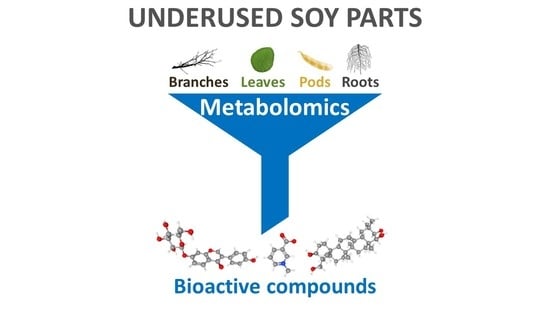
Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds
Abstract
:1. Introduction
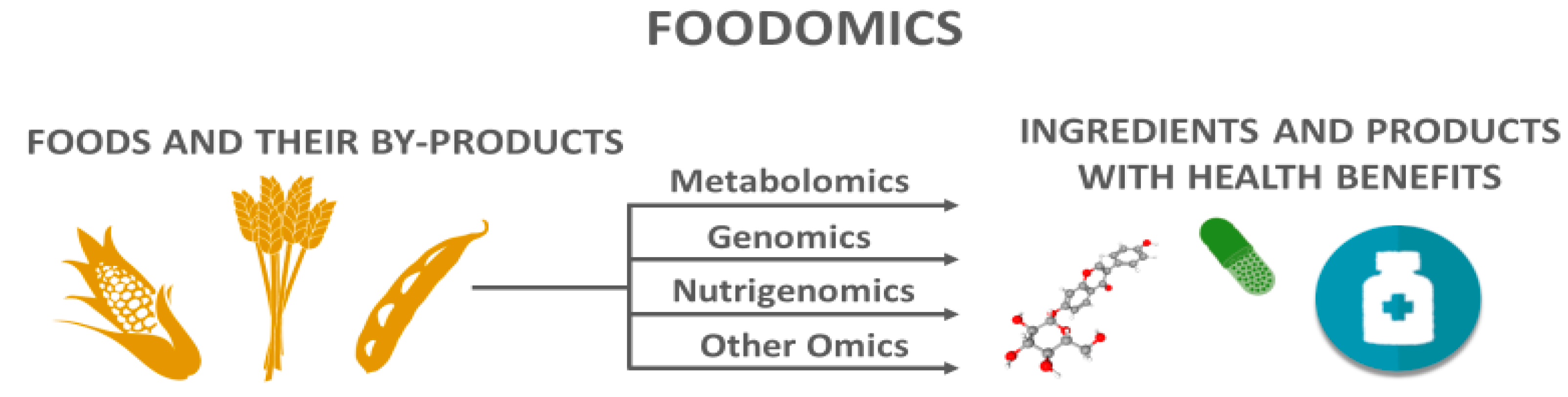
1.1. Metabolomics Applied to Agri-Foods and Their By-Products
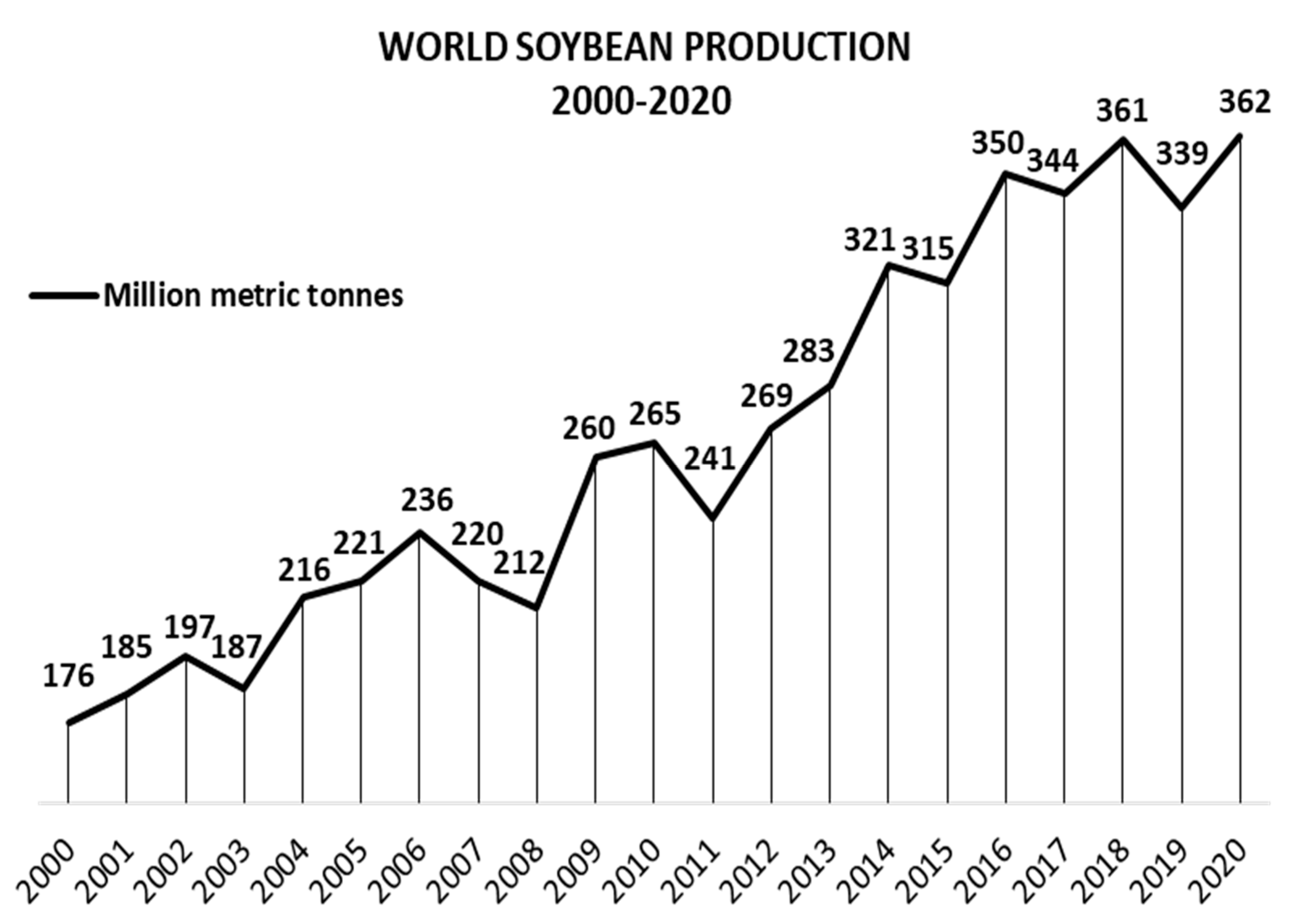
1.2. Glycine max: More Than Beans
2. Metabolomics and Soy
An Overview
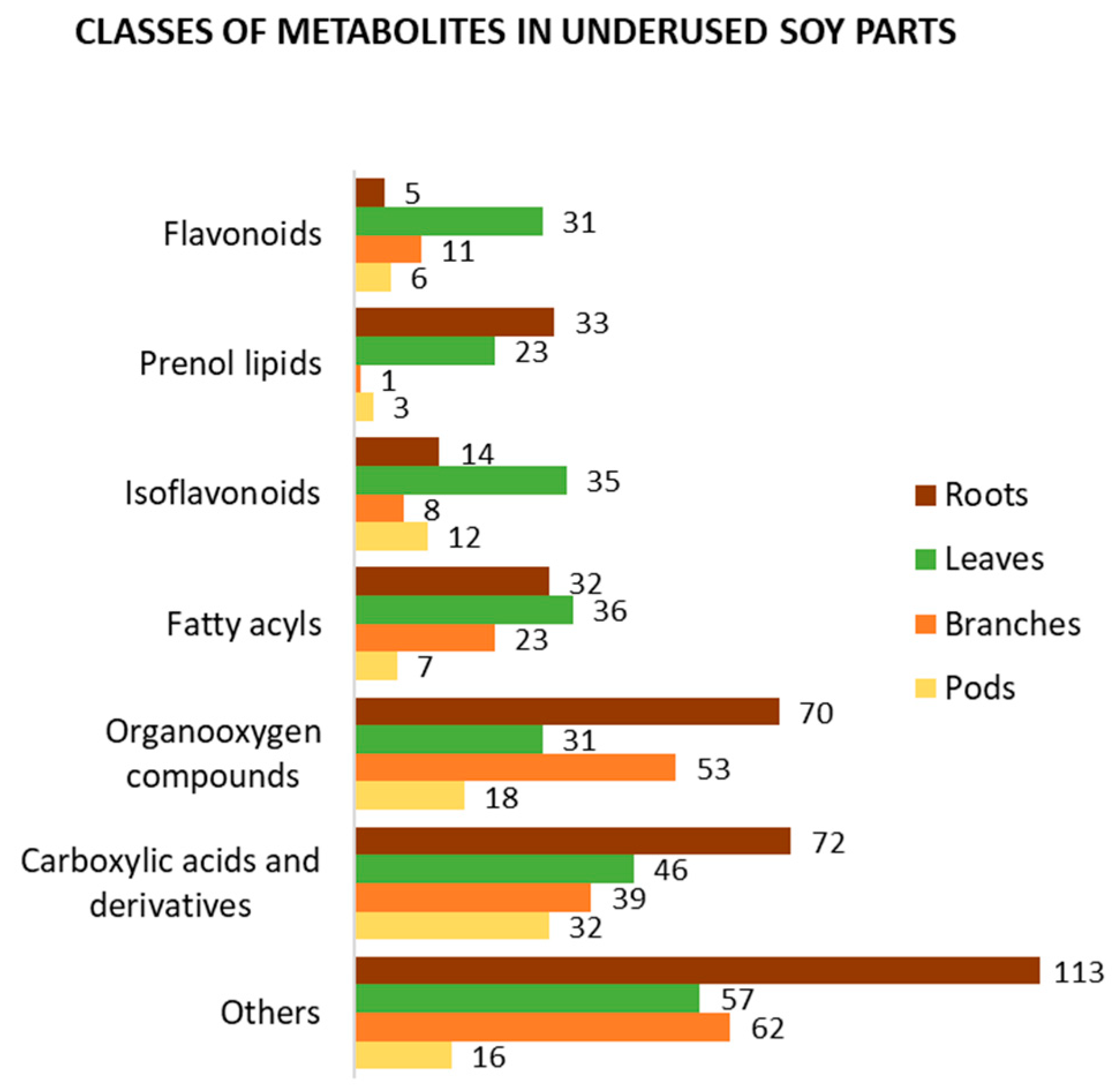
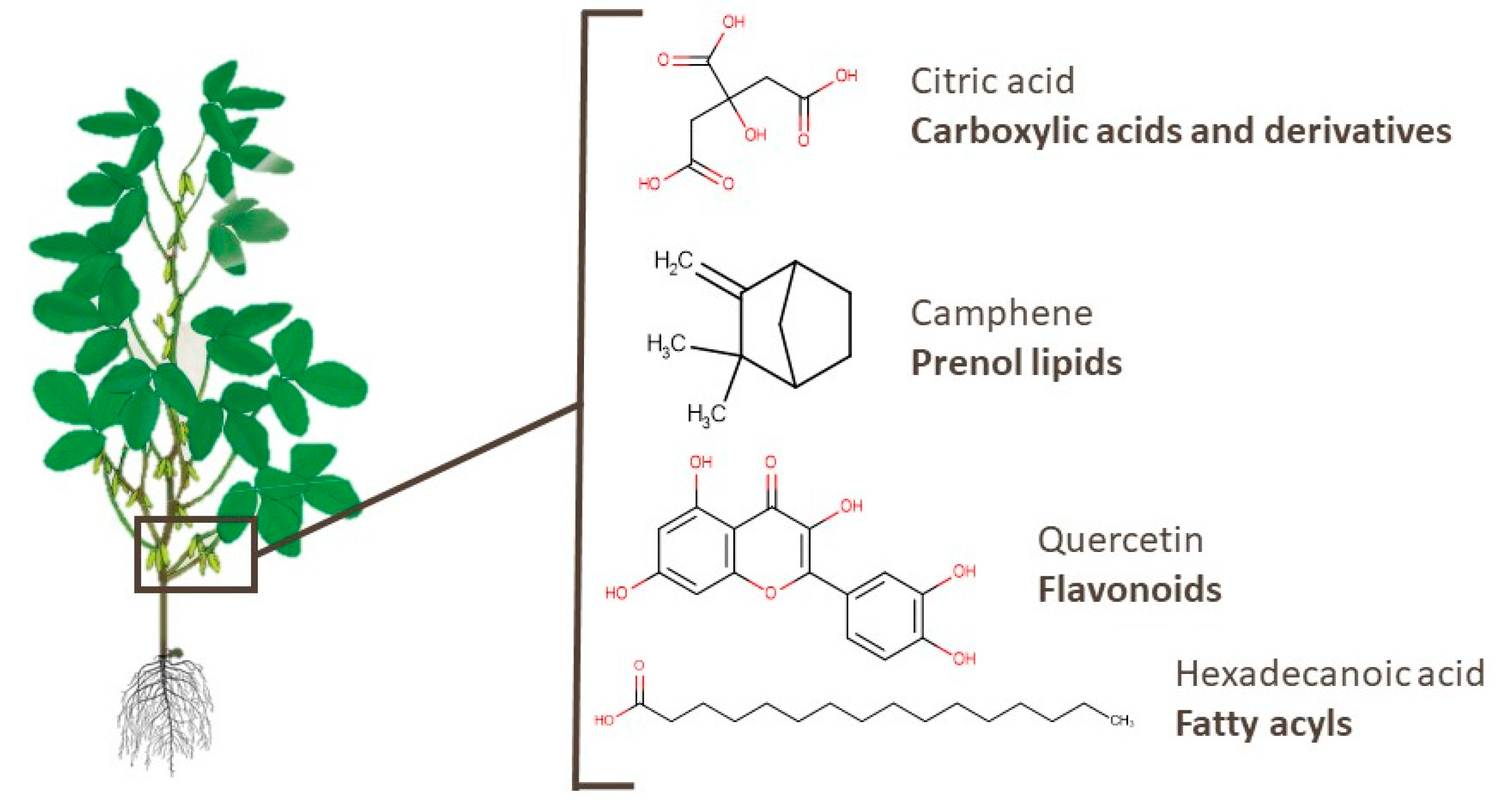
3. Bioactive Compounds in Underused Soy Parts
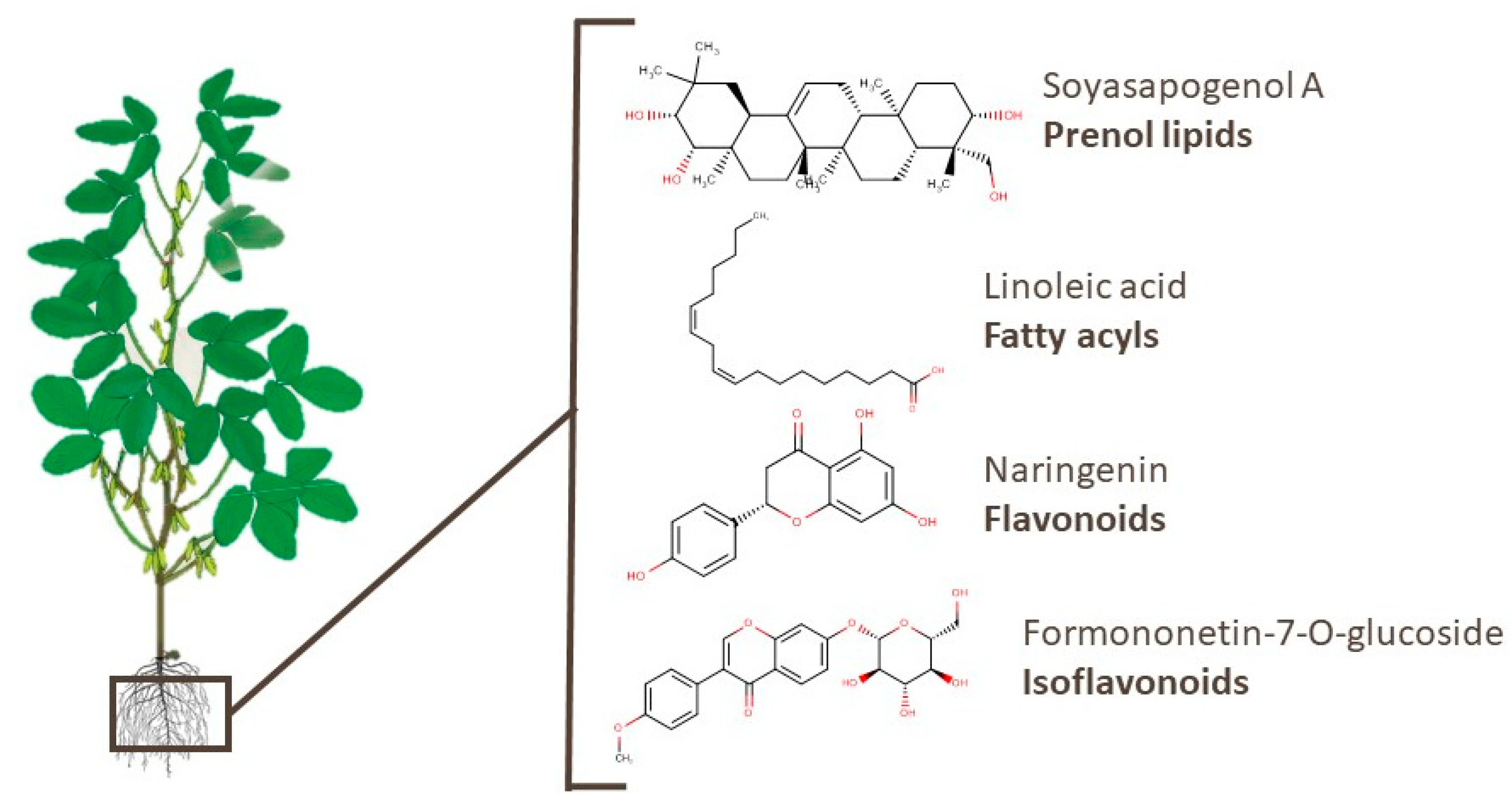
3.1. Roots
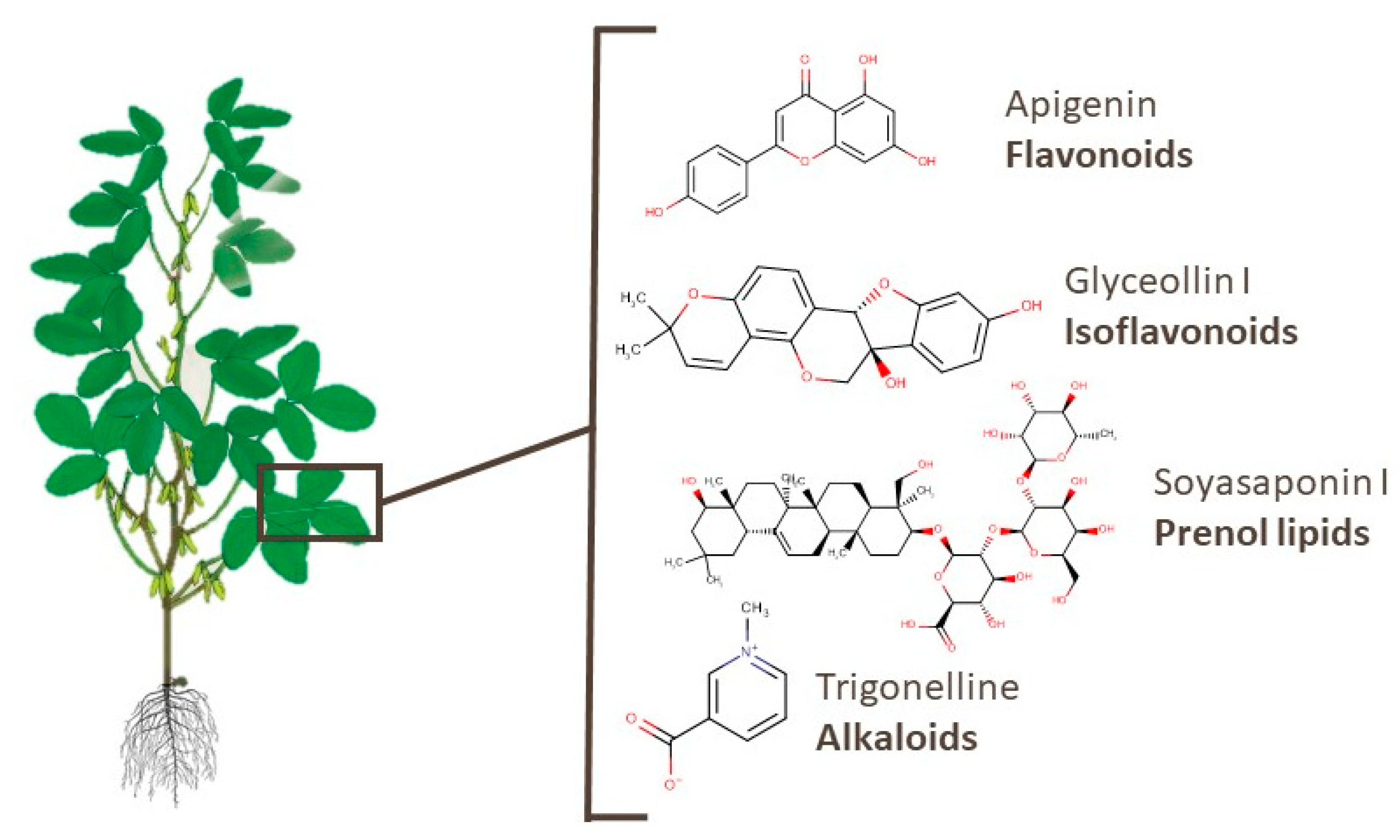
3.2. Leaves
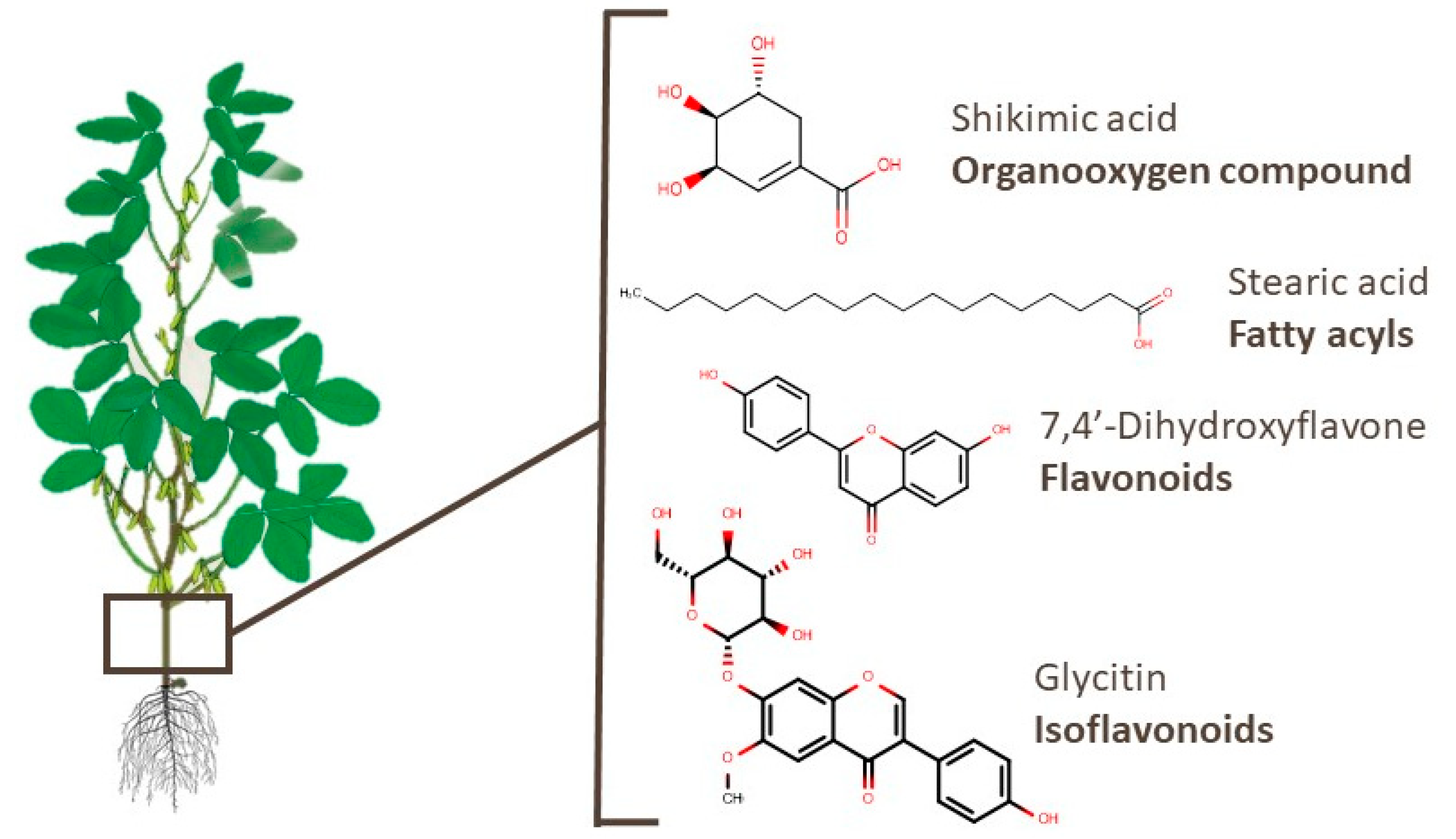
3.3. Branches
3.4. Pods
