59] | |||||
| 8 | 100 | 12.5 | 100 | 100 | 100 | [87] | |||||||||
| Rissen | 6 | 100 | 33.3 | 50.0 | 33.3 | 100 | [87] | ||||||||
| 81 | 77.8 | 2.5 | 19.8 | 1.2 | 8.6 | 7.4 | 77.8 | 77.8 | 8.6 | 95.1 | [92] | ||||
| 18 | 55.6 | 5.6 | 5.6 | 22.2 | 5.6 | 5.6 | 77.8 | 5.6 | 77.8 | [97] | |||||
| Schwarzengrund | 2 | 100 | 100 | 100 | 100 | 100 | [56] | ||||||||
| 7 | 100 | 100 | 14.3 | 100 | 14.3 | [87] | |||||||||
| Stanley | 6 | 83.3 | 50.0 | 83.3 | [87] | ||||||||||
| 3 | 33.3 | 33.3 | 33.3 | 33.3 | [97] | ||||||||||
| Thompson | 83 | 2.4 | 54.2 | 96.4 | 77.1 | 81.9 | [61] | ||||||||
| 54 | 13.0 | 31.5 | 59.3 | 96.3 | 72.2 | 75.9 | 3.7 | 90.7 | [81] | ||||||
| 3 | 33.3 | 33.3 | 33.3 | 33.3 | 33.3 | 33.3 | 33.3 | [97] | |||||||
| Typhimurium | 8 | 62.5 | 12.5 | 12.5 | 50.0 | 12.5 | 37.5 | 37.5 | 87.5 | [59] | |||||
| 3 | 33.3 | 66.7 | 66.7 | 100 | [61] | ||||||||||
| 3 | 100 | [69] | |||||||||||||
| 40 | 100 | 27.5 | 45.0 | 95.0 | 67.5 | 82.5 | 92.5 | [17] | |||||||
| 14 | 14.3 | 35.7 | 57.1 | 85.7 | 64.3 | 78.6 | 85.7 | [81] | |||||||
| 5 | 40.0 | 100 | 80.0 | 40.0 | 80.0 | 100 | 60.0 | 100 | [83] | ||||||
| 84 | 83.3 | 15.5 | 1.2 | 59.5 | 13.1 | 29.8 | 29.8 | 27.4 | 89.3 | 40.5 | 82.1 | [92] | |||
| 22 | 72.7 | 9.1 | 4.5 | 36.4 | 36.4 | 4.5 | 81.8 | 50.0 | 50.0 | 63.6 | [97] | ||||
3.7. Serotype Resistance Determined by MIC and Disc Diffusion Methods
4. Discussion
4.1. Number of Articles, Countries, and Standard
4.2. Salmonella Enterica Reported Serotypes
4.3. Antibiotic Concentration Analysis According to Each Method Used
4.4. Salmonella Serotype Resistance Evaluated Using MIC
4.5. Salmonella Serotype Resistance Evaluated Using Disc Diffusion
4.6. Salmonella Serotype Resistance Determined by MIC and Disc Diffusion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Antimicrobial Agent | Abbreviations in the Reports | Abbreviations in This Work |
| Penicillins | ||
| Ampicillin | AM, AMP, A, Amp, Ap | AMP |
| β-Lactam/β-Lactamase inhibitor combinations | ||
| Amoxicillin/clavulanate | AUG, AMC, Amc, AC | AMC |
| Ampicillin-sulbactam | SAM, AS | SAM |
| Piperacillin-tazobactam | PPC-TAZ, TZP | TZP |
| Cephems | ||
| Cefazolin | CFZ, KZ, CZ, CF, CZD | CFZ |
| Cephalothin | CF, CEP, CEF, KF | CEP |
| Cefepime | CPM, FEP | FEP |
| Cefotaxime | CTX, TAX, CT | CTX |
| Ceftriaxone | AXO, CRO, Co, CTR | CRO |
| Cefoxitin | FOX | FOX |
| Cefuroxime | FUR, CXM | CXM |
| Ceftazidime | CAZ, CTZ, CF | CAZ |
| Cefoperazone | CFP | CFP |
| Cefaclor | CEC, CFC | CFC |
| Cefpodoxime | CPD | CPD |
| Monobactams | ||
| Aztreonam | ATM, AZT, AM | ATM |
| Ertapenem | ETP | ETP |
| Imipenem | IPM, IMP, IMI | IMI |
| Meropenem | MEM | MEM |
| Aminoglycosides | ||
| Gentamicin | GM, G, CN, GE, Gm, GN | GEN |
| Tobramycin | TOB, To | TOB |
| Amikacin | AMI, AM, AMK, AN, Ak | AMK |
| Kanamycin | KAN, K | KAN |
| Streptomycin | S, STR, SM, EST | STR |
| Fluoroquinolones | ||
| Ciprofloxacin | CIP, Cp, CI, CPF, CPX | CIP |
| Levofloxacin | Lvx | Lvx |
| Ofloxacin | OFX | OFX |
| Norfloxacin | NOR | NOR |
| Quinolones | ||
| Nalidixic acid | NA, NAL, Nx, N | NAL |
| Folate pathway inhibitors | ||
| Trimethoprim- sulfamethoxazole | SXT, COT, ST, TMP—SLF, TS | SXT |
| Sulfonamides | SSS, SMX, sul, SUL, SMX | SUL |
| Trimethoprim | TMP, TRIM, TP, W | TMP |
| Phenicols | ||
| Chloramphenicol | C, CHL, CM, CLF, CLO, CRO | CHL |
| Nitrofurans | ||
| Nitrofurantoin | FT, NIT | NIT |
| Tetracyclines | ||
| Tetracycline | TE, TET, T, TCY | TET |
References
- World Health Organization. Initiative to estimate the global burden of foodborne diseases. In Proceedings of the Fourth Formal Meeting of the Foodborne Disease Burden Epidemiology Reference Group (FERG): Sharing New Results, Making Future Plans and Preparing Ground for the Countries, Geneva, Switzerland, 8–12 November 2010; World Health Organization: Geneva, Switzerland, 2014; p. 108. [Google Scholar]
- Destro, M.T.; Ribeiro, V.B. Foodborne zoonoses. In Encyclopedia of Meat Sciences; Devine, C., Dikeman, M., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 17–21. [Google Scholar]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management and prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef]
- Löfström, C.; Hansen, T.; Maurischat, S.; Malorny, B. Salmonella: Salmonellosis. In Encyclopedia of Food and Health; Caballero, B., Finglas, P., Toldra, F., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 701–705. [Google Scholar]
- Ellermeier, C.; Slauch, J. The genus Salmonella. Prokaryotes 2006, 6, 123–158. [Google Scholar]
