MicroRNAs Involved in the Therapeutic Functions of Noni (Morinda citrifolia L.) Fruit Juice in the Treatment of Acute Gouty Arthritis in Mice Induced with Monosodium Urate
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Production of the Noni Fruit Juice
2.3. Measurement of the Paw Swelling in Mice
2.4. Measurement of the Contents of NALP3 and TNF-α
2.5. High-Throughput Sequencing and Analysis of microRNAs
2.6. Identification and Analysis of Differentially Expressed Genes of microRNAs
3. Results and Discussion
3.1. Paw Swelling Degree in Mice
3.2. Contents of NALP3 and TNF-α
3.3. Small RNA Sequencing
3.3.1. Characteristics of Small RNAs
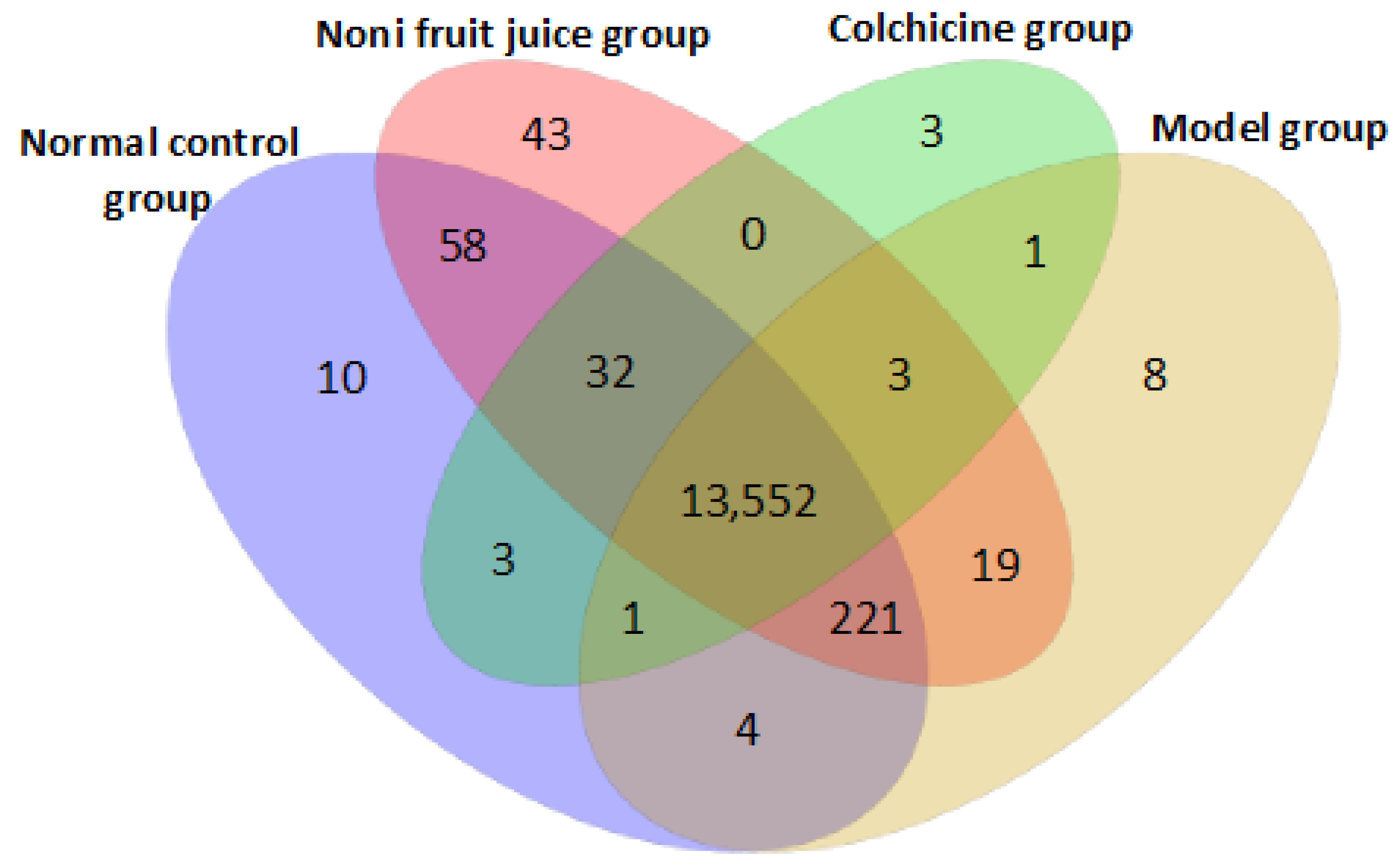
3.3.2. Differentially Expressed microRNAs
3.3.3. Target Genes of microRNAs in Mice
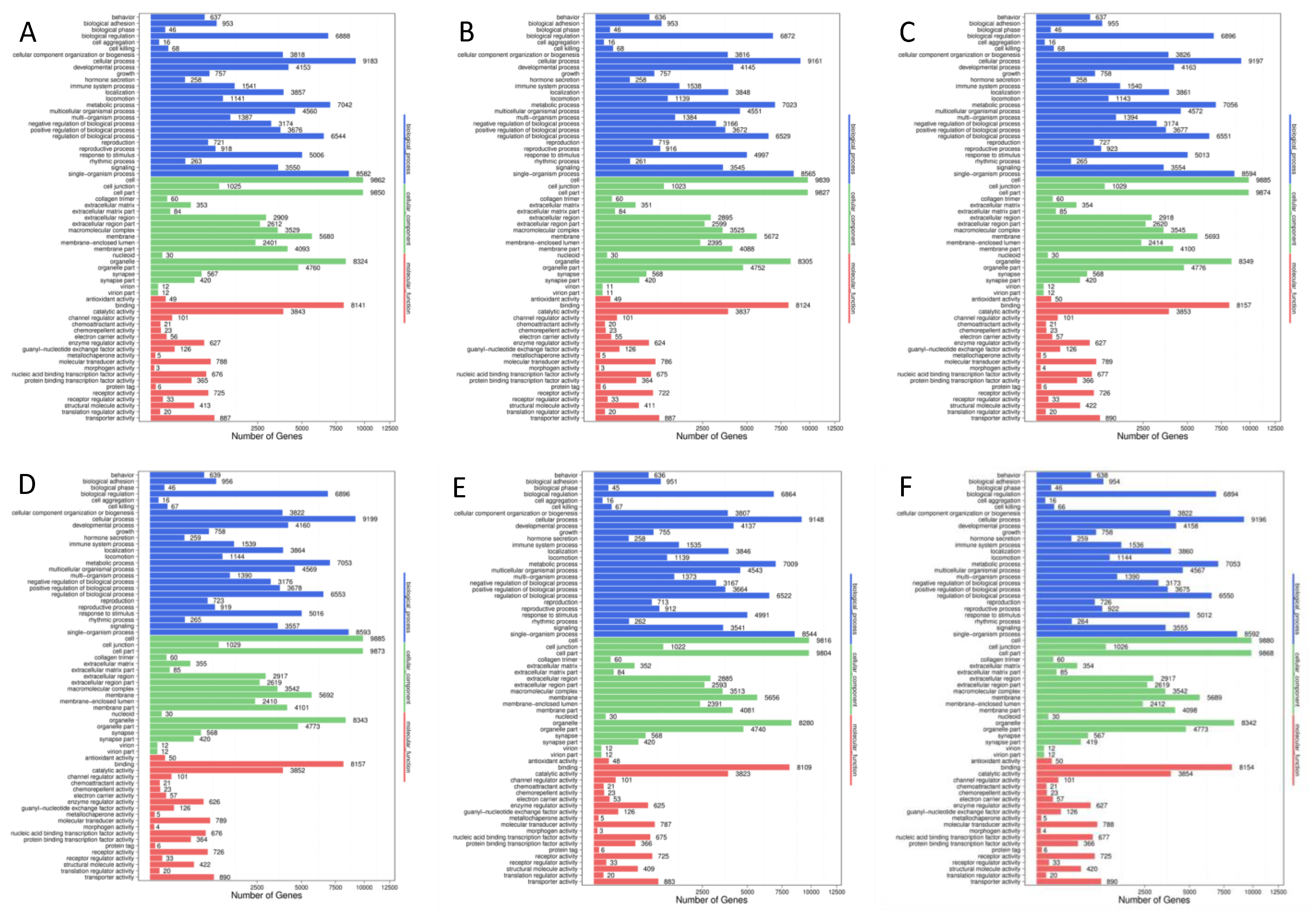
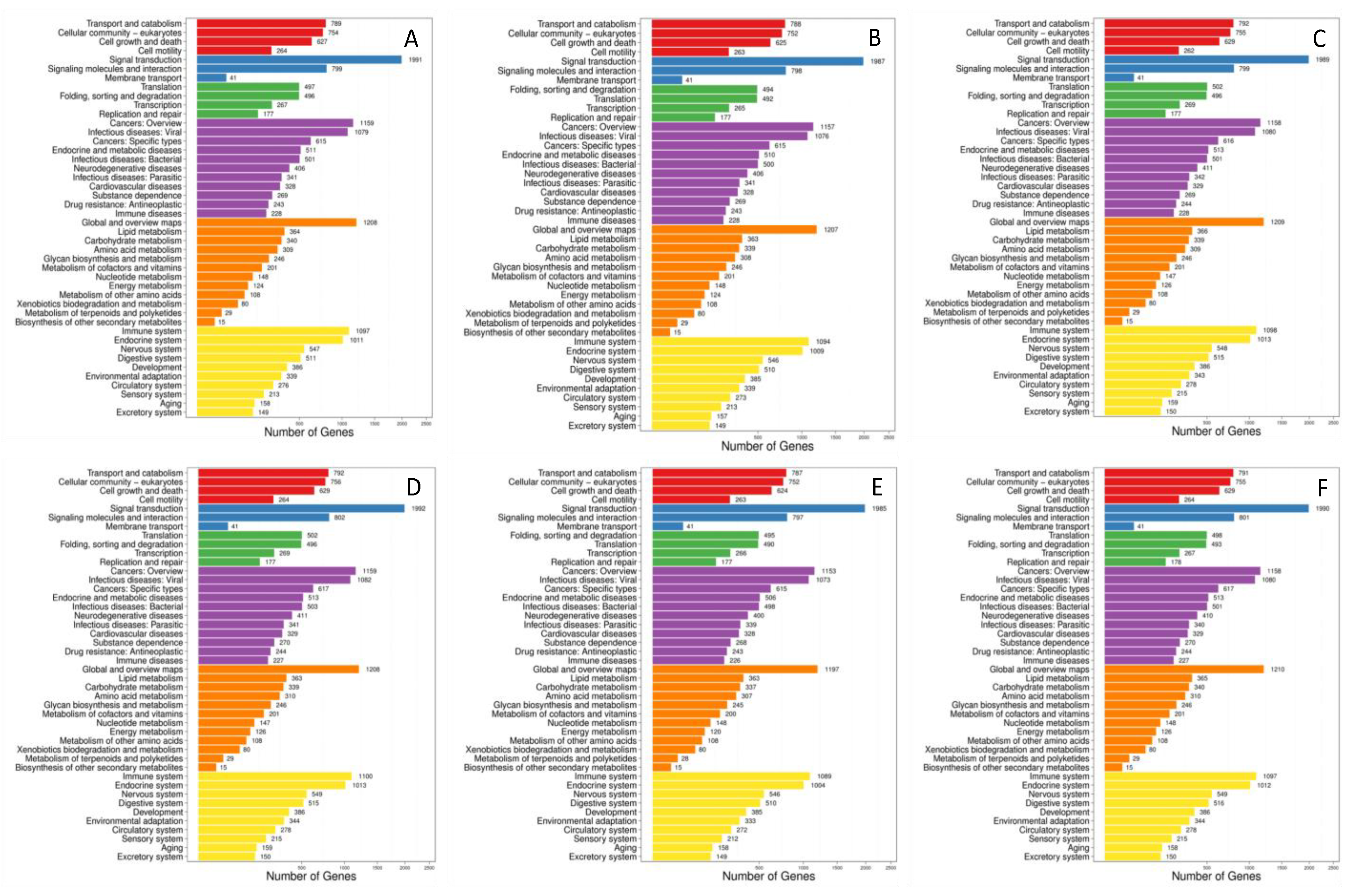
3.3.4. Enrichment Analyses of Target Genes of differentially Expressed microRNAs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dalbeth, N.; Choi, H.K.; Joosten, L.A.B.; Khanna, P.P.; Matsuo, H.; Perez-Ruiz, F.; Stamp, L.K. Gout. Nat. Rev. Dis. Primers 2019, 5, 69. [Google Scholar] [CrossRef]
- Wilson, L.; Saseen, J.J. Gouty Arthritis: A Review of Acute Management and Prevention. Pharmacotherapy 2016, 36, 906–922. [Google Scholar] [CrossRef]
- Pascart, T.; Lioté, F. Gout: State of the art after a decade of developments. Rheumatology 2019, 58, 27–44. [Google Scholar] [CrossRef]
- Richette, P.; Doherty, M.; Pascual, E.; Barskova, V.; Becce, F.; Castaneda, J.; Coyfish, M.; Guillo, S.; Jansen, T.; Janssens, H.; et al. 2018 updated European League Against Rheumatism evidence-based recommendations for the diagnosis of gout. Ann. Rheum. Dis. 2020, 79, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Pathmanathan, K.; Robinson, P.C.; Hill, C.L.; Keen, H.I. The prevalence of gout and hyperuricaemia in Australia: An updated systematic review. Semin. Arthritis Rheum. 2020, 51, 121–128. [Google Scholar] [CrossRef]
- Mattiuzzi, C.; Lippi, G. Recent updates on worldwide gout epidemiology. Clin. Rheumatol. 2019, 39, 1061–1063. [Google Scholar] [CrossRef]
- Choi, H.K.; Curhan, G. Gout: Epidemiology and lifestyle choices. Curr. Opin. Rheumatol. 2005, 17, 341–345. [Google Scholar] [PubMed]
- Gamala, M.; Jacobs, J.W.G. Gout and hyperuricaemia: A worldwide health issue of joints and beyond. Rheumatology 2019, 58, 2083–2085. [Google Scholar] [CrossRef]
- Yao, R.; Geng, Z.; Miao, X.; Bao, Y.; Guo, S.; Bao, L.; Sun, J.; Gao, Y.; Guo, B.; Meng, F.; et al. Tu-Teng-Cao Extract Alleviates Monosodium Urate-Induced Acute Gouty Arthritis in Rats by Inhibiting Uric Acid and Inflammation. Evid. Based Complement. Alternat. Med. 2020, 2020, 3095624. [Google Scholar] [CrossRef] [PubMed]
- So, A.; Dumusc, A.; Nasi, S. The role of IL-1 in gout: From bench to bedside. Rheumatology 2018, 57 (Suppl. S1), i12–i19. [Google Scholar] [PubMed]
- Lee, M.S.; Kim, Y.J. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007, 76, 447–480. [Google Scholar] [CrossRef]