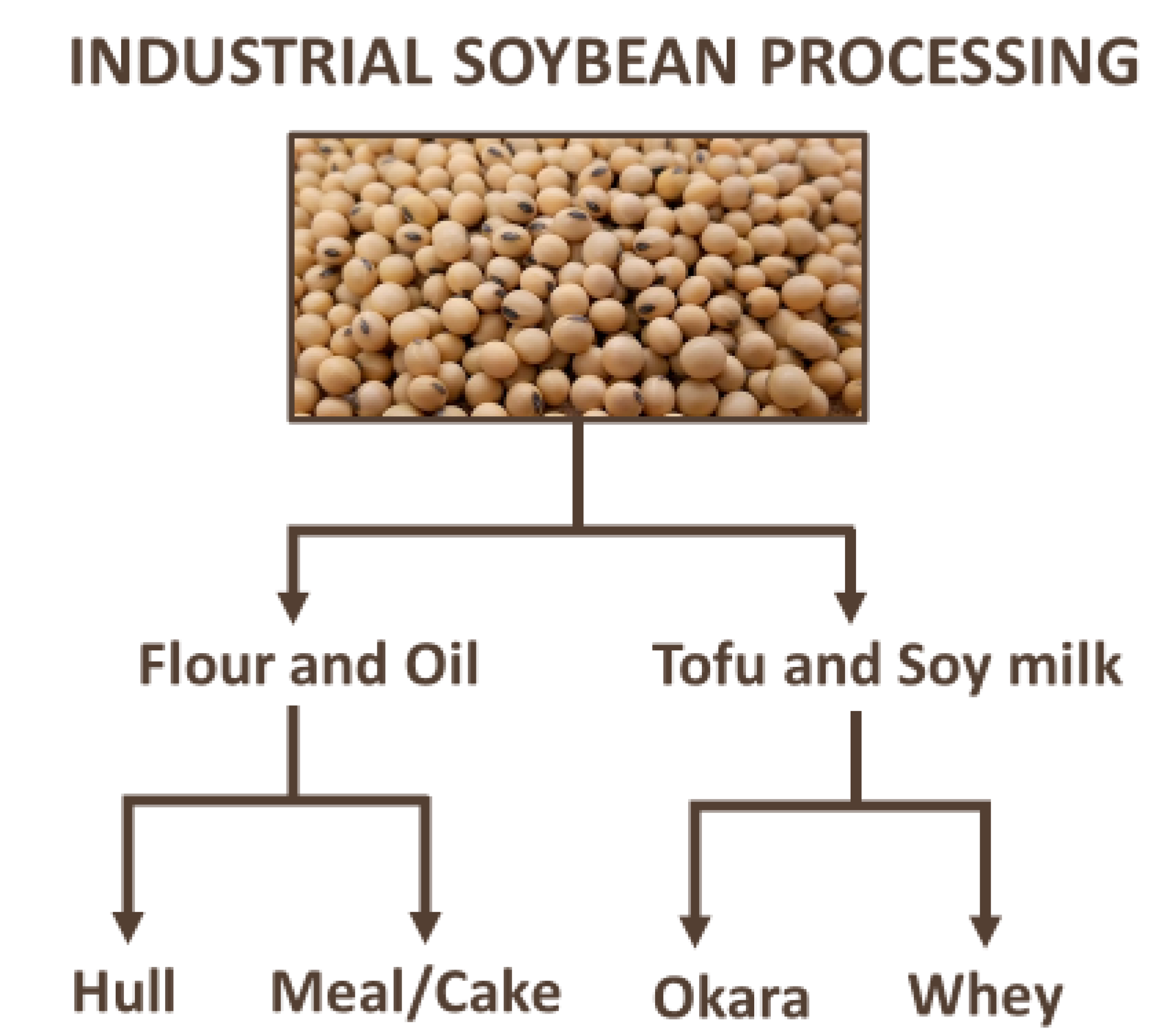
4. Bioactive Compounds in Industrial By-Products from Soybean Processing—An Overview and Trends
5. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Atanasov, A.G.; the International Natural Product Sciences Taskforce; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Chemat, F.; Vian, M.A.; Fabiano-Tixier, A.-S.; Nutrizio, M.; Jambrak, A.R.; Munekata, P.E.S.; Lorenzo, J.M.; Barba, F.J.; Binello, A.; Cravotto, G. A review of sustainable and intensified techniques for extraction of food and natural products. Green Chem. 2020, 22, 2325–2353. [Google Scholar] [CrossRef] [Green Version]
