Low-Temperature Virus vB_EcoM_VR26 Shows Potential in Biocontrol of STEC O26:H11
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Lytic Spectrum Determination
2.3. pH Tolerance of VR26
2.4. Bacterial Challenge Assay
2.5. Treatment of Inoculated Fresh-Cut Lettuce with VR26
2.6. Statistical Analysis
3. Results and Discussion
3.1. Lytic Spectrum and pH Tolerance of VR26
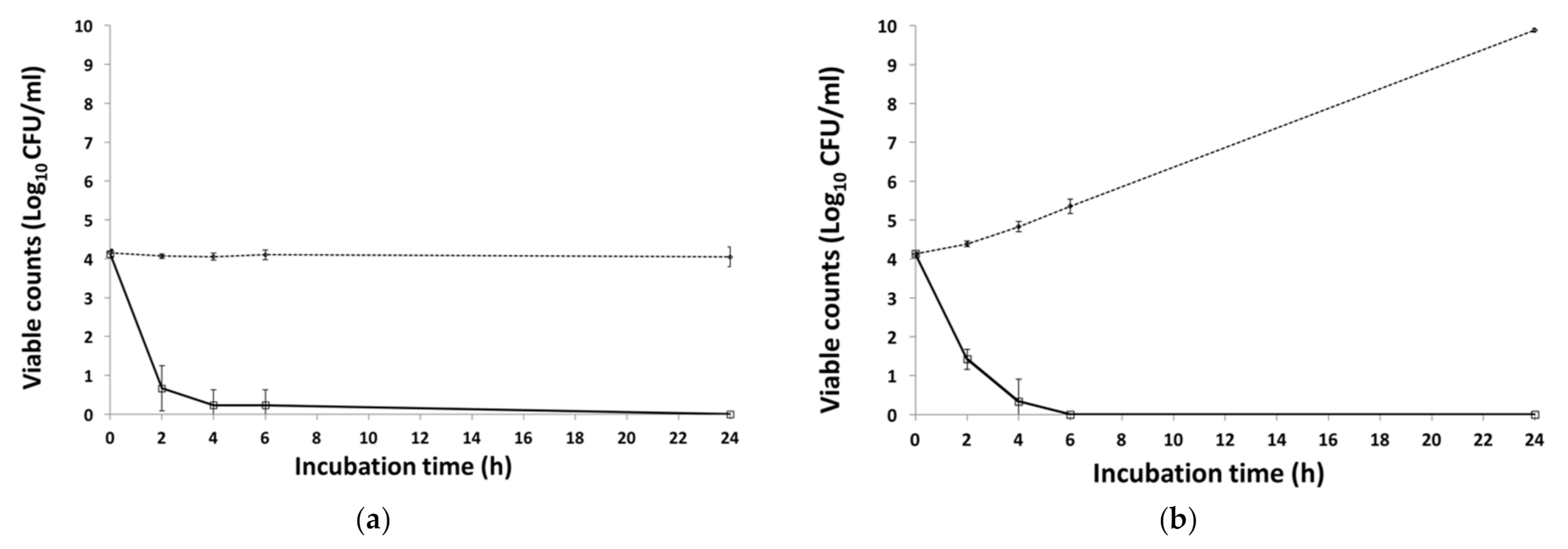
3.2. Bacterial Challenge Assay
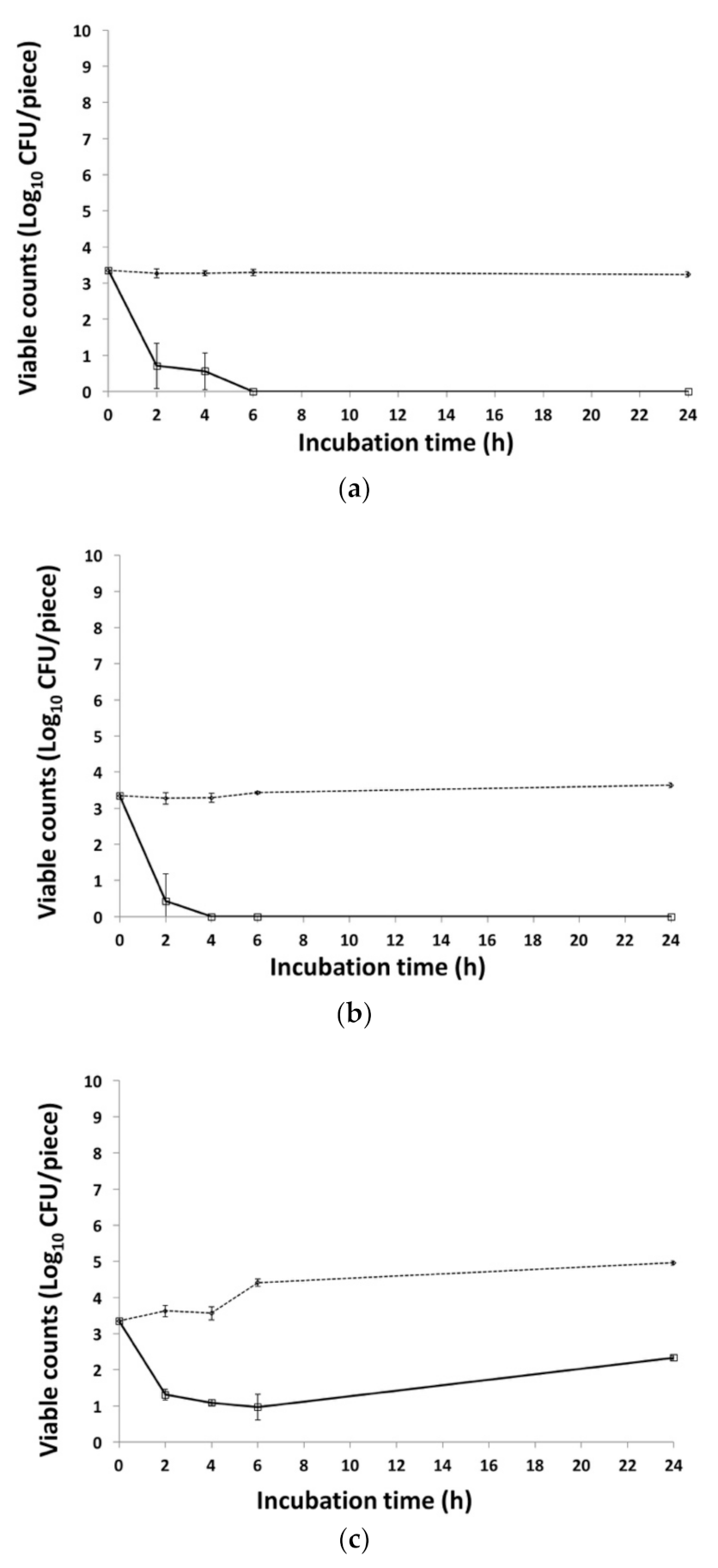
3.3. Effect of VR26 on O26:H11 on Fresh Lettuce
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scheutz, F.; Strockbine, N.A. Escherichia. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Bhunia, A.K. Foodborne Microbial Pathogens: Mechanisms and Pathogenesis, 2nd ed.; Heldman, D.R., Ed.; Springer: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- World Health Organization; Food and Agriculture Organization of the United Nations. Shiga Toxin-Producing Escherichia coli (STEC) and Food: Attribution, Characterization, and Monitoring: Report 2018. Available online: https://apps.who.int/iris/handle/10665/272871 (accessed on 30 May 2021).
- World Health Organization; Food and Agriculture Organization of the United Nations. Attributing Illness Caused by Shiga Toxin-Producing Escherichia coli (STEC) to Specific Foods: Report 2019. Available online: https://apps.who.int/iris/handle/10665/326923 (accessed on 30 May 2021).