- Liu-Bryan, R. Intracellular innate immunity in gouty arthritis: Role of NALP3 inflammasome. Immunol. Cell Biol. 2010, 88, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.J.; Tsai, P.C.; Chen, C.J.; Lai, H.M.; Ko, Y.C. The polymorphism-863C/A in tumour necrosis factor-alpha gene contributes an independent association to gout. Rheumatology 2007, 46, 1662–1666. [Google Scholar] [CrossRef][Green Version]
- Amaral, F.A.; Bastos, L.F.S.; Oliveira, T.H.C.; Dias, A.C.F.; Oliveira, V.L.S.; Tavares, L.D.; Costa, V.V.; Galvão, I.; Soriani, F.M.; Szymkowski, D.E.; et al. Transmembrane TNF-α is sufficient for articular inflammation and hypernociception in a mouse model of gout. Eur. J. Immunol. 2006, 46, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, H.; Belmadani, S.; Wu, J.; Xu, X.; Elford, H.; Potter, B.J.; Zhang, C. Role of TNF-alpha-induced reactive oxygen species in endothelial dysfunction during reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H2242–H2249. [Google Scholar] [CrossRef] [PubMed]
- Papanagnou, P.; Stivarou, T.; Tsironi, M. The Role of miRNAs in Common Inflammatory Arthropathies: Osteoarthritis and Gouty Arthritis. Biomolecules 2016, 6, 44. [Google Scholar] [CrossRef]
- Xu, Y.-T.; Leng, Y.-R.; Liu, M.-M.; Dong, R.-F.; Bian, J.; Yuan, L.-L.; Zhang, J.-G.; Xia, Y.-Z.; Kong, L.-Y. MicroRNA and long noncoding RNA involvement in gout and prospects for treatment. Int. Immunopharmacol. 2020, 87, 106842. [Google Scholar] [CrossRef] [PubMed]
- Tai, V.; Merriman, T.R.; Dalbeth, N. Genetic advances in gout: Potential applications in clinical practice. Curr. Opin. Rheumatol. 2019, 31, 144–151. [Google Scholar] [CrossRef]
- Jin, H.M.; Kim, T.-J.; Choi, J.-H.; Kim, M.-J.; Cho, Y.-N.; Nam, K.-I.; Kee, S.-J.; Moon, J.B.; Choi, S.-Y.; Park, D.-J.; et al. MicroRNA-155 as a proinflammatory regulator via SHIP-1 down-regulation in acute gouty arthritis. Arthritis Res. Ther. 2014, 16, R88. [Google Scholar] [CrossRef]
- Ma, T.; Liu, X.; Cen, Z.; Xin, C.; Guo, M.; Zou, C.; Song, W.; Xie, R.; Wang, K.; Zhou, H.; et al. MicroRNA-302b negatively regulates IL-1β production in response to MSU crystals by targeting IRAK4 and EphA2. Arthritis Res. Ther. 2018, 20, 34. [Google Scholar] [CrossRef]
- Liu, P.; Chen, Y.; Wang, B.; Wang, Z.; Li, C.; Wang, Y. Expression of microRNAs in the plasma of patients with acute gouty arthritis and the effects of colchicine and etoricoxib on the differential expression of microRNAs. Arch. Med. Sci. 2019, 15, 1047–1055. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Wu, R.; He, Y.; Su, Q.; Shi, G. MicroRNA-488 and -920 regulate the production of proinflammatory cytokines in acute gouty arthritis. Arthritis Res. Ther. 2017, 19, 203. [Google Scholar] [CrossRef]