- Octavia, S.; Lan, R. The family Enterobacteriaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.-X. Supplement 2008–2010 (no. 48) to the Whitee-Kauffmann-Le minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Achtman, M.; Wain, J.; Weill, F.-X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PloS Pathog. 2012, 8, e1002776. [Google Scholar] [CrossRef] [PubMed]
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Vos, T.; Lozano, R.; Naghavi, M.; Flaxman, A.D.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2197–2223. [Google Scholar] [CrossRef]
- de Jong, A.; Bywater, R.; Butty, P.; Deroover, E.; Godinho, K.; Klein, U.; Marion, H.; Simjee, S.; Smets, K.; Thomas, V.; et al. A pan-European survey of antimicrobial susceptibility towards human-use antimicrobial drugs among zoonotic and commensal enteric bacteria isolated from healthy food-producing animals. J. Antimicrob. Chemother. 2009, 63, 733–744. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef]
- Higgs, J.D. The changing nature of red meat: 20 years of improving nutritional quality. Trends Food Sci. Tech. 2000, 11, 85–95. [Google Scholar] [CrossRef]
- Charlebois, S.; McCormick, M.; Juhasz, M. Meat consumption and higher prices: Discrete determinants affecting meat reduction or avoidance amidst retail price volatility. Brit. Food. J. 2016, 118, 2251–2270. [Google Scholar] [CrossRef]
- Panea, B.; Ripoll, G. Quality and safety of meat products. Foods 2018, 7, 118. [Google Scholar] [CrossRef]
- Domenech, E.; Jimenez-Belenguer, A.; Amoros, J.A.; Ferrus, M.A.; Escriche, I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in Eastern Spain. Food Cont. 2015, 47, 120–125. [Google Scholar] [CrossRef]
- Sallam, K.I.; Mohammed, M.A.; Hassan, M.A.; Tamura, T. Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Cont. 2014, 38, 209–214. [Google Scholar] [CrossRef]
- Abd-Elghany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 2015, 143, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.A.; Kanki, M.; Nguyen, P.D.; Le, H.T.; Ngo, P.T.; Tran, D.N.M.; Le, N.H.; Van Dang, C.; Kawai, T.; Kawahara, R.; et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016, 236, 115–122. [Google Scholar] [CrossRef]
- Arslan, S.; Eyi, A. Occurrence and antimicrobial resistance profiles of Salmonella species in retail meat products. J. Food Prot. 2010, 73, 1613–1617. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. Characterization of integrons and resistance genes inmultidrug-resistant Salmonella enterica isolated from meat and dairy products in Egypt. Int. J. Food Microbiol. 2014, 189, 39–44. [Google Scholar] [CrossRef]
- Maka, Ł.; Mackiw, E.; Sciezyn’ska, H.; Modzelewska, M.; Popowska, M. Resistance to sulfonamides and dissemination of sul genes among Salmonella spp. isolated from food in Poland. Foodborne Pathog. Dis. 2015, 12, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Maka, Ł.; Mackiw, E.; Sciezyn´ska, H.; Paw1owska, K.; Popowska, M. Antimicrobial susceptibility of Salmonella strains isolated from retail meat products in Poland between 2008 and 2012. Food Cont. 2014, 36, 199–204. [Google Scholar] [CrossRef]
- Doménech, E.; Jiménez-Belenguer, A.; Pérez, R.; Ferrús, M.A.; Escriche, I. Risk characterization of antimicrobial resistance of Salmonella in meat products. Food Cont. 2015, 57, 18–23. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Ajariyakhajorn, K.; Koowatananukul, C.; Wannaprasat, W.; Khemtong, S.; Samngamnim, S. Antimicrobial resistance and virulence genes in Salmonella enterica isolates from dairy cows. Foodborne Pathog. Dis. 2010, 7, 63–69. [Google Scholar] [CrossRef]
- Donado-Godoy, P.; Byrne, B.A.; Leon, M.; Castellanos, R.; Vanegas, C.; Coral, A.; Arevalo, A.; Clavuo, V.; Vargas, M.; Romero Zuniga, J.J.; et al. Prevalence, resistance patterns, and risk factors for antimicrobial resistance in bacteria from retail chicken meat in Colombia. J. Food Prot. 2015, 78, 751–759. [Google Scholar] [CrossRef]
- Mattiello, S.P.; Drescher, G.; Barth, V.C., Jr.; Ferreira, C.A.S.; Oliveira, S.D. Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Antonie Leeuwenhoek 2015, 108, 1227–1238. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar]
- Kiska, D.L. In vitro testing of antimicrobial agents. Semin. Pediatr. Infect. Dis. 1998, 9, 281–291. [Google Scholar] [CrossRef]
- Rincón-Gamboa, S.M.; Poutou-Piñales, R.A.; Carrascal-Camacho, A.K. Analysis of the assessment of antimicrobial susceptibility. Non-typhoid Salmonella in meat and meat products as model (systematic review). BMC Microbiol. 2021, in press. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Haddaway, N.R.; McGuinness, L.A. PRISMA2020: R Package and ShinyApp for Producing PRISMA 2020 Compliant Flow Diagrams, version 0.0.1; Zenodo: Geneva, Switzerland, 2020; Available online: https://zenodo.org/record/4287835#.YOLLCUxRVhE (accessed on 20 January 2021).
