The Natural Antimicrobial trans-Cinnamaldehyde Interferes with UDP-N-Acetylglucosamine Biosynthesis and Cell Wall Homeostasis in Listeria monocytogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
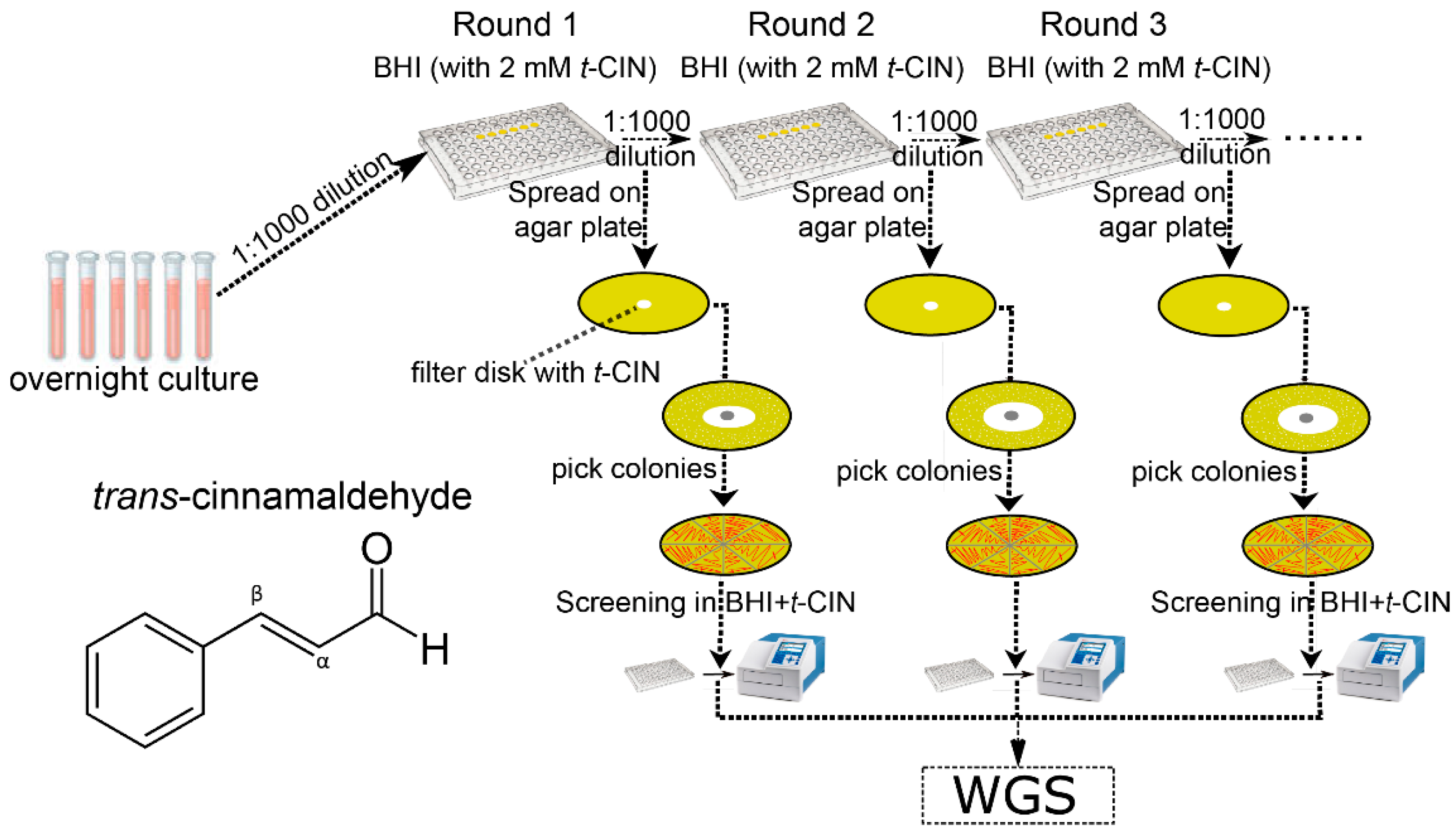
2.2. Evolutionary Study to Isolate yvcK::Himar1 Suppression Mutants with Regained t-CIN Tolerance
2.3. Whole Genome Sequencing
2.4. Growth Assay
2.5. Microscopy and Cell Dimension Measurement
2.6. Genetic Complementation of Mutant Strains
2.7. Construction of nagR Deletion Mutant
2.8. Statistical Analysis
3. Results
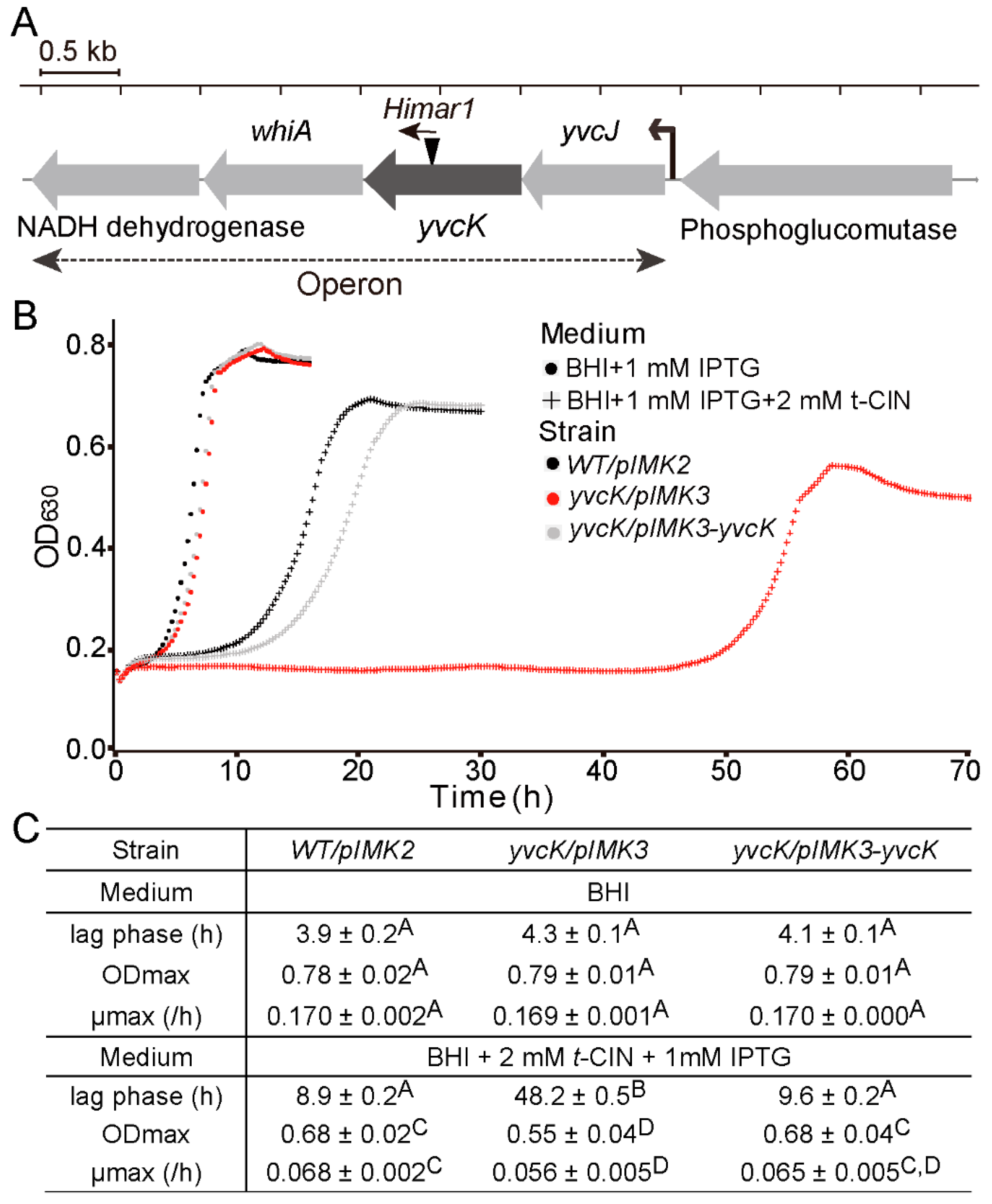
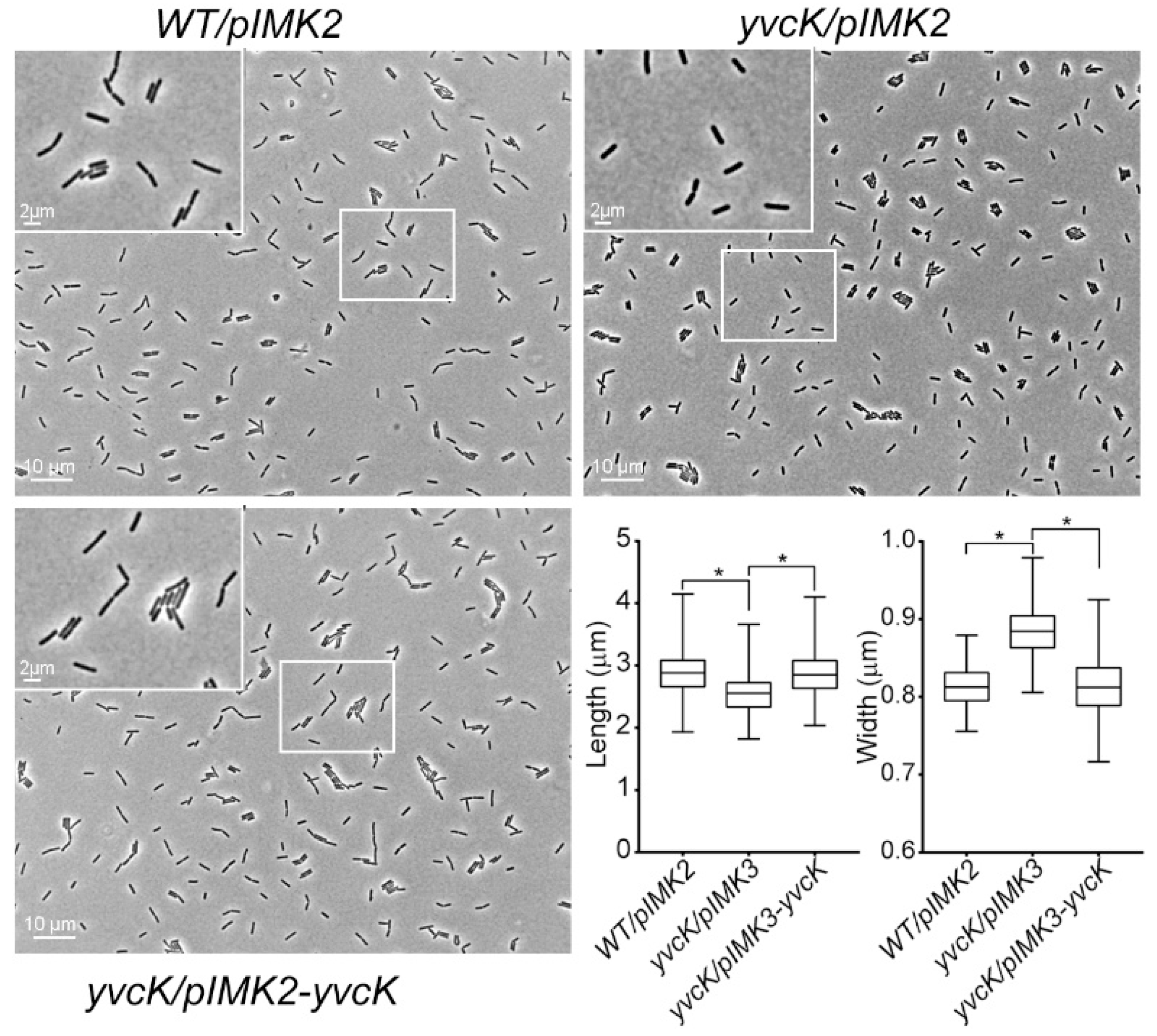
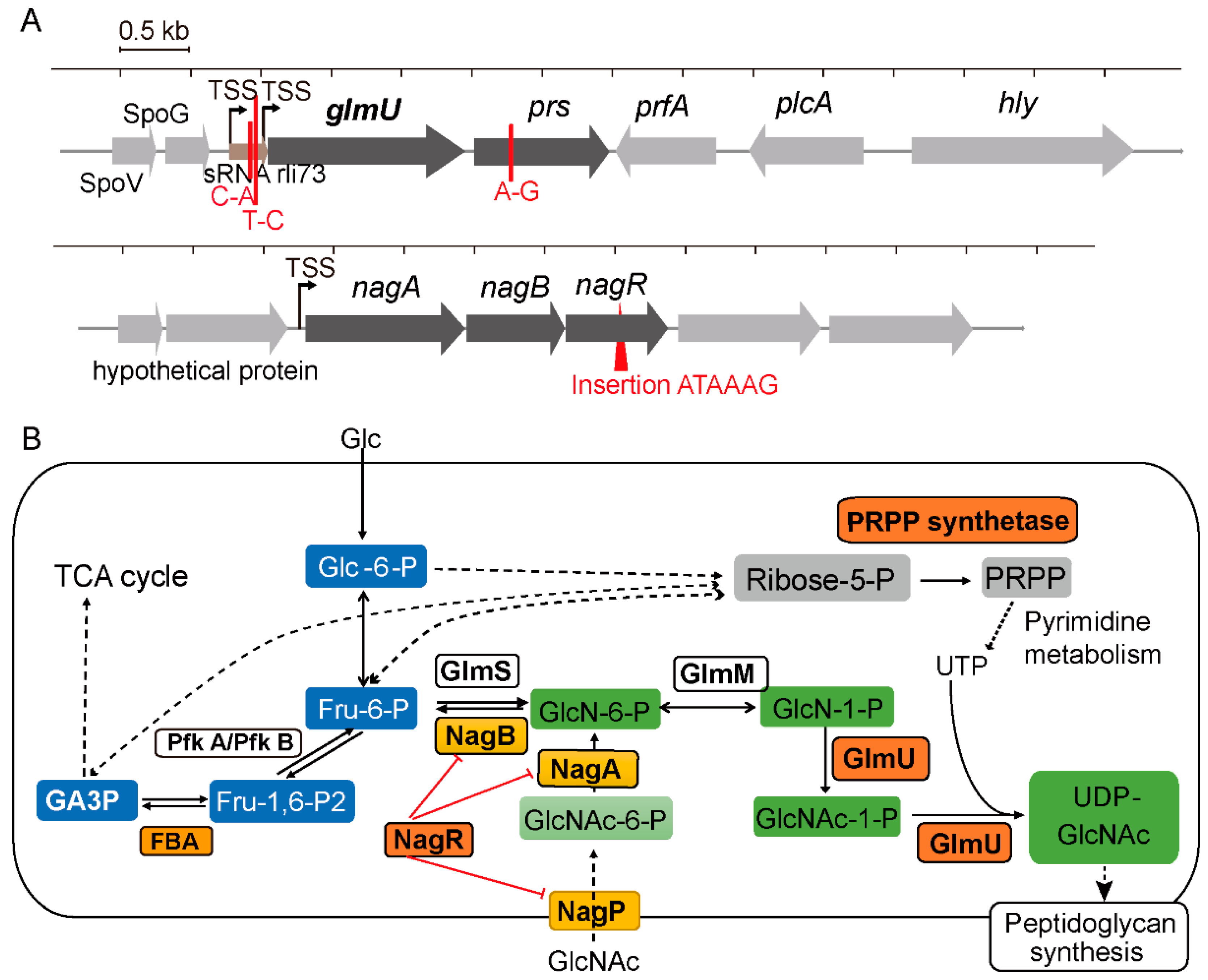
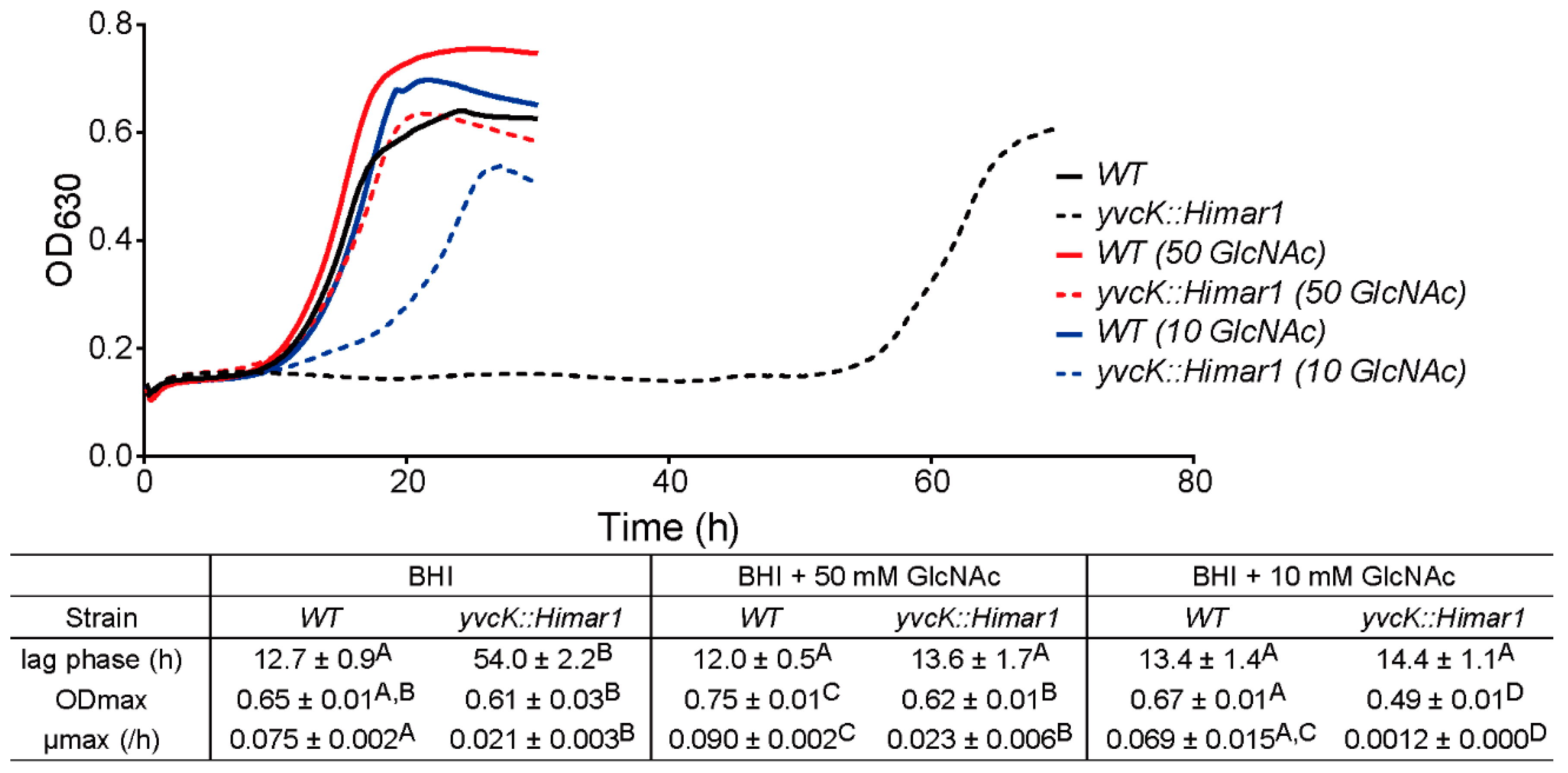
3.1. Characterization of the t-CIN Hypersensitive yvcK::Himar1 Mutant
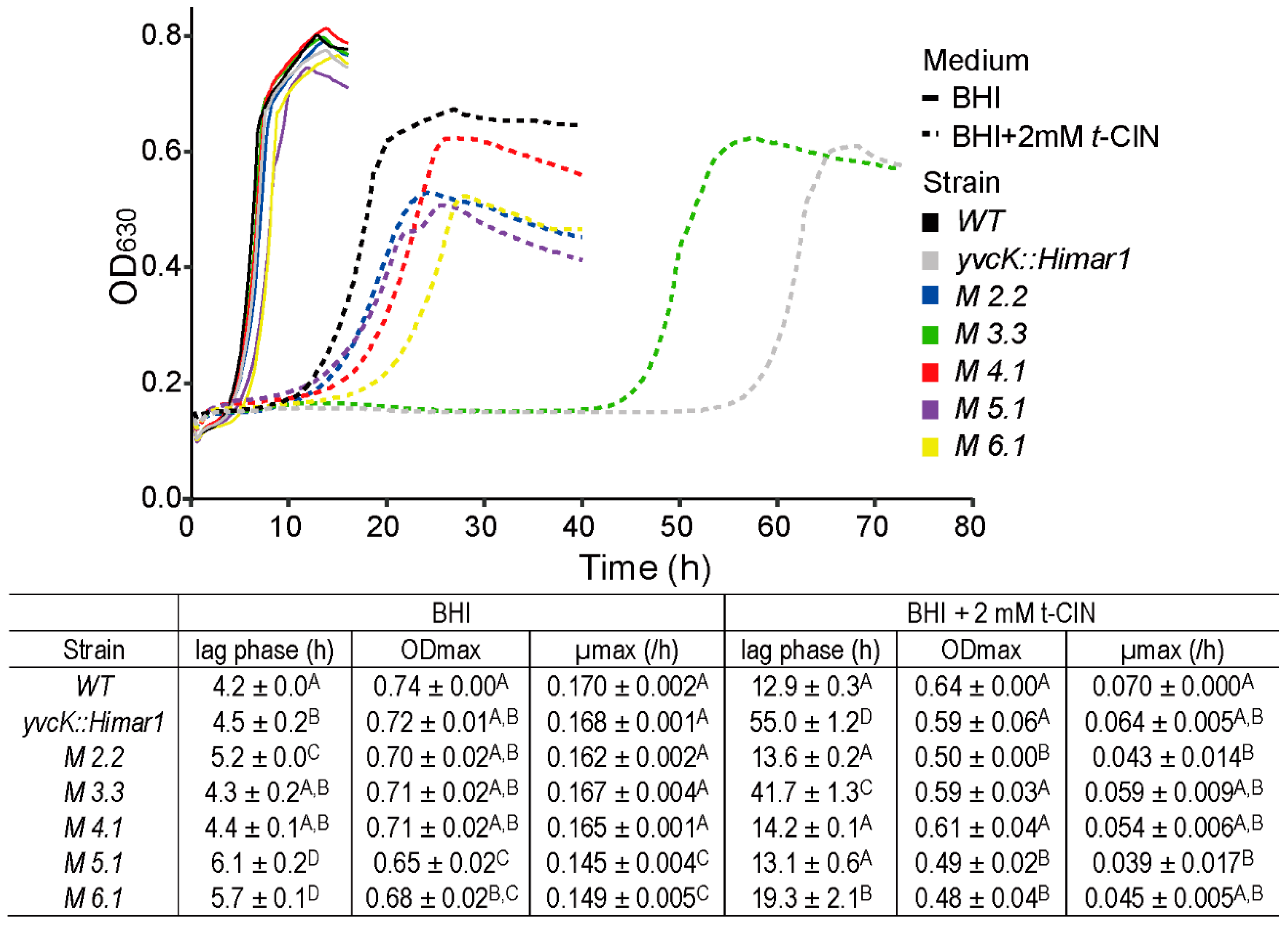
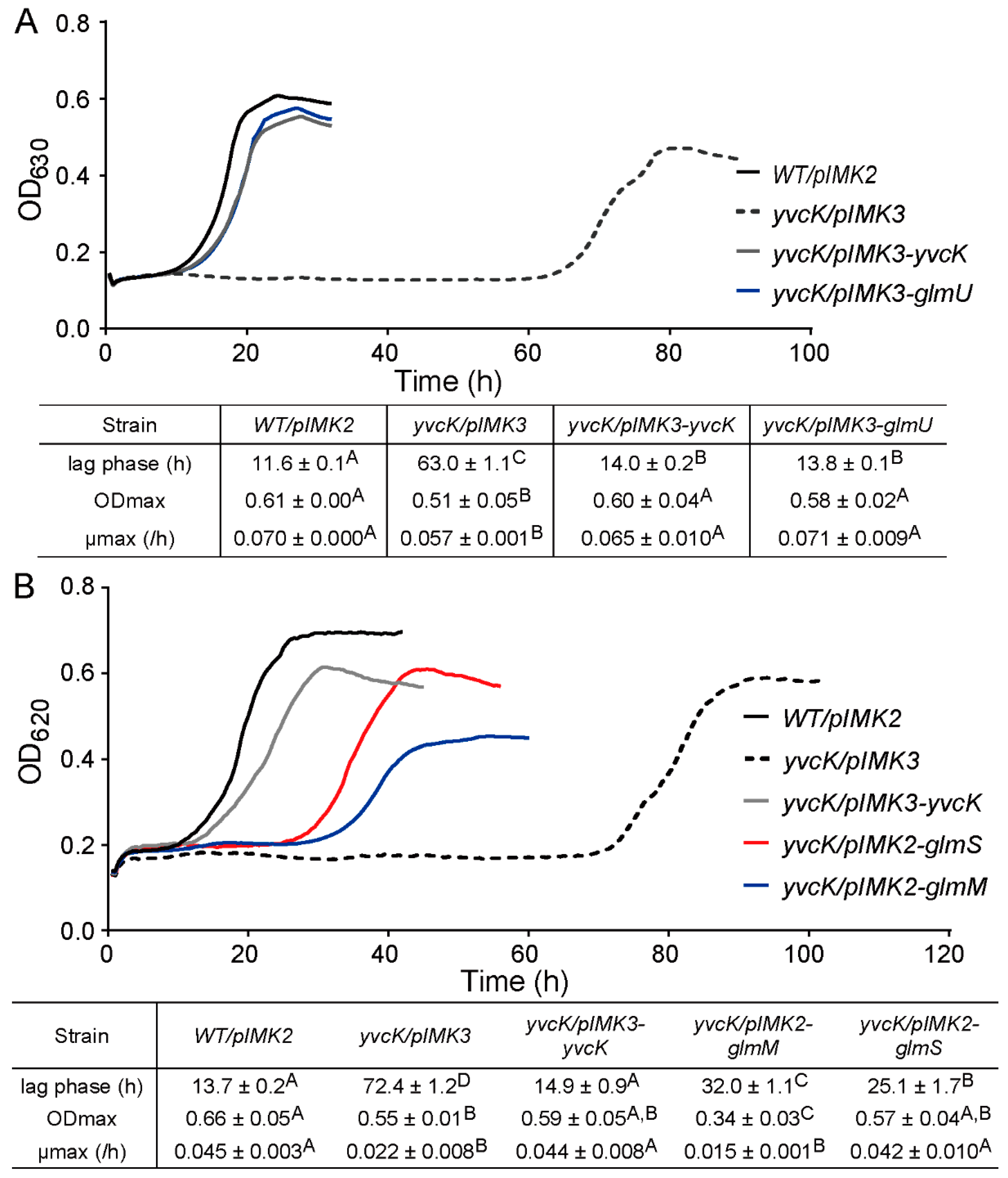
3.2. Suppressor Mutations Reverse the Sensitivity of yvcK::Himar1 to t-CIN
3.3. GlcNAc Supplementation Reverses the t-CIN Sensitivity of the yvcK::Himar1 Mutant
3.4. Overexpression of UDP-GlcNAc Biosynthetic Enzymes Reduce the t-CIN Sensitivity of the yvcK Mutant
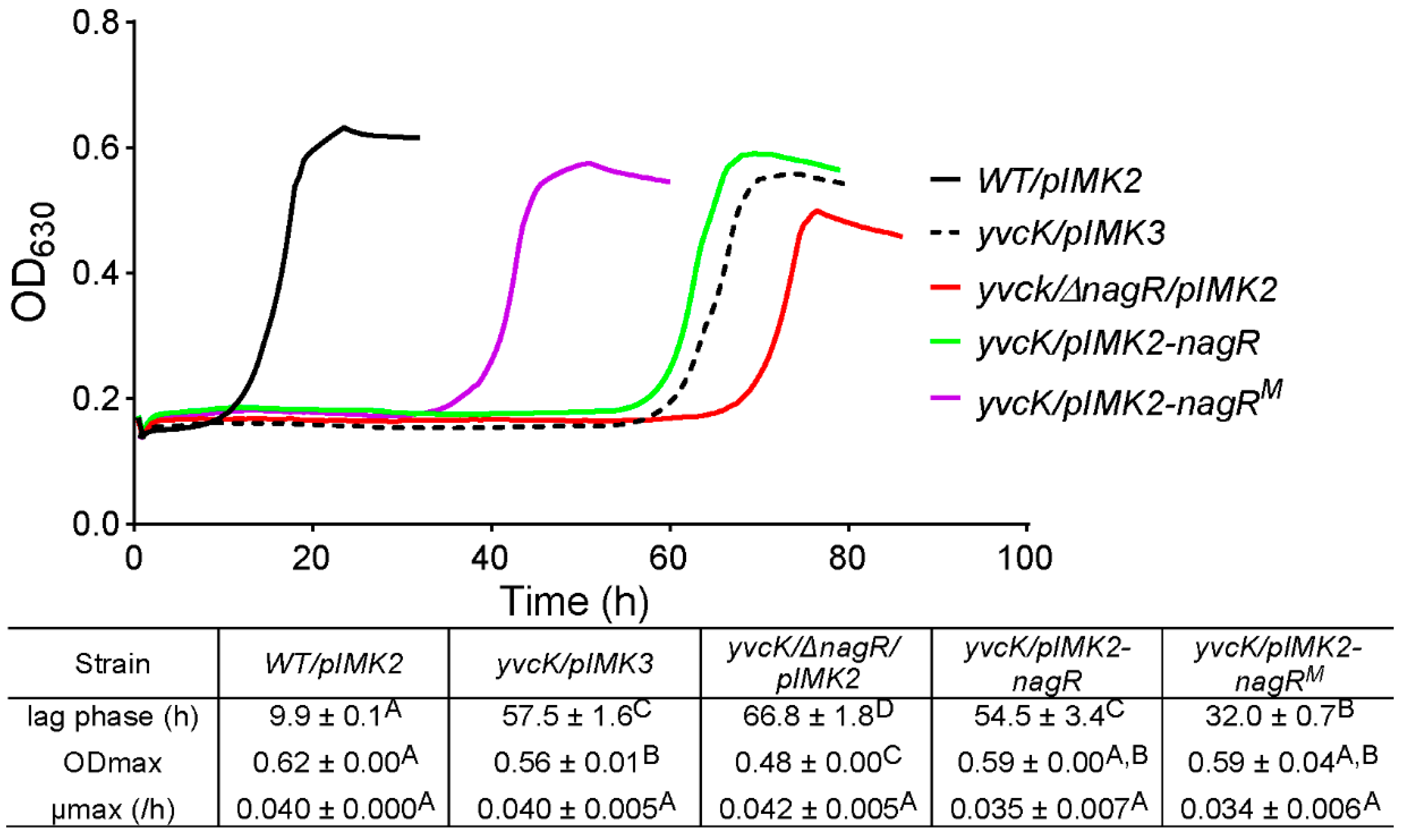
3.5. Complementation with Mutated nagR Allele from Strain M 2.2 Partially Restores Sensitivity of yvcK::Himar1 to t-CIN
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamont, R.F.; Sobel, J.; Mazaki-Tovi, S.; Kusanovic, J.P.; Vaisbuch, E.; Kim, S.K.; Uldbjerg, N.; Romero, R. Listeriosis in human pregnancy: A systematic review. J. Perinat. Med. 2011, 39, 227–236. [Google Scholar] [CrossRef]
- de Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef]
- Sauders, B.D.; Overdevest, J.; Fortes, E.; Windham, K.; Schukken, Y.; Lembo, A.; Wiedmann, M. Diversity of Listeria Species in Urban and Natural Environments. Appl. Environ. Microbiol. 2012, 78, 4420–4433. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and Food Processing Environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, N.G.; Croda, J.; Simionatto, S. Antibacterial mechanisms of cinnamon and its constituents: A review. Microb. Pathog. 