- Armenta, S.; Garrigues, S.; Esteve-Turrillas, F.A.; de la Guardia, M. Green extraction techniques in green analytical chemistry. TrAC Trends Anal. Chem. 2019, 116, 248–253. [Google Scholar] [CrossRef]
- Alvarez-Rivera, G.; Bueno, M.; Ballesteros-Vivas, D.; Mendiola, J.A.; Ibañez, E. Pressurized liquid extraction. In Liquid-Phase Extraction; Poole, C.F., Ed.; Handbooks in Separation Science; Elsevier: Amsterdam, The Netherlands, 2019; pp. 375–398. ISBN 9780128169117. [Google Scholar]
- Chai, Y.; Yusup, S.; Kadir, W.; Wong, C.; Rosli, S.; Ruslan, M.; Chin, B.; Yiin, C. Valorization of Tropical Biomass Waste by Supercritical Fluid Extraction Technology. Sustainability 2020, 13, 233. [Google Scholar] [CrossRef]
- Sánchez-Camargo, A.D.P.; Parada-Alonso, F.; Ibáñez, E.; Cifuentes, A. Recent applications of on-line supercritical fluid extraction coupled to advanced analytical techniques for compounds extraction and identification. J. Sep. Sci. 2019, 42, 243–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keppler, E.A.H.; Jenkins, C.; Davis, T.J.; Bean, H.D. Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. TrAC Trends Anal. Chem. 2018, 109, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Pirok, B.W.J.; Stoll, D.R.; Schoenmakers, P.J. Recent Developments in