- National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing (NCCLS M100-S9); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 1999; p. 320. [Google Scholar]
- National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing (NCCLS M100-S11); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2000. [Google Scholar]
- National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Testing (NCCLS M100-S13); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2003. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (M100-S15); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2005; p. 172. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (CLSI M100-S16); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2006; p. 188. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (CLSI M100-S17); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2007; p. 182. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S18); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2008; p. 188. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (CLSI M100-S19); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2009. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S20); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2010; p. 160. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S21); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2011; p. 172. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S22); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2012; p. 188. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S23); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2013; p. 206. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S24); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2014; p. 230. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S25); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2015; p. 240. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial SusceptibilityTesting (CLSI M100-S28); National Committee for Clinical Laboratory Standards: Albany, NY, USA, 2018; p. 296. [Google Scholar]
- Comité de l’Antibiogramme de la Société Française de Microbiologie. Communiqué 1997. Pathologie Biologie (Paris); Société Française de Microbiologie: Paris, France, 1997. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 2.0; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2012. [Google Scholar]
- Department of Health and Human Services; Centers for Disease Control and Prevention; Food and Drug Administratio; United States Department of Agriculture. National Antimicrobial Resistance Monitoring System (NARMS). 2011 Executive Report; Department of Health and Human Services: Washington, DC, USA, 2011; p. 123.
- Public Health Agency of Canada. Canadian Integrated Program. for Antimicrobial Resistance Surveillance (CIPARS) 2003 Report; Public Health Agency of Canada: Guelph, ON, Canada, 2005.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for BacteriaIsolated from Animals (NCCLS M31-S1); Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2004; p. 40. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution susceptibility Tests for Bacteria Isolated from Animals (NCCLS M31-A2); Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2002; p. 80. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution SusceptibilityTests for Bacteria Isolated From Animals (CLSI M31-A3); Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2008; p. 116. [Google Scholar]
- Fakhr, M.K.; Sherwood, J.S.; Thorsness, J.; Logue, C.M. Molecular characterization and antibiotic resistance profiling of Salmonella isolated from retail Turkey meat products. Foodborne Pathog. Dis. 2006, 3, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Bada-Alambedji, R.; Fofana, A.; Seydi, M.; Akakpo, A.J. Antimicrobial resistance of Salmonella isolated from poultry carcasses in Dakar (Senegal). Braz. J. Microbiol. 2006, 37, 510–515. [Google Scholar] [CrossRef]
- Valdezate, S.; Arroyo, M.; González-Sanz, R.; Ramíro, R.; Herrera-León, Ç.; Usera, M.A.; De La Fuente, M.; Echeita, A. Antimicrobial resistance and phage and molecular typing of Salmonella strains isolated from food for human consumption in Spain. J. Food Prot. 2007, 70, 2741–2748. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Moutafis, G.; Istivan, T.; Tran, L.T.; Coloe, P.J. Detection of Salmonella spp. in retail raw food samples from Vietnam and Characterization of their antibiotic resistance. Appl. Environ. Microbiol. 2007, 73, 6885–6890. [Google Scholar] [CrossRef] [PubMed]
- Murmann, L.; dos Santos, M.C.; Cardoso, M. Prevalence, genetic characterization and antimicrobial resistance of Salmonella isolated from fresh pork sausages in Porto Alegre, Brazil. Food Cont. 2009, 20, 191–195. [Google Scholar] [CrossRef]
- Cook, A.; Reid-Smith, R.; Irwin, R.; Mcewen, S.A.; Valdivieso-Garcia, A.; Ribble, C. Antimicrobial resistance in Campylobacter, Salmonella, and Escherichia coli isolated from retail Turkey meat from Southern Ontario, Canada. J. Food Prot. 2009, 72, 473–481. [Google Scholar] [CrossRef]
- Soltan Dallal, M.M.; Doyle, M.P.; Rezadehbashi, M.; Dabiri, H.; Sanaei, M.; Modarresi, S.; Bakhtiari, R.; Sharifiy, K.; Taremi, M.; Zali, M.R.; et al. Prevalence and antimicrobial resistance profiles of Salmonella serotypes, Campylobacter and Yersinia spp. isolated from retail chicken and beef, Tehran, Iran. Food Cont. 2010, 21, 388–392. [Google Scholar] [CrossRef]
- Molina, N.; Millán, B.; Araque, M. Indicadores de calidad sanitaria y fenotipificación de Salmonella enterica aislada de pollo crudo comercializado en el área urbana de Mérida, Venezuela. Rev. Infect. 2010, 14, 174–185. [Google Scholar] [CrossRef]
- Sirichote, P.; Bangtrakulnonth, A.; Tianmanee, K.; Unahalekhaka, A.; Oulai, A.; Chittaphithakchai, P.; Kheowrod, W.; Hendriksen, R.S. Serotypes and antimicrobial resistance of Salmonella enterica ssp in central Thailand, 2001–2006. SE Asian J. Trop. Med. 2010, 41, 1405–1415. [Google Scholar]
- MIkanatha, N.M.; Sandt, C.H.; Localio, A.R.; Tewari, D.; Rankin, S.C.; Whichard, J.M.; Altekruse, S.F.; Lautenbach, E.; Folster, J.P.; Russo, A.; et al. Multidrug-resistant Salmonella isolates from retail chicken meat compared with human clinical isolates. Foodborne Pathog. Dis. 2010, 7, 929–934. [Google Scholar] [CrossRef]