2018, 120, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Rogiers, G.; Kebede, B.T.; Van Loey, A.; Michiels, C.W. Membrane fatty acid composition as a determinant of Listeria monocytogenes sensitivity to trans -cinnamaldehyde. Res. Microbiol. 2017, 168, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Domadia, P.; Swarup, S.; Bhunia, A.; Sivaraman, J.; Dasgupta, D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 2007, 74, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Di Pasqua, R.; Hoskins, N.; Betts, G.; Mauriello, G. Changes in membrane fatty acids composition of microbial cells induced by addiction of thymol, carvacrol, limonene, cinnamaldehyde, and eugenol in the growing media. J. Agric. Food Chem. 2006, 54, 2745–2749. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.O.; Holley, R.A. Inhibition of membrane bound ATPases of Escherichia coli and Listeria monocytogenes by plant oil aromatics. Int. J. Food Microbiol. 2006, 111, 170–174. [Google Scholar] [CrossRef]
- Pensinger, D.A.; Boldon, K.M.; Chen, G.Y.; Vincent, W.J.B.; Sherman, K.; Xiong, M.; Schaenzer, A.J.; Forster, E.R.; Coers, J.; Striker, R.; et al. The Listeria monocytogenes PASTA Kinase PrkA and Its Substrate YvcK Are Required for Cell Wall Homeostasis, Metabolism, and Virulence. PLoS Pathog. 2016, 12, e1006001. [Google Scholar] [CrossRef]
- Mir, M.; Prisic, S.; Kang, C.M.; Lun, S.; Guo, H.; Murry, J.P.; Rubin, E.J.; Husson, R.N. Mycobacterial gene cuvA is required for optimal nutrient utilization and virulence. Infect. Immun. 2014, 82, 4104–4117. [Google Scholar] [CrossRef]
- Patel, V.; Wu, Q.; Chandrangsu, P.; Helmann, J.D. A metabolic checkpoint protein GlmR is important for diverting carbon into peptidoglycan biosynthesis in Bacillus subtilis. PLoS Genet. 2018, 14, e1007689. [Google Scholar] [CrossRef]
- Görke, B.; Foulquier, E.; Galinier, A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 2005, 151, 3777–3791. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barreteau, H.; Kovač, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef] [PubMed]