- Zhang, Q.; Qing, Y.; Yin, C.; Zhou, L.; Liu, X.; Mi, Q.; Zhou, J. Mice with miR-146a deficiency develop severe gouty arthritis via dysregulation of TRAF 6, IRAK 1 and NALP3 inflammasome. Arthritis Res. Ther. 2018, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, H.; Ma, H.; Qu, S.; Xing, Q.; Wang, G. MiR-221-5p is involved in the regulation of inflammatory responses in acute gouty arthritis by targeting IL-1β. Int. J. Rheum. Dis. 2020, 24, 335–340. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, Q.; Qing, Y.; Zhou, L.; Mi, Q.; Zhou, J. miR-155 is dispensable in monosodium urate-induced gouty inflammation in mice. Arthritis Res. Ther. 2018, 20, 144. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wu, H.; Wang, D.; Yang, Z.; Dong, J. LncRNA ANRIL promotes NLRP3 inflammasome activation in uric acid nephropathy through miR-122-5p/BRCC3 axis. Biochimie 2019, 157, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhong, Q.; Feng, Y.; Tang, X.; Wang, Q.; Zou, Y.; Duan, J. TUG1 Long noncoding RNA is downregulated in sepsis and may sponge miR-27a to downregulate tumor necrosis factor-α. Int. Med. Res. 2020, 48, 300060520910638. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Hu, J.; Wang, Z.; Zong, H.; Zhang, L.; Zhang, R.; Sun, L. LncRNA H19 functions as an Aquaporin 1 com- petitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. Biomed. Pharmacother. 2018, 105, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhou, E. Long noncoding RNA LINC00662-miR-15b-5p mediated GPR120 dysregulation contributes to osteoarthritis. Pathol. Int. 2020, 70, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhu, Y.; Gao, G.; Zhang, Y. Long noncoding RNA SNHG16 targets miR-146a- 5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia. Life Sci. 2019, 228, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, Y.; Gao, G.; Zhou, Z. Knockdown XIST alleviates LPS-induced WI-38 cell apoptosis and in ammation injury via targeting miR-370-3p/TLR4 in acute pneumonia. Cell Biochem. Funct. 2019, 37, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Peng, Q.; Gu, L.; Xie, W.; Li, W. LncRNA-MEG3 alleviates high glucose induced in ammation and apoptosis of retina epithelial cells via regulating miR-34a/SIRT1 axis. Exp. Mol. Pathol. 2019, 107, 102–109. [Google Scholar] [CrossRef]
- Cao, D.; Liu, M.; Duan, R.; Tao, Y.; Zhou, J.; Fang, W.; Zhu, J.; Niu, L.; Sun, J. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol. Sin. 2020, 41, 22–33. [Google Scholar] [CrossRef]
- Yan, S.; Wang, P.; Wang, J.; Yang, J.; Lu, H.; Jin, C.; Cheng, M.; Xu, D. Long Non-coding RNA HIX003209 Promotes Inflammation by Sponging miR-6089 via TLR4/NF-κB Signaling Pathway in Rheumatoid Arthritis. Front. Immunol. 2019, 10, 2218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chi, X.; Liu, L.; Wang, Y.; Mei, X.; Yang, Y.; Jia, T. Long noncoding RNA maternally expressed gene 3 knockdown alleviates lipopolysaccharide-induced in ammatory injury by up- regulation of miR-203 in ATDC5 cells. Biomed. Pharmacother. 2018, 100, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, Q.; Zhou, L.; Wang, Y.; Sun, R.-R.; Zhang, Z.-Y. MiR-146a alleviates inflammation of acute gouty arthritis rats through TLR4/MyD88 signal transduction pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9230–9237. [Google Scholar] [PubMed]