- Hyeon, J.-Y.; Chon, J.-W.; Hwang, I.-G.; Kwak, H.-S.; Kim, M.-S.; Kim, S.-K.; Choi, I.-S.; Song, C.-S.; Park, C.; Seo, K.-H. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J. Food Prot. 2011, 74, 161–166. [Google Scholar] [CrossRef]
- Nunes Medeiros, M.A.; Nunes de Oliveira, D.C.; Rodrigues, D.d.P.; Coradi de Freitas, D.R. Prevalence and antimicrobial resistance of Salmonella in chicken carcasses at retail in 15 Brazilian cities. Rev. Panam. Salud Pública 2011, 30, 555–560. [Google Scholar] [CrossRef]
- Aslam, M.; Checkley, S.; Avery, B.; Chalmers, G.; Bohaychuk, V.; Gensler, G.; Reid-Smith, R.; Boerlin, P. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 2012, 32, 110–117. [Google Scholar] [CrossRef]
- Thai, T.H.; Lan, N.T.; Hirai, T.; Yamaguchi, R. Antimicrobial resistance in Salmonella serovars isolated from meat shops at the markets in North Vietnam. Foodborne Pathog. Dis. 2012, 9, 986–991. [Google Scholar] [CrossRef]
- Kim, M.-S.; Lim, T.-H.; Jang, J.-H.; Lee, D.-H.; Kim, B.-Y.; Kwon, J.-H.; Choi, S.-W.; Noh, J.-Y.; Hong, Y.-H.; Lee, S.-B.; et al. Prevalence and antimicrobial resistance of Salmonella species isolated from chicken meats produced by different integrated broiler operations in Korea. Poult. Sci. 2012, 91, 2370–2375. [Google Scholar] [CrossRef]
- Zdragas, A.; Mazaraki, K.; Vafeas, G.; Giantzi, V.; Papadopoulos, T.; Ekateriniadou, L. Prevalence, seasonal occurrence and antimicrobial resistance of Salmonella in poultry retail products in Greece. Lett. Appl. Microbiol. 2012, 55, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Fernández, E.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain: Comparison between 1993 and 2006. Int. J. Food Microbiol. 2012, 153, 281–287. [Google Scholar] [CrossRef]
- Cabrera-Diaz, E.; Barbosa-Cardenas, C.M.; Perez-Montaño, J.A.; Gonzalez-Aguilar, D.; Pacheco-Gallardo, C.; Barba, J. Occurrence, serotype diversity, and antimicrobial resistance of Salmonella in ground beef at retail stores in Jalisco State, Mexico. J. Food Prot. 2013, 76, 2004–2010. [Google Scholar] [CrossRef] [PubMed]
- Clemente, L.; Manageiro, V.; Ferreira, E.; Jones-Dias, D.; Correia, I.; Themudo, P.; Albuquerque, T.; Caniça, M. Occurrence of extended-spectrum β-lactamases among isolates of Salmonella enterica subsp. enterica from food-producing animals and food products, in Portugal. Int. J. Food Microbiol. 2013, 167, 221–228. [Google Scholar] [CrossRef]
- Li, Y.-C.; Pan, Z.-M.; Kang, X.-L.; Geng, S.-Z.; Liu, Z.-Y.; Cai, Y.-Q.; Jiao, X.-A. Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu Province, Eastern China. J. Food Prot. 2014, 77, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Jiang, X.; Zhou, Q.; Wu, J.; Wu, Z. Antimicrobial resistance, class 1 integrons, and horizontal transfer in Salmonella isolated from retail food in Henan, China. J. Infect. Dev. Ctries. 2014, 8, 705–711. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoon, R.-H.; Cha, S.-Y.; Wei, B.; Roh, J.-H.; Seo, H.-S.; Oh, J.-Y.; Jang, H.-K. Prevalence of Salmonella isolates and antimicrobial resistance in poultry meat from South Korea. J. Food Prot. 2014, 77, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Bacci, C.; Lanzoni, E.; Vismarra, A.; Alpigiani, I.; Nuvoloni, R.; Bonardi, S.; Brindani, F. Antibiotic resistance and resistance genes in Salmonella enterica isolated from pork meat and pig carcasses in Northern Italy. Large Anim. Rev. 2014, 20, 201–207. [Google Scholar]
- Donado-Godoy, P.; Clavijo, V.; León, M.; Arevalo, A.; Castellanos, R.; Bernal, J.; Tafur, M.A.; Ovalle, M.V.; Alali, W.Q.; Hume, M.; et al. Counts, serovars, and antimicrobial resistance phenotypes of Salmonella on raw chicken meat at retail in Colombia. J. Food Prot. 2014, 77, 227–235. [Google Scholar] [CrossRef]
- Ta, Y.T.; Nguyen, T.T.; To, P.B.; Pham, D.X.; Le, H.T.H.; Thi, G.N.; Alali, W.Q.; Walls, I.; Doyle, M.P. Quantification, serovars, and antibiotic resistance of Salmonella isolated from retail raw chicken meat in Vietnam. J. Food Prot. 2014, 77, 57–66. [Google Scholar] [CrossRef]
- Yang, B.; Cui, Y.; Shi, C.; Wang, J.; Xia, X.; Xi, M.; Wang, X.; Meng, J.; Alali, W.Q.; Walls, I.; et al. Counts, serotypes, and antimicrobial resistance of Salmonella isolates on retail raw poultry in the People’s Republic of China. J. Food Prot. 2014, 77, 894–902. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Mashak, Z.; Ghadimianazar, A. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J. Infect. Dev. Ctries. 2015, 9, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Chon, J.-W.; Kim, H.-S.; Kim, D.-H.; Lim, J.-S.; Yim, J.-H.; Seo, K.-H. Incidence, antimicrobial resistance, and molecular characteristics of nontyphoidal Salmonella including extended-spectrum p lactamase producers in retail chicken meat. J. Food Prot. 2015, 78, 1932–1937. [Google Scholar] [CrossRef]
- Gharieb, R.M.; Tartor, Y.H.; Khedr, M.H.E. Non-typhoidal Salmonella in poultry meat and diarrhoeic patients: Prevalence, antibiogram, virulotyping, molecular detection and sequencing of class I integrons in multidrug resistant strains. Gut Pathog. 2015, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Tirziu, E.; Lazar, R.; Sala, C.; Nichita, I.; Morar, A.; Ere, M.; Imre, K. Salmonella in raw chicken meat from the Romanian seaside: Frequency of isolation and antibiotic resistance. J. Food Prot. 2016, 78, 1003–1006. [Google Scholar] [CrossRef]
- Cai, Y.; Tao, J.; Jiao, Y.; Fei, X.; Zhou, L.; Wang, Y.; Zheng, H.; Pan, Z.; Jiao, X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 2016, 222, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Amin, H.S.; Abdelrahman, A.A.; Abdellrazeq, G.S. Occurrence of multidrug-resistant Salmonella enterica in retail chicken meat and development of a six genes-based multiplex PCR as alternative diagnostic method. J. Food Safe. 2016, 36, 459–466. [Google Scholar] [CrossRef]
- Niyomdecha, N.; Mungkornkaew, N.; Samosornsuk, W. Serotypes and antimicrobial resistance of Salmonella enterica isolated from pork, chicken meat and lettuce, Bangkok and central Thailand. Southeast. Asian J. Trop. Med. Public Health 2016, 47, 31–39. [Google Scholar]