- Hoagland, L.; Ximenes, E.; Ku, S.; Ladisch, M. Foodborne pathogens in horticultural production systems: Ecology and mitigation. Sci. Hortic. 2018, 236, 192–206. [Google Scholar] [CrossRef]
- Yu, K.; Newman, M.C.; Archbold, D.D.; Hamilton-Kemp, T.R. Survival of Escherichia coli O157:H7 on Strawberry Fruit and Reduction of the Pathogen Population by Chemical Agents. J. Food Prot. 2001, 64, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, B.V.; Kitchens, S.; Vijayakumar, P.P.; Price, S.; Morgan, M. Potential for Bacteriophage Cocktail to Complement Commercial Sanitizer Use on Produce against Escherichia coli O157:H7. Microorganisms 2020, 8, 1316. [Google Scholar] [CrossRef] [PubMed]
- Balali, G.I.; Yar, D.; Dela, V.G.A.; Adjei-Kusi, P. Microbial Contamination, an Increasing Threat to the Consumption of Fresh Fruits and Vegetables in Today’s World. Int. J. Microbiol. 2020, 2020, 3029295. [Google Scholar] [CrossRef] [PubMed]
- Hess, T.; Sutcliffe, C. The exposure of a fresh fruit and vegetable supply chain to global water-related risks. Water Int. 2018, 43, 746–761. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.A.; Gooneratne, S.R. Understanding the Fresh Produce Safety Challenges. Foods 2017, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- Luna-Guevara, J.J.; Arenas-Hernandez, M.M.P.; de la Peña, C.M.; Silva, J.L.; Luna-Guevara, M.L. The Role of PathogenicE. coliin Fresh Vegetables: Behavior, Contamination Factors, and Preventive Measures. Int. J. Microbiol. 2019, 2019, 1–10. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority. Urgent advice on the public health risk of Shiga-toxin producing Escherichia coli in fresh vegetables. EFSA J. 2011, 9, 2274. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [Green Version]
- Lang, L.H. FDA Approves Use of Bacteriophages to be Added to Meat and Poultry Products. Gastroenterology 2006, 131, 1370. [Google Scholar] [CrossRef]
- Pinto, G.; Almeida, C.; Azeredo, J. Bacteriophages to control Shiga toxin-producing E. coli—Safety and regulatory challenges. Crit. Rev. Biotechnol. 2020, 40, 1081–1097. [Google Scholar] [CrossRef]
- Ferguson, S.; Roberts, C.; Handy, E.; Sharma, M. Lytic bacteriophages reduce Escherichia coliO157: H7 on fresh cut lettuce introduced through cross-contamination. Bacteriophage 2013, 3, e24323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duc, H.M.; Son, H.M.; Yi, H.P.S.; Sato, J.; Ngan, P.H.; Masuda, Y.; Honjoh, K.-I.; Miyamoto, T. Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157:H7 in different food matrices. Food Res. Int. 2020, 131, 108977. [Google Scholar] [CrossRef]
- Marshall, K.E.; Hexemer, A.; Seelman, S.L.; Fatica, M.K.; Blessington, T.; Hajmeer, M.; Kisselburgh, H.; Atkinson, R.; Hill, K.; Sharma, D.; et al. Lessons Learned from a Decade of Investigations of Shiga Toxin–Producing Escherichia coli Outbreaks Linked to Leafy Greens, United States and Canada. Emerg. Infect. Dis. 2020, 26, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Mariani-Kurkdjian, P.; Bonacorsi, S.; Liguori, S.; Fach, P. Characteristics of Emerging Human-Pathogenic Escherichia coli O26:H11 Strains Isolated in France between 2010 and 2013 and Carrying thestx2dGene Only. J. Clin. Microbiol. 2014, 53, 486–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallman, T.J.; Greig, D.R.; Gharbia, S.E.; Jenkins, C. Phylogenetic context of Shiga toxin-producing Escherichia coli serotype O26:H11 in England. Microb. Genom. 2019, 7, 000551. [Google Scholar] [CrossRef]
- Ogura, Y.; Gotoh, Y.; Itoh, T.; Sato, M.P.; Seto, K.; Yoshino, S.; Isobe, J.; Etoh, Y.; Kurogi, M.; Kimata, K.; et al. Population structure of Escherichia coli O26:H11 with recent and repeated stx2 acquisition in multiple lineages. Microb. Genom. 2017, 3, 000141. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 14, 4634. [Google Scholar]