- Dalbeth, N.; Pool, B.; Shaw, O.M.; Harper, J.L.; Tan, P.; Franklin, C.; House, M.E.; Cornish, J.; Naot, D. Role of miR-146a in regulation of the acute inflammatory response to monosodium urate crystals. Ann. Rheum. Dis. 2015, 74, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xie, Y.; Yan, M.; Pan, Q.; Liang, Y.; Sun, X. Colchicine causes prenatal cell toxicity and increases tetraploid risk. BMC Pharmacol. Toxicol. 2019, 20, 66. [Google Scholar] [CrossRef]
- An, J.M.; Kim, E.H.; Lee, H.; Lee, H.J.; Hahm, K.B. Dietary walnut as food factor to rescue from NSAID-induced gastrointestinal mucosal damages. Arch. Biochem. Biophys. 2020, 689, 108466. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Zhuang, Y.; Gu, S.W.; Jiang, Z.Y.; Zhao, L. The efficacy and tolerability of febuxostat treatment in a cohort of Chinese Han population with history of gout. J. Int. Med. Res. 2020, 48, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Chen, H.; He, S.Y.; Xue, C.X.; Sui, H.; Chen, J. Evaluating the efficacy and adverse effects of clearing heat and removing dampness method of traditional Chinese medicine by comparison with western medicine in patients with gout. Evid. Based Complement. Alternat. Med. 2018, 2018, 8591349. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Zhang, H.; Zhang, S.; Ma, K. Chinese herbal medicine for gout: A review of the clinical evidence and pharmacological mechanisms. Chin. Med. 2020, 15, 17. [Google Scholar] [CrossRef]
- Chen, B.; Li, H.; Ou, G.; Ren, L.; Yang, X.; Zeng, M. Curcumin attenuates MSU crystal-induced inflammation by inhibiting the degradation of IκBα and blocking mitochondrial damage. Arthritis Res. Ther. 2019, 21, 193. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.-W.; Wang, C.-P.; Liu, L.; Liu, Y.-L.; Wang, X.; Hong, Y.; Zhen, L.; Kong, L.-D. Antihyperuricemic and nephroprotective effects of resveratrol and its analogues in hyperuricemic mice. Mol. Nutr. Food Res. 2012, 56, 1433–1444. [Google Scholar] [CrossRef]
- Oliviero, F.; Zamudio-Cuevas, Z.; Belluzzi, E.; Andretto, L.; Scanu, A.; Favero, M.; Ramonda, R.; Ravagnan, G.; Lopez-Reyes, A.; Spinella, P.; et al. Polydatin and resveratrol inhibit the inflammatory process induced by urate and pyrophosphate crystals in THP-1 Cells. Foods 2019, 8, 560. [Google Scholar] [CrossRef]
- El-Tantawy, W.H. Natural products for the management of hyperuricaemia and gout: A review. Arch. Physiol. Biochem. 2021, 127, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Baker, F.I.A.; Baker, M.F.A.; Rahmat, A.; Abdullah, N.; Sabran, S.F.; Endrini, S. Anti-gout potential of Malaysian medicinal plants. Front. Pharmacol. 2018, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Lanaspa, M.A.; Johnson, R.J. The effects of fruit consumption in patients with hyperuricaemia or gout. Rheumatology 2019, 58, 1133–1141. [Google Scholar] [CrossRef]
- Batista, J.A.; Magalhães, D.d.A.; Sousa, S.G.; Ferreira, J.d.S.; Pereira, C.M.C.; Lima, J.V.d.N.; Albuquerque, I.F.d.; Dutra Bezerra, N.L.S.; Monteiro, C.E.d.S.; Franco, A.X.; et al. Polysaccharides derived from Morinda citrifolia Linn reduce inflammatory markers during experimental colitis. J. Ethnopharmacol. 2020, 248, 112303. [Google Scholar] [CrossRef]