- Thung, T.Y.; Mahyudin, N.A.; Basri, D.F.; Wan Mohamed Radzi, C.W.J.; Nakaguchi, Y.; Nishibuchi, M.; Radu, S. Prevalence and antibiotic resistance of Salmonella enteritidis and Salmonella typhimurium in raw chicken meat at retail markets in Malaysia. Poult. Sci. 2016, 95, 1888–1893. [Google Scholar] [CrossRef]
- Nghiem, M.N.; Nguyen, V.T.; Nguyen, T.T.H.; Nguyen, T.D.; Vo, T.T.B. Antimicrobial resistance gene expression associated with multidrug resistant Salmonella spp. isolated from retail meat in Hanoi, Vietnam. Int. Microbiol. 2017, 20, 85–93. [Google Scholar] [PubMed]
- Moawad, A.A.; Hotzel, H.; Awad, O.; Tomaso, H.; Neubauer, H.; Hafez, H.M.; El-Adawy, H. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Gad, A.H.; Abo-Shama, U.H.; Harclerode, K.K.; Fakhr, M.K. Prevalence, serotyping, molecular typing, and antimicrobial resistance of Salmonella isolated from conventional and organic retail ground poultry. Front. Microbiol. 2018, 9, 2653. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, Y.; Xiong, Z.; Ma, Y.; Wei, Y.; Qu, X.; Zhang, H.; Zhang, J.; Liao, M. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 2018, 9, 2104. [Google Scholar] [CrossRef]
- Nhung, N.T.; Van, N.T.B.; Cuong, N.V.; Duong, T.T.Q.; Nhat, T.T.; Hang, T.T.T.; Nhi, N.T.H.; Kiet, B.T.; Hien, V.B.; Ngoc, P.T.; et al. Antimicrobial residues and resistance against critically important antimicrobials in non-typhoidal Salmonella from meat sold at wet markets and supermarkets in Vietnam. Int. J. Food Microbiol. 2018, 266, 301–309. [Google Scholar] [CrossRef]
- Thung, T.Y.; Radu, S.; Mahyudin, N.A.; Rukayadi, Y.; Zakaria, Z.; Mazlan, N.; Tan, B.H.; Lee, E.; Yeoh, S.L.; Chin, Y.Z.; et al. Prevalence, virulence genes and antimicrobial resistance profiles of Salmonella serovars from retail beef in Selangor, Malaysia. Front. Microbiol. 2018, 8, 2697. [Google Scholar] [CrossRef] [PubMed]
- Zwe, Y.H.; Tang, V.C.Y.; Aung, K.T.; Alikiiteaga Gutierrez, R.; Ng, L.C.; Yuk, H.-G. Prevalence, sequence types, antibiotic resistance and, gyrA mutations of Salmonella isolated from retail fresh chicken meat in Singapore. Food Cont. 2018, 90, 233–240. [Google Scholar] [CrossRef]
- Zhu, A.; Zhi, W.; Qiu, Y.; Wei, L.; Tian, J.; Pan, Z.; Kang, X.; Gu, W.; Duan, L. Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou, China. Food Cont. 2019, 98, 474–480. [Google Scholar] [CrossRef]
- Yang, X.; Wu, Q.; Zhang, J.; Huang, J.; Chen, L.; Wu, S.; Zeng, H.; Wang, J.; Chen, M.; Wu, H.; et al. Prevalence, bacterial load, and antimicrobial resistance of Salmonella serovars isolated from retail meat and meat products in China. Front. Microbiol. 2019, 10, 2121. [Google Scholar] [CrossRef]
- Cheng, R.A.; Eade, C.R.; Wiedmann, M. Embracing diversity: Differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Front. Microbiol. 2019, 10, 1368. [Google Scholar] [CrossRef]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emeg. Infect. Dis. 2015, 21, 941. [Google Scholar] [CrossRef]
- Wallis, T.S.; Barrow, P.A. Salmonella epidemiology and pathogenesis in food-producing animals. EcoSas Plus 2005, 1. [Google Scholar] [CrossRef] [PubMed]
- Putturu, R.; Eevuri, T.; Ch, B.; Nelapati, K. Salmonella enteritidis—Food borne pathogen—A review. Int. J. Pharm. Biol. Sci. 2015, 5, 86–95. [Google Scholar]
- Quan, G.; Xia, P.; Zhao, J.; Zhu, C.; Meng, X.; Yang, Y.; Wang, Y.; Tian, Y.; Ding, X.; Zhu, G. Fimbriae and related receptors for Salmonella enteritidis. Microb. Pathog. 2019, 126, 357–362. [Google Scholar] [CrossRef]
- Chiou, C.-S.; Hong, Y.-P.; Liao, Y.-S.; Wang, Y.-W.; Tu, Y.-H.; Chen, B.-H.; Chen, Y.-S. New multidrug-resistant Salmonella enterica serovar Anatum clone, Taiwan, 2015–2017. Emerg. Infect. Dis. 2019, 25, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Valdezate, S.; Vidal, A.; Herrera-León, S.; Pozo, J.; Rubio, P.; Usera, M.A.; Carvajal, A.; Echeita, M.A. Salmonella derby clonal spread from pork. Emerg. Infect. Dis. 2005, 11, 694–698. [Google Scholar] [CrossRef]
- Fernandes Inagaki, J.M.; Jagnow Sereno, M.; Pegoraro, K.; Zanatta Waz, M.; Mendonça Soares, V.; Gonçalves Pereira, J.; Bersot, L.D.S. Effect of organic matter and pH on the resistance of Salmonella typhimurium and Salmonella derby in scalding water from pig slaughter. J. Food Safe. 2018, 38, e12504. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Surveillance Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Surveillance Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2014.
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/tularemia/index.html (accessed on 10 July 2013).
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2012.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2011.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2010.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2009.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2008.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2007.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2006.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2005.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2004.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. Human Isolates Final Report; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2003.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. National Antimicrobial Resistance Monitoring System (NARMS): Enteric Bacteria; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2002.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. National Antimicrobial Resistance Monitoring System (NARMS) 2001 Annual Report Summary; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2001.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. National Antimicrobial Resistance Monitoring System (NARMS): Enteric Bacteria; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2000.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. National Antimicrobial Resistance Monitoring System (NARMS) 1999 Annual Report Summary; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 1999.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. National Antimicrobial Resistance Monitoring System (NARMS) 1998 Annual Report Summary; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 1998.