- Bumunang, E.W.; McAllister, T.A.; Stanford, K.; Anany, H.; Niu, Y.D.; Ateba, C.N. Characterization of Non-O157 STEC Infecting Bacteriophages Isolated from Cattle Faeces in North-West South Africa. Microorganisms 2019, 7, 615. [Google Scholar] [CrossRef] [Green Version]
- Litt, P.K.; Saha, J.; Jaroni, D. Characterization of Bacteriophages Targeting Non-O157 Shiga Toxigenic Escherichia coli. J. Food Prot. 2018, 81, 785–794. [Google Scholar] [CrossRef]
- Wang, J.; Niu, Y.D.; Chen, J.; Anany, H.; Ackermann, H.-W.; Johnson, R.P.; Ateba, C.N.; Stanford, K.; McAllister, T.A. Feces of feedlot cattle contain a diversity of bacteriophages that lyse non-O157 Shiga toxin-producing Escherichia coli. Can. J. Microbiol. 2015, 61, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.-T.; Sun, X.; Quintela, I.A.; Bridges, D.F.; Liu, F.; Zhang, Y.; Salvador, A.; Wu, V.C.H. Discovery of Shiga Toxin-Producing Escherichia coli (STEC)-Specific Bacteriophages from Non-fecal Composts Using Genomic Characterization. Front. Microbiol. 2019, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.D.; Liu, H.; Johnson, R.P.; McAllister, T.A.; Stanford, K. Effect of a bacteriophage T5virus on growth of Shiga toxigenic Escherichia coli and Salmonella strains in individual and mixed cultures. Virol. J. 2020, 17, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangieri, N.; Picozzi, C.; Cocuzzi, R.; Foschino, R. Evaluation of a Potential Bacteriophage Cocktail for the Control of Shiga-Toxin Producing Escherichia coli in Food. Front. Microbiol. 2020, 11, 1801. [Google Scholar] [CrossRef] [PubMed]
- Kaliniene, L.; Zajančkauskaite, A.; Šimoliūnas, E.; Truncaitė, L.; Meškys, R. Low-temperature bacterial viruses VR—A small but diverse group of E. coli phages. Arch. Virol. 2015, 160, 1367–1370. [Google Scholar] [CrossRef]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef]
- E Selick, H.; Kreuzer, K.N.; Alberts, B.M. The bacteriophage T4 insertion/substitution vector system. A method for introducing site-specific mutations into the virus chromosome. J. Biol. Chem. 1988, 263, 11336–11347. [Google Scholar] [CrossRef]
- Kutter, E.; Sulakvelidze, A. Bacteriophages: Biology and Applications, 1st ed.; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; pp. 69–76. [Google Scholar]
- Hiramatsu, R.; Matsumoto, M.; Sakae, K.; Miyazaki, Y. Ability of Shiga Toxin-Producing Escherichia coli and Salmonella spp. to Survive in a Desiccation Model System and in Dry Foods. Appl. Environ. Microbiol. 2005, 71, 6657–6663. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Choi, I.Y.; Park, D.H.; Park, M.-K. Isolation and characterization of a novel Escherichia coli O157:H7-specific phage as a biocontrol agent. J. Environ. Health Sci. Eng. 2020, 18, 189–199. [Google Scholar] [CrossRef]
- Ramirez, K.; Cazarez-Montoya, C.; Lopez-Moreno, H.S.; Campo, N.C.-D. Bacteriophage cocktail for biocontrol of Escherichia coli O157:H7: Stability and potential allergenicity study. PLoS ONE 2018, 13, e0195023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Ku, H.-J.; Lee, D.-H.; Kim, Y.-T.; Shin, H.; Ryu, S.; Lee, J.-H. Characterization and Genomic Study of the Novel Bacteriophage HY01 Infecting Both Escherichia coli O157:H7 and Shigella flexneri: Potential as a Biocontrol Agent in Food. PLoS ONE 2016, 11, e0168985. [Google Scholar] [CrossRef] [Green Version]
- Filho, R.L.A.; Higgins, J.P.; Higgins, S.E.; Gaona, G.; Wolfenden, A.D.; Tellez, G.; Hargis, B.M. Ability of Bacteriophages Isolated from Different Sources to Reduce Salmonella enterica Serovar Enteritidis In Vitro and In Vivo. Poult. Sci. 2007, 86, 1904–1909. [Google Scholar] [CrossRef] [PubMed]