- Potterat, O.; Hamburger, M. Morinda citrifolia (Noni) fruit--phytochemistry, pharmacology, safety. Planta Med. 2007, 73, 191–199. [Google Scholar] [CrossRef]
- Assi, R.A.; Darwis, Y.; Abdulbaqi, I.M.; Khan, A.A.; Vuanghao, L.; Laghari, M.H. Morinda citrifolia (Noni): A comprehensive review of its industrial uses, pharmacological activities, and clinical trials. Arab. J. Chem. 2017, 10, 691–707. [Google Scholar] [CrossRef]
- Niu, Y.; Liu, J. The complete chloroplast genome of Morinda citrifolia (noni). Mitochondrial DNA B Resour. 2020, 5, 377–378. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Niu, D.; Zhang, Y.; Zeng, X.A. Physicochemical properties, antioxidant and antiproliferative activities of polysaccharides from Morinda citrifolia L. (Noni) based on different extraction methods. Int. J. Biol. Macromol. 2020, 150, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Su, B.-N.; Pawlus, A.D.; Jung, H.-A.; Keller, W.J.; McLaughlin, J.L.; Kinghorn, A.D. Chemical constituents of the fruits of Morinda citrifolia (noni) and their antioxidant activity. J. Nat. Prod. 2005, 68, 592–595. [Google Scholar] [CrossRef] [PubMed]
- Nerurkar, P.V.; Hwang, P.W.; Saksa, E. Anti-Diabetic Potential of Noni: The Yin and the Yang. Molecules 2015, 20, 17684–17719. [Google Scholar] [CrossRef]
- West, B.J.; Su, C.X.; Jensen, C.J. Hepatotoxicity and subchronic toxicity tests of Morinda citrifolia (noni) fruit. J. Toxicol. Sci. 2009, 34, 581–585. [Google Scholar] [PubMed]
- Palu, A.; Deng, S.; West, B.; Jensen, J. Xanthine oxidase inhibiting effects of noni (Morinda citrifolia) fruit juice. Phytother. Res. 2009, 23, 1790–1791. [Google Scholar] [CrossRef]
- Pachauri, S.D.; Verma, P.R.P.; Dwivedi, A.K.; Tota, S.; Khandelwal, K.; Saxena, J.K.; Nath, C. Ameliorative effect of Noni fruit extract on streptozotocin-induced memory impairment in mice. Behav. Pharmacol. 2013, 24, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Lohani, M.; Majrashi, M.; Govindarajulu, M.; Patel, M.; Ramesh, S.; Bhattacharya, D.; Joshi, S.; Fadan, M.; Nadar, R.; Darien, B.; et al. Immunomodulatory actions of a Polynesian herb Noni (Morinda citrifolia) and its clinical applications. Complement. Ther. Med. 2019, 47, 102206. [Google Scholar] [CrossRef]
- Clafshenkel, W.P.; King, T.L.; Kotlarczyk, M.P.; Cline, J.M.; Foster, W.G.; Davis, V.L.; Witt-Enderby, P.A. Morinda citrifolia (Noni) Juice Augments Mammary Gland Differentiation and Reduces Mammary Tumor Growth in Mice Expressing the Unactivated c-erbB2 Transgene. Evid.-Based Complement Alternat. Med. 2012, 2012, 487423. [Google Scholar] [CrossRef]