- Centers for Disease Control and Prevention; National Center for Infectious Diseases; Division of Bacterial and Mycotic Diseases; Foodborne and Diarrheal Diseases Branch. National Antimicrobial Resistance Monitoring System (NARMS) 1997 Annual Report Summary; Enteric Diseases Epidemiology Branch, Division of Foodborne, Bacterial, and Mycotic Diseases, National Center for Zoonotic, Vector-Borne, and Enteric Diseases, Centers for Disease Control and Prevention: Atlanta, GA, USA, 1997.
- European Food Safety Authority (EFSA). Trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA J. 2010, 8, 1496. [Google Scholar]
- European Food Safety Authority (EFSA). The community summary report on trends and sources of zoonoses and zoonotic agents in the European Union in 2007. EFSA J. 2009, 7, 233r. [Google Scholar]
- European Food Safety Authority (EFSA). The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and fooborne outbreaks in the European Union in 2005. EFSA J. 2006, 94, 288. [Google Scholar]
- European Food Safety Authority (EFSA). Trends and sources of zoonoses, zoonotic agents and antimicrobial resistance in the European Union in 2004. EFSA J. 2006, 310, 280. [Google Scholar]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 2014, 12, 3547. [Google Scholar]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J. 2013, 11, 3129. [Google Scholar]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J. 2012, 10, 2597. [Google Scholar]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. EFSA J. 2011, 9, 2090. [Google Scholar]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 2015, 13, 3991. [Google Scholar] [CrossRef]
- de Jong, A.; Moyaert, H.; Simjee, S. Antimicrobial susceptibility testing of foodborne bacteria related to national and international resistance-monitoring programs. In Antimicrobial Food Packaging; Barros-Velázquez, J., Ed.; Academic Press: Brussels, Belgium, 2016; pp. 117–128. [Google Scholar]
- Nassar, M.S.M.; Hazzah, W.A.; Bakr, W.M.K. Evaluation of antibiotic susceptibility test results: How guilty a laboratory could be? Egypt. Public Health Assoc. 2019, 94, 4. [Google Scholar] [CrossRef]
- World Health Organization Advisory Group on Integrated Surveillance of Antimicrobial Resistance; Food and Agriculture Organization of the United Nations; World Organisation for Animal Health. Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a one Health Approach; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Wasyl, D.; Kern-Zdanowicz, I.; Doman´ska-Blicharz, K.; Zajac, M.; Hoszowski, A. High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying blaCTX-M-25 gene. Vet. Microbiol. 2015, 175, 85–91. [Google Scholar] [CrossRef]
- Louden, B.C.; Haarmann, D.; Han, J.; Foley, S.L.; Lynne, A.M. Characterization of antimicrobial resistance in Salmonella enterica serovar typhimurium isolates from food animals in the U.S. Food Res. Int. 2012, 45, 968–972. [Google Scholar] [CrossRef]
- Dessie, H.K.; Bae, D.H.; Lee, Y.J. Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea. Poult. Sci. 2013, 92, 3036–3043. [Google Scholar] [CrossRef] [PubMed]
- Samanta, I.; Bandyopadhyay, S. Characteristics of antimicrobial resistance among microorganisms of concern to animal, fish and human health: Salmonella. In Antimicrobial Resistance in Agriculture: Perspective, Policy and Mitigation; Academic Press: London, UK, 2020; pp. 136–150. [Google Scholar]
- Antunes, P.; Mourao, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Grossman, T.H. Tetracycline antibiotics and resistance. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Doran, G.; NiChulain, M.; DeLappe, N.; O’Hare, C.; Corbett-Feeney, G.; Cormican, M. Interpreting streptomycin susceptibility test results for Salmonella enterica serovar typhimurium. Int. J. Antimicrob. Agents 2006, 27, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Council Regulation (EEC). Laying down a community procedure for the establishment of maximum residue limits of veterinary medicinal products in foodstuffs of animal origin (No. 2377/90). Off. J. Eur. Comunities 1990, 224, 1–8. [Google Scholar]
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.-M.; Cole, L.; Daignault, D.; Desruisseau, A.; et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010, 16, 48–55. [Google Scholar] [CrossRef]
- Otto, S.J.G.; Carson, C.A.; Finley, R.L.; Thomas, M.K.; Reid-Smith, R.J.; McEwen, S.A. Estimating the number of human cases of ceftiofur-resistant Salmonella enterica serovar Heidelberg in Québec and Ontario, Canada. Clin. Infect. Dis. 2014, 59, 1–90. [Google Scholar] [CrossRef][Green Version]