Two-Dimensional Liquid Chromatography: Fundamental Improvements for Practical Applications. Anal. Chem. 2019, 91, 240–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, G.; Montero, L.; Llorens, L.; Castro-Puyana, M.; Cifuentes, A. Recent advances in the application of capillary electromigration methods for food analysis and Foodomics. Electrophoresis 2018, 39, 136–159. [Google Scholar] [CrossRef]
- Alvarez-Rivera, G.; Ballesteros-Vivas, D.; Parada-Alfonso, F.; Ibañez, E.; Cifuentes, A. Recent applications of high resolution mass spectrometry for the characterization of plant natural products. TrAC Trends Anal. Chem. 2019, 112, 87–101. [Google Scholar] [CrossRef]
- Aksenov, A.A.; Da Silva, R.; Knight, R.; Lopes, N.P.; Dorrestein, P.C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017, 1, 0054. [Google Scholar] [CrossRef]
- McAlpine, J.B.; Chen, S.-N.; Kutateladze, A.; MacMillan, J.B.; Appendino, G.; Barison, A.; Beniddir, M.A.; Biavatti, M.W.; Bluml, S.; Boufridi, A.; et al. The value of universally available raw NMR data for transparency, reproducibility, and integrity in natural product research. Nat. Prod. Rep. 2019, 36, 35–107. [Google Scholar] [CrossRef] [Green Version]
- Sorokina, M.; Steinbeck, C. Review on natural products databases: Where to find data in 2020. J. Chemin 2020, 12, 1–51. [Google Scholar] [CrossRef] [Green Version]
- Medema, M.H. The year 2020 in natural product bioinformatics: An overview of the latest tools and databases. Nat. Prod. Rep. 2021, 38, 301–306. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Reher, R.; Kim, H.W.; Zhang, C.; Mao, H.H.; Wang, M.; Nothias, L.-F.; Caraballo-Rodriguez, A.M.; Glukhov, E.; Teke, B.; Leao, T.; et al. A Convolutional Neural Network-Based Approach for the Rapid Annotation of Molecularly Diverse Natural Products. J. Am. Chem. Soc. 2020, 142, 4114–4120. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 2 February 2021).