- Yang, T.-H.; Yan, D.-X.; Huang, X.-Y.; Hou, B.; Ma, Y.-B.; Peng, H.; Zhang, X.-M.; Chen, J.-J.; Geng, C.-A. Termipaniculatones A-F, chalcone-flavonone heterodimers from Terminthia paniculate, and their protective effects on hyperuricemia and acute gouty arthritis. Phytochemistry 2019, 164, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013, 41, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Evers, M.; Huttner, M.; Dueck, A.; Meiser, G.; Engelmann, J.C. miRA: Adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinform. 2015, 16, 370. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. 1997. The significance of digital gene expression profiles. Genome Res. 1997, 10, 986–995. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA Targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Zhou, Z.G.; Chou, X.Y.; Fei, H.X.; Han, Y.S.; Liao, T.; Zhang, Y.B.; Wang, J.K.; Du, H.; Piao, C.Y.; Bai, Y.; et al. Experimental study of Danxi Tongfeng Recipe on acute gouty arthritis in mice. Acta Chin. Med. Pharmacol. 2015, 43, 59–62. [Google Scholar]
- Jiang, Y.; Lin, Y.; Hu, Y.J.; Song, X.J.; Pan, H.H.; Zhang, H.J. Caffeoylquinic acid derivatives rich extract from Gnaphalium pensylvanicum willd, Ameliorates hyperuricemia and acute gouty arthritis in animal model. BMC Complement. Altern. Med. 2017, 17, 320. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.H.; Sheng, T.; Tian, C.M.; Xing, M.Y.; Yan, L.J.; Xia, D.Z. Effect of ethanolic extract of Polygonum cuspidatum on acute gouty arthritis in mice through NLRP3/ASC/caspase-1 axis. China J. Chin. Mat. Med. 2019, 44, 546–552. [Google Scholar]
- Zhang, K.H.; Wang, M.Q.; Wei, L.L.; Feng, C.J.; Zhang, Y.S.; Teng, J.B. Investigation of the effects and mechanisms of Dendrobium loddigesii Rolfe extract on the treatment of gout. Evid.-Based Complement Alternat. Med. 2020, 2020, 4367347. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.G.; Oliveira, L.A.; Magalhães, D.A.; Brito, T.V.; Batista, J.A.; Pereira, C.M.C.; Costa, M.S.; Mazulo, J.C.R.; Filgueiras, M.C.; Vasconselos, D.F.P.; et al. Chemical structure and anti-inflammatory effect of polysaccharide extracted from Morinda citrifolia Linn (Noni). Carbohydr. Polym. 2018, 197, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Chunhieng, T.; Hay, L.; Montet, D. Detailed study of the juice composition of noni (Morinda citrifolia) fruits from Cambodia. Fruits 2005, 60, 13–24. [Google Scholar] [CrossRef][Green Version]
- Bauernfeind, F.; Rieger, A.; Schildberg, F.A.; Knolle, P.A.; Schmid-Burgk, J.L.; Hornung, V. NLRP3 inflammasome activity is negatively controlled by miR-223. J. Immunol. 2012, 189, 4175–4181. [Google Scholar] [CrossRef]
- Xie, Y.F.; Shu, R.; Jiang, S.Y.; Liu, D.L.; Ni, J.; Zhang, X.L. MicroRNA-146 inhibits pro-inflammatory cytokine secretion through IL-1 receptor-associated kinase 1 in human gingival fibroblasts. J. Inflamm. 2013, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Qing, Y.; Zhang, Q. Research progress of regulation of microRNA on acute gouty arthritis. Chin. J. Clin. (Electron. Ed.) 2017, 11, 104–108. [Google Scholar]
- Zhang, Y.; Ma, L.; Wang, C.; Wang, L.; Guo, Y.; Wang, G. Long noncoding RNA LINC00461 induced osteoarthritis progression by inhibiting miR-30a-5p. Aging 2020, 12, 4111–4123. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, W.; Cai, Y. NEAT1 Promotes LPS-induced Inflammatory Injury in Macrophages by Regulating MiR-17-5p/TLR4. Open. Med. 2020, 15, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, D.; Wang, B.; Hou, X. Could MicroRNAs be Regulators of Gout Pathogenesis? Cell Physiol. Biochem. 2015, 36, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