- Shigemura, H.; Matsui, M.; Sekizuka, T.; Onozuka, D.; Noda, T.; Yamashita, A.; Kuroda, M.; Suzuki, S.; Kimura, H.; Fujimoto, S.; et al. Decrease in the prevalence of extended-spectrum cephalosporin-resistant Salmonella following cessation of ceftiofur use by the Japanese poultry industry. Int. J. Food Microbiol. 2018, 274, 45–51. [Google Scholar] [CrossRef]
- Ohta, N.; Norby, B.; Loneragan, G.H.; Vinasco, J.; den Bakker, H.C.; Lawhon, S.D.; Norman, K.N.H.M. Scott Quantitative dynamics of Salmonella and E. coli in feces of feedlot cattle treated with ceftiofur and chlortetracycline. PLoS ONE 2019, 14, e0225697. [Google Scholar] [CrossRef]
- Collineau, L.; Chapman, B.; Bao, X.; Sivapathasundaram, B.; Carson, C.A.; Fazil, A.; Reid-Smith, R.J.; Smith, B.A. A farm-to-fork quantitative risk assessment model for Salmonella Heidelberg resistant to third-generation cephalosporins in broiler chickens in Canada. Int. J. Food Microbiol. 2020, 330, 108559. [Google Scholar] [CrossRef]
- Choi, J.; Kim, W.K. Dietary application of tannins as a potential mitigation strategy for current challenges in poultry production: A review. Animals 2020, 10, 2389. [Google Scholar] [CrossRef] [PubMed]
- Seventy-third World Health Assembly. (WHA 73.5) Strengthening Efforts on Food Safety; World Health Organization: Geneva, Switzerland, 2020; p. 5. [Google Scholar]
| Ref. | Pattern | Serovar/ (Total Serovar) | Total Isolates with the Pattern | % Isolates with the Pattern |
|---|---|---|---|---|
| [67] | AMP SXTTET | Rissen (1) | 1 | 100 |
| Worthington (1) | 1 | 100 | ||
| [91] | AMC CEP GEN KAN STR | Ouakam (1) | 1 | 100 |
| Tennessee (7) | 2 | 29 | ||
| [60] | AMP AMC CEP FOX | Agona (3) | 3 | 100 |
| Heidelberg (13) | 2 | 15 | ||
| Litchfield (2) | 2 | 100 | ||
| [91] | AMP AMC CEP GEN KAN STR | Anatum (1) | 1 | 100 |
| Tennessee (7) | 4 | 57 | ||
| [91] | AMP AMC CEP GEN KAN STR NAL TET | Enteritidis (3) | 1 | 33 |
| Saintpaul (3) | 1 | 33 | ||
| [91] | AMP AMC CEP GEN KAN STR TET | Montevideo (1) | 1 | 100 |
| Anatum_var-15+ (1) | 1 | 100 | ||
| Saintpaul (3) | 2 | 67 | ||
| Seftenberg (4) | 2 | 50 | ||
| [67] | AMP AMC CRO FOX | Heidelberg (13) | 11 | 85 |
| I:ROUGH-O:r:1,2 (1) | 1 | 100 | ||
| Infantis (4) | 1 | 25 | ||
| Kentucky (2) | 2 | 100 | ||
| [21] | AMP AMC CTX CRO FOX CPD ATM GEN KAN STR CIP NAL SXT CHL TET | Typhimurium (4) | 1 | 25 |
| Enteritidis (3) | 1 | 33 | ||
| [91] | AMP AMC GEN KAN STR | Seftenberg (4) | 1 | 25 |
| Tennessee (7) | 1 | 14 | ||
| [60,91] | AMP CEP GEN | Enteritidis (3) | 1 | 33 |
| Seftenberg (4) | 1 | 25 | ||
| [21] | AMP KAN NAL SXT CHL TET | Typhimurium (4) | 2 | 50 |
| Typhimurium (4) | 1 | 25 | ||
| [86] | GEN TOB AMK CIP SXT | Enteritidis | ND | 0 |
| Kentucky | ND | 0 | ||
| [84] | STR CIP NAL SUL TET | Infantis (4) | 3 | 75 |
| Ref. | Pattern | Serovar/ (Total Serovar) | Total Isolates with the Pattern | % of Isolates with the Pattern |
|---|---|---|---|---|
| [71] | AMC CEP NAL | Typhimurium (34) | 1 | 3 |
| [69] | Montevideo (3) | 1 | 33 | |
| [70] | AMP CEP NAL TET | Hadar (1) | 1 | 100 |
| [56] | Tshiongwe (1) | 1 | 100 | |
| [70] | AMP GEN KAN STR CIP SXT TET | Enteritidis (25) | 2 | 8 |
| [21] | Infantis (4) | 1 | 25 | |
| [74] | AMP GEN NAL SXT TET | Anatum (20) | 1 | 5 |
| [70] | Typhimurium (34) | 1 | 3 | |
| [74] | AMP GEN STR SXT CHL TET | Derby (70) | 1 | 1 |
| [85] | London (3) | 1 | 33 | |
| [72] | Panama (9) | 1 | 11 | |
| [89] | Typhimurium (34) | 1 | 3 | |
| [74] | AMP KAN STR NAL SXT CHL TET | Meleagridis (9) | 1 | 11 |
| [21] | Typhimurium (34) | 1 | 3 | |
| [56] | AMP NAL CHL | Kentucky (7) | 2 | 29 |
| [87] | Schwarzengrund (7) | 6 | 86 | |
| [87] | AMP NAL CHL TET | Albany (5) | 1 | 20 |
| Anatum (20) | 1 | 5 | ||
| [18] | Derby (70) | 1 | 1 | |
| [76] | AMP NAL SXT | Enteritidis (25) | 1 | 4 |
| [18] | Kentucky (7) | 4 | 57 | |
| Muenster (6) | 6 | 100 | ||
| Virchow (2) | 2 | 100 | ||
| [87] | AMP NAL SXT CHL | Schwarzengrund (7) | 1 | 14 |
| [74] | Anatum (20) | 1 | 5 | |
| Infantis (4) | 1 | 25 | ||
| [74] | AMP NAL SXT CHL TET | Derby (70) | 1 | 1 |
| Infantis (4) | 1 | 25 | ||
| [87] | Albany (5) | 4 | 80 | |
| Anatum (20) | 2 | 10 | ||
| Panama (9) | 1 | 11 | ||
| Rissen (4) | 1 | 17 | ||
| [95] | Brancaster (3) | 1 | 33 | |
| Stanley (8) | 1 | 13 | ||
| [87] | AMP NAL SXT TET | Agona (3) | 1 | 33 |
| Anatum (20) | 1 | 5 | ||
| [74] | Derby (70) | 1 | 1 | |
| [70] | Typhimurium (34) | 1 | 3 | |
| [74] | AMP NAL TET | Derby (70) | 4 | 6 |
| [87] | Rissen (4) | 1 | 17 | |
| S.4.5.12:I: −(2) | 2 | 100 | ||
| Stanley (8) | 3 | 38 | ||
| [81] | Enteritidis (25) | 1 | 4 | |