- Foulquier, E.; Pompeo, F.; Byrne, D.; Fierobe, H.P.; Galinier, A. Uridine diphosphate N-acetylglucosamine orchestrates the interaction of GlmR with either YvcJ or GlmS in Bacillus subtilis. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fugett, E.; Fortes, E.; Nnoka, C.; Wiedmann, M. International Life Sciences Institute North America Listeria monocytogenes strain collection: Development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 2006, 69, 2929–2938. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.G.N.; Jessee, J.; Bloom, F.R.; Hanahan, D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 1990, 87, 4645–4649. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Monk, I.R.; Gahan, C.G.M.; Hill, C. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 3921–3934. [Google Scholar] [CrossRef]
- Smith, K.; Youngman, P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 1992, 74, 705–711. [Google Scholar] [CrossRef]
- Camilli, A.; Tilney, L.G.; Portnoy, D.A. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 1993, 8, 143–157. [Google Scholar] [CrossRef]
- Sliusarenko, O.; Heinritz, J.; Emonet, T.; Jacobs-Wagner, C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011, 80, 612–627. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sesto, N.; Mellin, J.R.; Karunker, I.; Edelheit, S.; Bécavin, C.; Archambaud, C.; Cossart, P.; Sorek, R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 2012, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.J.; Leonard, G.A.; McSweeney, S.M.; Thompson, A.W.; Naismith, J.H.; Qamar, S.; Plater, A.; Berry, A.; Hunter, W.N. The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold. Structure 1996, 4, 1303–1315. [Google Scholar] [CrossRef]
- Hove-Jensen, B.; Andersen, K.R.; Kilstrup, M.; Martinussen, J.; Switzer, R.L.; Willemoës, M. Phosphoribosyl Diphosphate (PRPP): Biosynthesis, Enzymology, Utilization, and Metabolic Significance. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Bertram, R.; Rigali, S.; Wood, N.; Lulko, A.T.; Kuipers, O.P.; Titgemeyer, F. Regulon of the N-acetylglucosamine utilization regulator NagR in Bacillus subtilis. J. Bacteriol. 2011, 193, 3525–3536. [Google Scholar] [CrossRef]
- Gaugué, I.; Oberto, J.; Plumbridge, J. Regulation of amino sugar utilization in Bacillus subtilis by the GntR family regulators, NagR and GamR. Mol. Microbiol. 2014, 92, 100–115. [Google Scholar] [CrossRef]