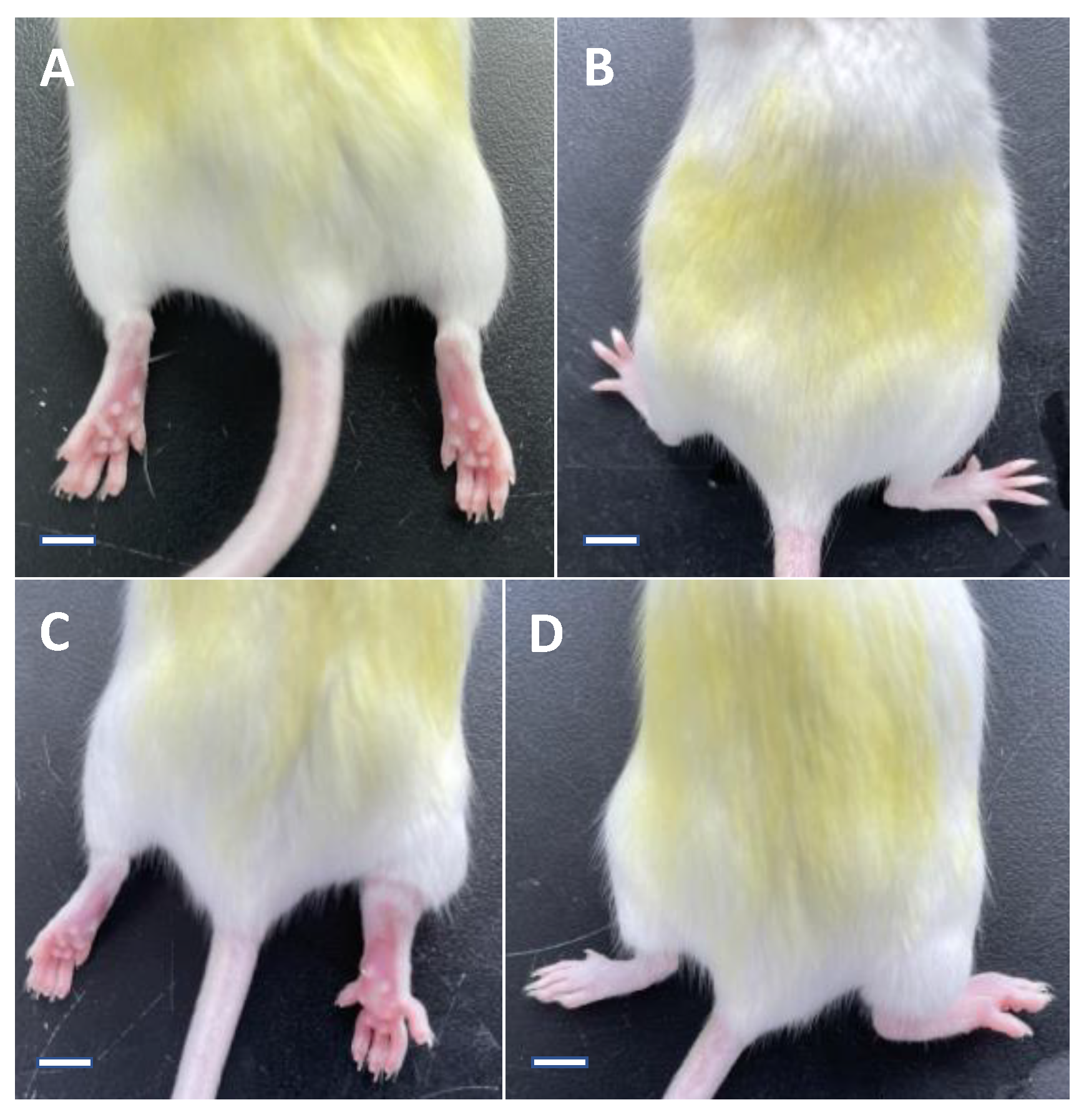

| Group of Mice | Day 0 | Day 1 | Day 5 |
|---|---|---|---|
| Normal control group | 1.69 ± 0.08 | 1.72 ± 0.12 (0.12 ± 0.08) ** | 1.71 ± 0.08 (0.04 ± 0.02) ** |
| Model group | 1.70 ± 0.10 | 3.70 ± 0.25 (2.01 ± 0.22) | 2.89 ± 0.27 (1.19 ± 0.24) |
| Colchicine group | 1.70 ± 0.11 | 3.50 ± 0.21 (1.80 ± 0.16) * | 2.31 ± 0.25 (0.61 ± 0.27) ** |
| Noni fruit juice group | 1.74 ± 0.07 | 3.65 ± 0.16 (1.91 ± 0.16) | 2.68 ± 0.20 (0.94 ± 0.21) * |
| Group of Mice | NALP3 (pg/mL) | TNF-α (pg/mL) |
|---|---|---|
| Normal control group | 86.09 ± 6.92 ** | 37.07 ± 4.49 ** |
| Model group | 128.90 ± 9.31 | 71.94 ± 9.76 |
| Colchicine group | 102.73 ± 7.25 ** | 43.09 ± 3.76 ** |
| Noni fruit juice group | 110.19 ± 8.25 * | 62.06 ± 7.29 * |
| Sample | Raw Tag | Clean Tag (%) | Q20 of Clean Tag (%) | Mapped Tag (%) |
|---|---|---|---|---|
| Normal control group | 29,268,292 | 23,957,783 (81.86) | 99.20 | 21,749,074 (90.78) |
| Model group | 26,562,125 | 18,683,543 (70.34) | 99.30 | 15,904,741 (85.13) |
| Colchicine group | 27,814,896 | 25,478,925 (91.60) | 99.20 | 24,180,100 (94.90) |
| Noni fruit juice group | 28,571,428 | 24,142,747 (84.50) | 99.30 | 22,911,510 (94.90) |
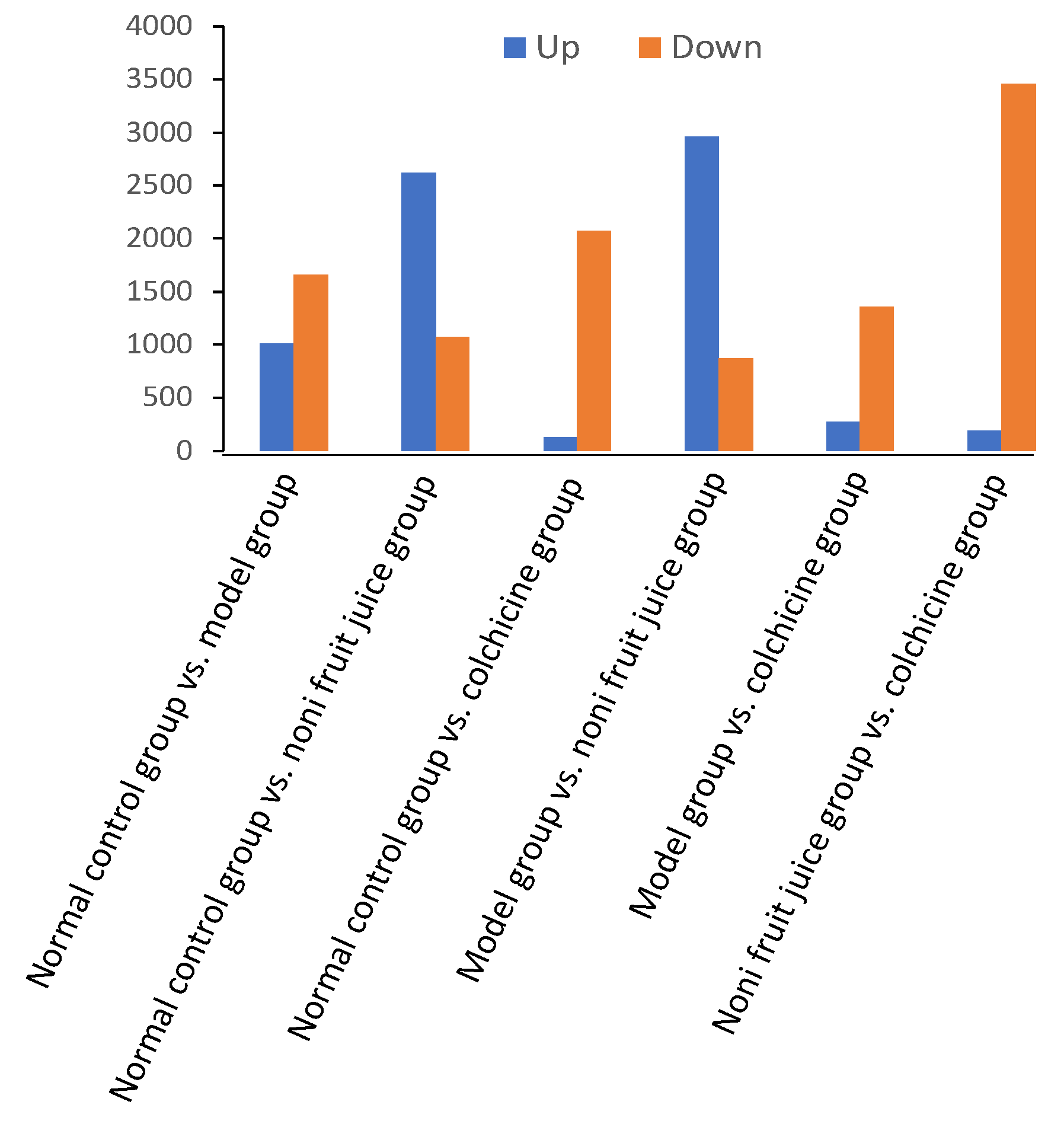
| GO Term and Gene | Control vs. Model Groups | Control vs. Noni Fruit Juice Groups | Control vs. Colchicine Groups | Model vs. Noni Fruit Juice Groups | Noni Fruit Juice vs. Colchicine Groups | Model vs. Colchicine Groups |
|---|---|---|---|---|---|---|
| Formation of NLRP3 inflammasome complex (GO: 0072559) | ||||||
| NM_026960.4 | (2↑)/(9↑, 13↓) | (3↓)/(21↑, 8↓) | (3↓)/(2↑, 17↓) | (3↓)/(26↑, 8↓) | (1↑)/(28↓) | (3↓)/(2↑, 11↓) |
| NM_009807.2 | (0) /(1↑, 1↓) | (0)/(5↑) | (0)/(1↓) | (0)/(6↑, 1↓) | (0)/(6↓) | (0)/(1↓) |
| XM_006532857.1 | (2↑, 1↓)/(36↑, 95↓) | (2↓)/(117↑, 53↓) | (5↓)/(10↑, 115↓) | (1↑, 3↓)/(147↑, 27↓) | (4↓)/(13↑, 174↓) | (4↓)/(17↑, 63↓) |
| Positive regulation of NLRP3 inflammasome complex assembly (GO: 1900226) | ||||||
| XM_006503857.2 | (0)/(5↑, 3↓) | (0)/(9↑, 2↓) | (0)/(1↑, 8↓) | (0)/(10↑, 4↓) | (0)/(14↓) | (0)/(7↓) |
| XM_006535624.3 | (1↑, 1↓)/(16↑, 21↓) | (1↑, 1↓)/(41↑, 12↓) | (1↓)/(27↓) | (0)/(44↑, 12↓) | (1↓)/(1↑, 57↓) | (1↓)/(4↑, 20↓) |
| NM_153564.2 | (0)/(3↑, 13↓) | (1↑)/(21↑, 6↓) | (0)/(15↓) | (1↑, 1↓)/(28↑, 3↓) | (1↓)/(1↑, 27↓) | (1↓)/(2↑, 5↓) |
| NM_021297.3 | (5↑)/(72↑, 125↓) | (3↑, 3↓)/(193↑, 71↓) | (5↓)/(8↑, 139↓) | (1↑, 4↓)/(226↑, 57↓) | (1↑, 4↓)/(14↑, 253↓) | (6↓)/(23↑, 86↓) |
| Negative regulation of NLRP3 inflammasome complex assembly (GO: 1900227) | ||||||
| NM_022432.4 | (3↑, 2↓)/(54↑, 99↓) | (1↓)/(143↑, 59↓) | (3↓)/(9↑, 132↓) | (1↑, 3↓)/(170↑, 39↓) | (1↓)/(11↑, 200↓) | (3↓)/(14↑, 78↓) |
| NM_019453.2 | (5↑, 1↓)/(45↑, 73↓) | (2↑)/(115↑, 46↓) | (5↓)/(10↑, 88↓) | (1↑, 4↓)/(124↑, 35↓) | (4↓)/(11↑, 148↓) | (6↓)/(15↑, 59↓) |