- Bach, S.J.; McAllister, T.A.; Veira, D.M.; Gannon, V.P.; Holley, R.A. Effect of bacteriophage DC22 on Escherichia coli O157:H7 in an artificial rumen system (Rusitec) and inoculated sheep. Anim. Res. 2003, 52, 89–101. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, A.B.; Perry, J.J.; Yousef, A.E. Developing and optimizing bacteriophage treatment to control enterohemorrhagic Escherichia coli on fresh produce. Int. J. Food Microbiol. 2016, 236, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Patel, J.R.; Conway, W.S.; Ferguson, S.; Sulakvelidze, A. Effectiveness of Bacteriophages in Reducing Escherichia coli O157:H7 on Fresh-Cut Cantaloupes and Lettuce†. J. Food Prot. 2009, 72, 1481–1485. [Google Scholar] [CrossRef] [Green Version]
- Minh, D.H.; Minh, S.H.; Honjoh, K.-I.; Miyamoto, T. Isolation and bio-control of Extended Spectrum Beta-Lactamase (ESBL)-producing Escherichia coli contamination in raw chicken meat by using lytic bacteriophages. LWT 2016, 71, 339–346. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Q.; Zhang, C.; Yang, J.; Lu, Z.; Lu, F.; Bie, X. Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 2017, 236, 14–23. [Google Scholar] [CrossRef]
- Guenther, S.; Herzig, O.; Fieseler, L.; Klumpp, J.; Loessner, M.J. Biocontrol of Salmonella Typhimurium in RTE foods with the virulent bacteriophage FO1-E2. Int. J. Food Microbiol. 2012, 154, 66–72. [Google Scholar] [CrossRef]
- Bigot, B.; Lee, W.-J.; McIntyre, L.; Wilson, T.; Hudson, J.; Billington, C.; Heinemann, J. Control of Listeria monocytogenes growth in a ready-to-eat poultry product using a bacteriophage. Food Microbiol. 2011, 28, 1448–1452. [Google Scholar] [CrossRef]
- Hagens, S.; Offerhaus, M.L. Bacteriophages-New weapons for food safety. Food Technol. 2008, 62, 46–54. Available online: https://www.ift.org/news-and-publications/food-technology-magazine/issues/2008/april/features/bacteriophages-new-weapons-for-food-safety (accessed on 30 May 2021).
- Hudson, J.; Billington, C.; Cornelius, A.; Wilson, T.; On, S.; Premaratne, A.; King, N. Use of a bacteriophage to inactivate Escherichia coli O157:H7 on beef. Food Microbiol. 2013, 36, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Phage therapy dosing: The problem(s) with multiplicity of infection (MOI). Bacteriophage 2016, 6, e1220348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulakvelidze, A. Using lytic bacteriophages to eliminate or significantly reduce contamination of food by foodborne bacterial pathogens. J. Sci. Food Agric. 2013, 93, 3137–3146. [Google Scholar] [CrossRef] [PubMed]

| Strain/Serotype | Spot Test | EOP | ||
|---|---|---|---|---|
| 8 °C | 22 °C | 8 °C | 22 °C | |
| O26:H11 | + | + | 0.89 | 1.0 |
| O157:H7 | - | - | - | - |
| O103:H2 | - | - | - | - |
| O111:H8 | - | - | - | - |
| O145:H128 | - | - | - | - |
| MH1 1 | + | + | 0.84 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zajančkauskaitė, A.; Noreika, A.; Rutkienė, R.; Meškys, R.; Kaliniene, L. Low-Temperature Virus vB_EcoM_VR26 Shows Potential in Biocontrol of STEC O26:H11. Foods 2021, 10, 1500. https://doi.org/10.3390/foods10071500
Zajančkauskaitė A, Noreika A, Rutkienė R, Meškys R, Kaliniene L. Low-Temperature Virus vB_EcoM_VR26 Shows Potential in Biocontrol of STEC O26:H11. Foods. 2021; 10(7):1500. https://doi.org/10.3390/foods10071500
Chicago/Turabian StyleZajančkauskaitė, Aurelija, Algirdas Noreika, Rasa Rutkienė, Rolandas Meškys, and Laura Kaliniene. 2021. "Low-Temperature Virus vB_EcoM_VR26 Shows Potential in Biocontrol of STEC O26:H11" Foods 10, no. 7: 1500. https://doi.org/10.3390/foods10071500
APA StyleZajančkauskaitė, A., Noreika, A., Rutkienė, R., Meškys, R., & Kaliniene, L. (2021). Low-Temperature Virus vB_EcoM_VR26 Shows Potential in Biocontrol of STEC O26:H11. Foods, 10(7), 1500. https://doi.org/10.3390/foods10071500