- Zeng, L.; Burne, R.A. NagR differentially regulates the expression of the glmS and nagAB genes required for amino sugar metabolism by Streptococcus mutans. J. Bacteriol. 2015, 197, 3533–3544. [Google Scholar] [CrossRef]
- Reizer, J.; Bachem, S.; Reizer, A.; Arnaud, M.; Saier, M.H.; Stülke, J. Novel phosphotransferase system genes revealed by genome analysis-the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 1999, 145 Pt 1, 3419–3429. [Google Scholar] [CrossRef]
- Vincent, F.; Yates, D.; Garman, E.; Davies, G.J.; Brannigan, J.A. The Three-dimensional Structure of the N-Acetylglucosamine-6-phosphate Deacetylase, NagA, from Bacillus subtilis. J. Biol. Chem. 2004, 279, 2809–2816. [Google Scholar] [CrossRef]
- Vincent, F.; Davies, G.J.; Brannigan, J.A. Structure and kinetics of a monomeric glucosamine 6-phosphate deaminase: Missing link of the NagB superfamily? J. Biol. Chem. 2005, 280, 19649–19655. [Google Scholar] [CrossRef]
- Huang, T.C.; Fu, H.Y.; Ho, C.T.; Tan, D.; Huang, Y.T.; Pan, M.H. Induction of apoptosis by cinnamaldehyde from indigenous cinnamon Cinnamomum osmophloeum Kaneh through reactive oxygen species production, glutathione depletion, and caspase activation in human leukemia K562 cells. Food Chem. 2007, 103, 434–443. [Google Scholar] [CrossRef]
- Du, G.F.; Yin, X.F.; Yang, D.H.; He, Q.Y.; Sun, X. Proteomic Investigation of the Antibacterial Mechanism of trans-Cinnamaldehyde against Escherichia coli. J. Proteome Res. 2021, 20, 2319–2328. [Google Scholar] [CrossRef]
- Visvalingam, J.; Hernandez-Doria, J.D.; Holley, R.A. Examination of the genome-wide transcriptional response of Escherichia coli O157: H7 to cinnamaldehyde exposure. Appl. Environ. Microbiol. 2013, 79, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Seng, Z.J.; Kohli, G.S.; Yang, L.; Yuk, H.G. Stress resistance development and genome-wide transcriptional response of Escherichia coli O157:H7 adapted to sublethal thymol, carvacrol, and trans-cinnamaldehyde. Appl. Environ. Microbiol. 2018, 84, e01616–e01618. [Google Scholar] [CrossRef]