| Medical Condition | Control vs. Model Groups | Control vs. Noni Fruit Juice Groups | Control vs. Colchicine Groups | Model vs. Noni Fruit Juice Groups | Model vs. Colchicine Groups | Noni Fruit Juice vs. Colchicine Groups |
|---|---|---|---|---|---|---|
| Osteoarthritis | miR-30a-5p ↑ | miR-30a-5p ↓ | miR-30a-5p ↓ | miR-30a-5p ↓ | miR-30a-5p ↓ | |
| Osteoarthritis | miR-15b-5p ↑ | miR-15b-5p ↑ | miR-15b-5p ↓ | miR-15b-5p ↓ | miR-15b-5p ↓ | |
| Uric acid nephropathy | miR-122-5p ↑ | miR-122-5p ↑ | miR-122-5p ↑ | miR-122-5p ↓ | ||
| Acute pneumonia | miR-146a-5p ↑ | miR-146a-5p ↓ | ||||
| LPS-induced inflammatory injury | miR-17-5p ↓ | miR-17-5p ↓ | miR-17-5p ↓ | miR-17-5p ↓ | miR-17-5p ↓ | |
| Poststroke inflammation | miR-181c-5p ↓ | miR-181c-5p ↓ | miR-181c-5p ↓ | miR-181c-5p ↓ |
| Cytokine | Control vs. Model Groups | Control vs. Noni Fruit Juice Groups | Control vs. Colchicine Groups | Model vs. Noni Fruit Juice Groups | Noni Fruit Juice vs. Colchicine Groups | Model vs. Colchicine Groups |
|---|---|---|---|---|---|---|
| Interleukin-6 (IL-6) | (2↑)/(4↑, 2↓) | (2↓)/(9↑, 1↓) | (2↓)/(1↑, 4↓) | (3↓)/(9↑, 4↓) | (1↑)/(11↓) | (3↓)/(4↓) |
| TNF-α | (4↑)/(52↑, 89↓) | (3↓)/(98↑, 354↓) | (4↓)/(3↑, 119↓) | (5↓)/(116↑, 43↓) | (2↓)/(5↑, 150↓) | (5↓)/(12↑, 81↓) |
| NALP3 | (2↑, 1↓)/(37↑, 99↓) | (2↓)/(118↑, 57↓) | (5↓)/(10↑, 120↓) | (1↑, 3↓)/(149↑, 27↓) | (4↓)/(13↑, 177↓) | (4↓)/(17↑, 64↓) |
| Interleukin-1β (IL-1β) | (3↑, 1↓)/(18↑, 39↓) | (1↑)/(49↑, 21↓) | (2↓)/(2↑, 41↓) | (1↑, 3↓)/(57↑, 11↓) | (2↓)/(3↑, 65↓) | (3↓)/(8↑, 21↓) |
| Monocyte chemoattractant protein-1 (MCP-1) | (1↑)/(11↑, 18↓) | (1↓)/(27↑, 10↓) | (1↓)/(1↑, 20↓) | (2↓)/(31↑, 8↓) | (0)/(34↓) | (2↓)/(11↓) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liu, Y.; Shan, Y.; Wang, Y.; Li, Z.; Bi, Y.; Zhao, W.; Yin, Y.; Wang, T.; Li, S.; et al. MicroRNAs Involved in the Therapeutic Functions of Noni (Morinda citrifolia L.) Fruit Juice in the Treatment of Acute Gouty Arthritis in Mice Induced with Monosodium Urate. Foods 2021, 10, 1638. https://doi.org/10.3390/foods10071638
Li X, Liu Y, Shan Y, Wang Y, Li Z, Bi Y, Zhao W, Yin Y, Wang T, Li S, et al. MicroRNAs Involved in the Therapeutic Functions of Noni (Morinda citrifolia L.) Fruit Juice in the Treatment of Acute Gouty Arthritis in Mice Induced with Monosodium Urate. Foods. 2021; 10(7):1638. https://doi.org/10.3390/foods10071638
Chicago/Turabian StyleLi, Xiaohong, Yue Liu, Yaming Shan, Yukun Wang, Zhandong Li, Yingxin Bi, Weihao Zhao, Yuhe Yin, Tianlong Wang, Shuang Li, and et al. 2021. "MicroRNAs Involved in the Therapeutic Functions of Noni (Morinda citrifolia L.) Fruit Juice in the Treatment of Acute Gouty Arthritis in Mice Induced with Monosodium Urate" Foods 10, no. 7: 1638. https://doi.org/10.3390/foods10071638
APA StyleLi, X., Liu, Y., Shan, Y., Wang, Y., Li, Z., Bi, Y., Zhao, W., Yin, Y., Wang, T., Li, S., Sun, F., Chen, C., & Li, H. (2021). MicroRNAs Involved in the Therapeutic Functions of Noni (Morinda citrifolia L.) Fruit Juice in the Treatment of Acute Gouty Arthritis in Mice Induced with Monosodium Urate. Foods, 10(7), 1638. https://doi.org/10.3390/foods10071638