| Typhimurium (34) | 2 | 6 | ||
| [58] | Anatum | ND | ND | |
| [70] | Typhimurium (34) | 2 | 6 | |
| [72] | AMP STR NAL SXT TET | Anatum (20) | 1 | 5 |
| Derby (70) | 2 | 3 | ||
| [85] | Derby (70) | 2 | 3 | |
| [85] | AMP STR SXT CHL TET | Meleagridis (9) | 2 | 22 |
| [74] | Typhimurium (34) | 1 | 3 | |
| Derby (70) | 1 | 1 | ||
| [89] | Rissen (4) | 1 | 17 | |
| Give (1) | 1 | 100 | ||
| Typhimurium (34) | 1 | 3 | ||
| [72] | AMP STR SXT TET | Agona (3) | 1 | 33 |
| Brandenburg (1) | 1 | 100 | ||
| Saintpaul (2) | 1 | 50 | ||
| [74] | Derby (70) | 1 | 1 | |
| [72] | AMP STR TET | Derby (70) | 7 | 10 |
| Typhimurium (34) | 4 | 12 | ||
| [85] | Derby (70) | 7 | 10 | |
| Typhimurium (34) | 4 | 12 | ||
| [74] | Derby (70) | 7 | 10 | |
| Meleagridis (9) | 1 | 11 | ||
| Newport (5) | 1 | 20 | ||
| [89] | Derby (70) | 7 | 10 | |
| Typhimurium (34) | 4 | 12 | ||
| [70] | Bredeney (3) | 1 | 33 | |
| [74] | AMP SXT CHL TET | Typhimurium (34) | 3 | 9 |
| [87] | Anatum (20) | 1 | 5 | |
| Panama (9) | 7 | 78 | ||
| [81] | Bovismorbificans (1) | 1 | 100 | |
| [95] | Brancaster (3) | 1 | 33 | |
| Stanley (8) | 4 | 50 | ||
| Typhimurium (34) | 3 | 9 | ||
| [77] | AMP SXT TET | Agona (3) | 1 | 33 |
| Anatum (20) | 5 | 25 | ||
| Bredeney (3) | 2 | 67 | ||
| Coeln (1) | 1 | 100 | ||
| Derby (70) | 8 | 11 | ||
| London (3) | 1 | 33 | ||
| Senftenberg (1) | 1 | 100 | ||
| Typhimurium (34) | 1 | 3 | ||
| [85] | Derby (70) | 8 | 11 | |
| [74] | Derby (70) | 8 | 11 | |
| [87] | Anatum (20) | 5 | 25 | |
| Rissen (4) | 1 | 17 | ||
| [71] | CEP STR NAL | Enteritidis (25) | 4 | 16 |
| Newport (5) | 1 | 20 | ||
| [69] | Enteritidis (25) | 4 | 16 | |
| Montevideo (3) | 2 | 67 | ||
| [70] | KAN STR NAL TET | Blockey (4) | 4 | 100 |
| [81] | Enteritidis (25) | 3 | 12 | |
| Newport (5) | 2 | 40 | ||
| [85] | NAL CHL TET | Derby (70) | 1 | 1 |
| Typhimurium (34) | 4 | 12 | ||
| [87] | Indiana (1) | 1 | 100 | |
| [74] | NAL SXT TET | Infantis (4) | 1 | 25 |
| Meleagridis (9) | 5 | 56 | ||
| [81] | Thompson (1) | 1 | 100 | |
| [71] | STR NAL TET | Enteritidis (25) | 5 | 20 |
| [72] | Anatum (20) | 1 | 5 | |
| Derby (70) | 1 | 1 | ||
| Reading (1) | 1 | 100 | ||
| [81] | Enteritidis (25) | 5 | 20 | |
| Newport (5) | 1 | 20 | ||
| [58] | Hadar | ND | ND | |
| [56] | STR SUL TET | Brancaster (3) | 1 | 33 |
| [58] | Anatum | ND | ND | |
| London | ND | ND | ||
| [72] | STR SXT CHL TET | Anatum (20) | 1 | 5 |
| Derby (70) | 1 | 1 | ||
| Saintpaul (2) | 1 | 50 | ||
| [74] | London (3) | 1 | 33 | |
| [75] | STR SXT SUL TET | Derby (70) | 1 | 1 |
| [56] | Kentucky (7) | 1 | 14 |
| Reference | Multiresistance Patterns | Serovar (Total Isolates) | Total Isolateswith the Multiresistance Pattern | % Frequency of Multiresistance Pattern |
|---|---|---|---|---|
| [62] | AMP CTX SXT | Enteritidis (3) | 1 | 33.3 |
| [26] | CIP NAL TET | Heidelberg (11) | 2 | 25.0 |
| STR SXT TET | Typhimurium (1) | 1 | 100 | |
| STR CIP NAL TET | Heidelberg (11) | 1 | 9.1 | |
| [82] | AMP AMC CFZ CEP CTX TET | Bareilly (1) | 1 | 100 |
| [62] | AMP CFZ CTX ATM CIP SXT | Heidelberg (11) | 2 | 18.2 |
| AMP TZP CFZ CTX ATM SXT | Heidelberg (11) | 1 | 9.1 | |
| [82] | AMP CFZ CEP CTX STR NAL TET | Infantis (8) | 8 | 100 |
| Virchow (16) | 8 | 50 | ||
| [82] | AMP CFZ CEP CTX GEN STR NAL TET | Enteritidis (3) | 1 | 33.3 |
| Richmond (3) | 3 | 100 | ||
| [26] | AMP AMC CFZ CTX CRO FOX NAL TET | Heidelberg (11) | 3 | 27.3 |
| [82] | AMP AMC CFZ CEP CTX STR NAL TET | Virchow (16) | 7 | 43.8 |
| [82] | AMP AMC CFZ CEP CTX GEN STR NAL TET | Enteritidis (3) | 1 | 33.3 |
| Virchow (16) | 1 | 6.3 | ||
| [26] | AMP AMC CFZ CTX CRO FOX CIP NAL TET | Heidelberg (11) | 2 | 18.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rincón-Gamboa, S.M.; Poutou-Piñales, R.A.; Carrascal-Camacho, A.K. Antimicrobial Resistance of Non-Typhoid Salmonella in Meat and Meat Products. Foods 2021, 10, 1731. https://doi.org/10.3390/foods10081731
Rincón-Gamboa SM, Poutou-Piñales RA, Carrascal-Camacho AK. Antimicrobial Resistance of Non-Typhoid Salmonella in Meat and Meat Products. Foods. 2021; 10(8):1731. https://doi.org/10.3390/foods10081731
Chicago/Turabian StyleRincón-Gamboa, Sandra M., Raúl A. Poutou-Piñales, and Ana K. Carrascal-Camacho. 2021. "Antimicrobial Resistance of Non-Typhoid Salmonella in Meat and Meat Products" Foods 10, no. 8: 1731. https://doi.org/10.3390/foods10081731
APA StyleRincón-Gamboa, S. M., Poutou-Piñales, R. A., & Carrascal-Camacho, A. K. (2021). Antimicrobial Resistance of Non-Typhoid Salmonella in Meat and Meat Products. Foods, 10(8), 1731. https://doi.org/10.3390/foods10081731