- Staerck, C.; Gastebois, A.; Vandeputte, P.; Calenda, A.; Larcher, G.; Gillmann, L.; Papon, N.; Bouchara, J.P.; Fleury, M.J.J. Microbial antioxidant defense enzymes. Microb. Pathog. 2017, 110, 56–65. [Google Scholar] [CrossRef]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Wamelink, M.M.; Kowald, A.; Gerisch, B.; Heeren, G.; Struys, E.A.; Klipp, E.; Jakobs, C.; Breitenbach, M.; Lehrach, H.; et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Fuhrer, T.; Chavarría, M.; Sánchez-Pascuala, A.; Sauer, U.; de Lorenzo, V. Reconfiguration of metabolic fluxes in Pseudomonas putida as a response to sub-lethal oxidative stress. ISME J. 2021, 15, 1751–1766. [Google Scholar] [CrossRef]
- Christodoulou, D.; Link, H.; Fuhrer, T.; Kochanowski, K.; Gerosa, L.; Sauer, U. Reserve Flux Capacity in the Pentose Phosphate Pathway Enables Escherichia coli’s Rapid Response to Oxidative Stress. Cell Syst. 2018, 6, 569–578.e7. [Google Scholar] [CrossRef]
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020, 18, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Oliva, G.; Fontes, M.R.; Garratt, R.C.; Altamirano, M.M.; Calcagno, M.L.; Horjales, E. Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 å resolution. Structure 1995, 3, 1323–1332. [Google Scholar] [CrossRef]
- Isupov, M.N.; Obmolova, G.; Butterworth, S.; Badet-Denisot, M.A.; Badet, B.; Polikarpov, I.; Littlechild, J.A.; Teplyakov, A. Substrate binding is required for assembly of the active conformation of the catalytic site in Ntn amidotransferases: Evidence from the 1.8 Å crystal structure of the glutaminase domain of glucosamine 6- phosphate synthase. Structure 1996, 4, 801–810. [Google Scholar] [CrossRef]
- Badet, B.; Vermoote, P.; Le Goffic, F.; Haumont, P.Y.; Lederer, F. Glucosamine Synthetase from Escherichia coli: Purification, Properties, and Glutamine-Utilizing Site Location. Biochemistry 1987, 26, 1940–1948. [Google Scholar] [CrossRef] [PubMed]
- Tinsley, R.A.; Furchak, J.R.W.; Walter, N.G. Trans-acting glmS catalytic riboswitch: Locked and loaded. RNA 2007, 13, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286. [Google Scholar] [CrossRef]
- Foulquier, E.; Galinier, A. YvcK, a protein required for cell wall integrity and optimal carbon source utilization, binds uridine diphosphate-sugars. Sci. Rep. 2017, 7, 4139. [Google Scholar] [CrossRef]
- Gonzalez, G.M.; Durica-Mitic, S.; Hardwick, S.W.; Moncrieffe, M.C.; Resch, M.; Neumann, P.; Ficner, R.; Görke, B.; Luisi, B.F. Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism. Nucleic Acids Res. 2017, 45, 10845–10860. [Google Scholar] [CrossRef]
- Göpel, Y.; Papenfort, K.; Reichenbach, B.; Vogel, J.; Görke, B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 2013, 27, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Resch, M.; Schiltz, E.; Titgemeyer, F.; Muller, Y.A. Insight into the induction mechanism of the GntR/HutC bacterial transcription regulator YvoA. Nucleic Acids Res. 2010, 38, 2485–2497. [Google Scholar] [CrossRef] [PubMed]
- Fillenberg, S.B.; Grau, F.C.; Seidel, G.; Muller, Y.A. Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR. Nucleic Acids Res. 2015, 43, 1283–1296. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Tauqeer Alam, M.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.; Aymerich, S. Regulation of the central glycolytic genes in Bacillus subtilis: Binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 2003, 47, 1709–1721. [Google Scholar] [CrossRef]
- Zamboni, N.; Fischer, E.; Laudert, D.; Aymerich, S.; Hohmann, H.P.; Sauer, U. The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J. Bacteriol. 2004, 186, 4528–4534. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Species | Designation in This Work | Description | Reference |
|---|---|---|---|
| L. monocytogenes | WT | wild-type strain Scott A; WGS accession number at NCBI: CM001159 | [17] |
| WT/pIMK2 | WT with pIMK2 integrated, KmR | ||
| yvcK::Himar1 | Transposon insertion in yvcK, EmR | ||
| yvcK/pIMK3 | yvcK::Himar1 with pIMK3 integrated, KmR EmR | ||
| yvcK/pIMK3-yvcK | yvcK::Himar1 with pIMK3-yvcK integrated, KmR EmR | ||
| yvcK/pIMK3-glmU | yvcK::Himar1 with pIMK3-glmU integrated, KmR EmR | ||
| yvcK/pIMK2-glmM | yvcK::Himar1 with pIMK2-glmM integrated, KmR EmR | ||
| yvcK/pIMK2-glmS | yvcK::Himar1 with pIMK2-glmS integrated, KmR EmR | ||
| yvcK::Himar1 ΔnagR | In-frame deletion of nagR in yvcK::Himar1 strain, EmR | ||
| yvcK::Himar1 ΔnagR/pIMK2 | yvcK::Himar1 ΔnagR with pIMK2 integrated, KmR EmR | ||
| yvcK/pIMK2-nagR | yvcK::Himar1 with pIMK2-nagR (WT allele) integrated, KmR EmR | ||
| yvcK/pIMK2-nagRM | yvcK::Himar1 with pIMK2-nagR (mutated allele from M 2.2 suppression mutant) integrated, KmR EmR | ||
| E. coli | S17-1 λpir | Donor strain for plasmid conjugation | [19] |
| DH5-α | Host strain for plasmid constructs | [18] | |
| Plasmids | Description | Reference | |
| pIMK2 | Site-specific listerial integrative vector, Phelp constitutive promoter, 6.2 kb, KmR | [21] | |
| pIMK3 | Site-specific listerial integrative vector, Phelp IPTG inducible promoter, 7.5 kb, KmR | [21] | |
| pIMK3-yvcK | pIMK3 with yvcK gene (locus tag: LMOSA_4390) from Scott A | ||
| pIMK3-glmU | pIMK3 with glmU gene (locus tag: LMOSA_10910) from Scott A | ||
| pIMK2-glmM | pIMK2 with glmM gene (locus tag: LMOSA_1060) from Scott A | ||
| pIMK2-glmS | pIMK2 with glmS gene (locus tag: LMOSA_16310) from Scott A | ||
| pIMK2-nagR | pIMK2 with nagR gene (locus tag: LMOSA_18480) from Scott A | ||
| pIMK2-nagRM | pIMK2 with mutated nagRM gene from M 2.2 suppression mutant strain | ||
| pKSV7-oriT | Temperature-sensitive shuttle vector for making gene deletion in L. monocytogenes, 6742 bp, AmpR, CmR | [22,23] | |
| pKSV7-oriT--∆nagR | pKSV7-oriT with 1 kb flanking fragments upstream and downstream of nagR inserted | ||
| Primer | Sequence (5′–3′) * | Reference |
|---|---|---|
| yvcK_NcoI | GCATCCATGGGAAAAAAGGAAATGAAACC | |
| yvcK_SalI | CACTGTCGACTCACTCCTTTTCAATAG | |
| glmU_NcoI | ATATCCATGGAATCAAAACGATATGCTGTAGTGC | |
| glmU_SalI | ATATGTCGACTTATTTACCGTGATTCAAATGTTTTGC | |
| glmM_NcoI | ATATATCCATGGGTAAATATTTTGGTACGGATGGAGT | |
| glmM_SalI | ATATATGTCGACTGTTGTTTTAATCGTTAAGTGCCAT | |
| glmS_NcoI | ATATATCCATGGAATGTGGAATCGTTGGATATATTGGAA | |
| glmS_SalI | ATATATGTCGACTTATTCTACTGTGACACTTTTTGCTA | |
| nagR-KO-A | ATATGGTACCGGCTGGTAAGGATGCAGATTT | |
| nagR-KO-B | CATTTTCCCGCCCTCTTCTT | |
| nagR-KO-C | AAGAAGAGGGCGGGAAAATGATGAAACTCAGGCAGATTACAACA | |
| nagR-KO-D | ATATCTGCAGCAAGTGTCCCAGCGATTAACA | |
| nagR_BspHI | ATATATTCATGATCGATAAACAATCAGGAATAC | |
| nagR_SalI | ATATATCTGCAGTTATTGTTTAATCCTAGCTACAAATTGAA | |
| pIMK_FW | GAGTCAGTGAGCGAGGAAGC | [7] |
| pIMK_REV | CCTATCACCTCAAATGGTTCG | [7] |
| NC16(II) | GTCAAAACATACGCTCTTATCGATTC | |
| pKSV7-CK-F | TAGCTCACTCATTAGGCAC | |
| pKSV7-CK-R | TAAGGAGAAAATACCGCATCA |
| Strain | Mutations | Coding Region Change | Location in Gene | Affected Gene | Encoded Product |
|---|---|---|---|---|---|
| M 3.3 | C-A | −108 b from glmU start codon | rli73 | Small RNA rli73 | |
| T-C | 243 b downstream of peroxide-responsive repressor gene perR | Noncoding region | |||
| M 6.1 | A-G | Y97C | +290 b from prs start codon | prs | Ribose-phosphate pyrophosphokinase |
| M 2.2 | In frame insertion ATAAAG | Insertion of I K between L165-Y166 | between +495 b–+496 b of nagR start codon | nagR | Transcriptional regulator NagR |
| M 4.1 | T-C | −111 bp from glmU start codon | rli73 | Small RNA rli73 | |
| C-A | W239L | +716 bp from fbaA start codon | fbaA | Fructose-bisphosphate aldolase | |
| M 5.1 | In frame deletion ACCACG | Deletion of R144G145 | from +430 b to +435 b of rpoA | rpoA | DNA-directed RNA polymerase subunit alpha |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Rogiers, G.; Michiels, C.W. The Natural Antimicrobial trans-Cinnamaldehyde Interferes with UDP-N-Acetylglucosamine Biosynthesis and Cell Wall Homeostasis in Listeria monocytogenes. Foods 2021, 10, 1666. https://doi.org/10.3390/foods10071666
Sun L, Rogiers G, Michiels CW. The Natural Antimicrobial trans-Cinnamaldehyde Interferes with UDP-N-Acetylglucosamine Biosynthesis and Cell Wall Homeostasis in Listeria monocytogenes. Foods. 2021; 10(7):1666. https://doi.org/10.3390/foods10071666
Chicago/Turabian StyleSun, Lei, Gil Rogiers, and Chris W. Michiels. 2021. "The Natural Antimicrobial trans-Cinnamaldehyde Interferes with UDP-N-Acetylglucosamine Biosynthesis and Cell Wall Homeostasis in Listeria monocytogenes" Foods 10, no. 7: 1666. https://doi.org/10.3390/foods10071666
APA StyleSun, L., Rogiers, G., & Michiels, C. W. (2021). The Natural Antimicrobial trans-Cinnamaldehyde Interferes with UDP-N-Acetylglucosamine Biosynthesis and Cell Wall Homeostasis in Listeria monocytogenes. Foods, 10(7), 1666. https://doi.org/10.3390/foods10071666