- Ishangulyyev, R.; Kim, S.; Lee, S.H. Understanding Food Loss and Waste—Why Are We Losing and Wasting Food? Foods 2019, 8, 297. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.S.; Bharadwaj, S. Prospects for Detecting the 326.5 MHz Redshifted 21-Cm HI Signal with the Ooty Radio Telescope (ORT); Food and Agriculture Organization of the United Nations: Rome, Italy, 2014; Volume 35, ISBN 9789251072059. [Google Scholar]
- UN DP Goal 12: Responsible Consumption and Production; United Nations Development Programme: New York, NY, USA, 2016.
- Cifuentes, A. Food analysis and Foodomics. J. Chromatogr. A 2009, 1216, 7109. [Google Scholar] [CrossRef] [Green Version]
- Valdés, A.; Cifuentes, A.; León, C. Foodomics evaluation of bioactive compounds in foods. TrAC Trends Anal. Chem. 2017, 96, 2–13. [Google Scholar] [CrossRef]
- Katsinas, N.; da Silva, A.B.; Enríquez-De-Salamanca, A.; Fernández, N.; Bronze, M.R.; Rodríguez-Rojo, S. Pressurized Liquid Extraction Optimization from Supercritical Defatted Olive Pomace: A Green and Selective Phenolic Extraction Process. ACS Sustain. Chem. Eng. 2021, 9, 5590–5602. [Google Scholar] [CrossRef]
- Assirati, J.; Rinaldo, D.; Rabelo, S.C.; Bolzani, V.D.S.; Hilder, E.F.; Funari, C.S. A green, simplified, and efficient experimental setup for a high-throughput screening of agri-food by-products—From polar to nonpolar metabolites in sugarcane solid residues. J. Chromatogr. A 2020, 1634, 461693. [Google Scholar] [CrossRef]
- Sánchez-Martínez, J.D.; Bueno, M.; Alvarez-Rivera, G.; Tudela, J.; Ibañez, E.; Cifuentes, A. In vitro neuroprotective potential of terpenes from industrial orange juice by-products. Food Funct. 2021, 12, 302–314. [Google Scholar] [CrossRef]
- Agrawal, M.; Ajazuddin; Tripathi, D.K.; Saraf, S.; Saraf, S.; Antimisiaris, S.G.; Mourtas, S.; Hammarlund-Udenaes, M.; Alexander, A. Recent advancements in liposomes targeting strategies to cross blood-brain barrier (BBB) for the treatment of Alzheimer’s disease. J. Control. Release 2017, 260, 61–77. [Google Scholar] [CrossRef]
- Anderson, E.J.; Ali, M.L.; Beavis, W.D.; Chen, P.; Clemente, T.E.; Diers, B.W.; Graef, G.L.; Grassini, P.; Hyten, D.L.; McHale, L.K.; et al. Soybean [Glycine max (L.) Merr.] Breeding: History, Improvement, Production and Future Opportunities. In Advances in Plant Breeding Strategies: Legumes; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 7, pp. 431–516. ISBN 978-3-030-23400-3. [Google Scholar]
- United Nations United Nations Statistics Division-Environment Statistics. Available online: https://unstats.un.org/unsd/environment/totalarea.htm (accessed on 20 March 2021).
- United States Department of Agriculture. Oilseeds: World Markets and Trade. 2021. Available online: https://downloads.usda.library.cornell.edu/usda-esmis/files/tx31qh68h/hq37wg66q/q237jm44p/oilseeds.pdf (accessed on 20 March 2021).
- United States Department of Agriculture. Oilseeds: World Markets and Trade. 2011. Available online: https://downloads.usda.library.cornell.edu/usda-esmis/files/tx31qh68h/w3763722n/db78tc388/oilseed-trade-03-10-2011.pdf (accessed on 20 March 2021).
- Krisnawati, A.; Adie, M.M. Variability of Biomass and Harvest Index from Several Soybean Genotypes as Renewable Energy Source. Energy Procedia 2015, 65, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Buffett, H.G. Conservation: Reaping the Benefits of No-Tillage Farming. Nature 2012, 484, 455. [Google Scholar] [CrossRef]
- Gawęda, D.; Haliniarz, M.; Bronowicka-Mielniczuk, U.; Łukasz, J. Weed Infestation and Health of the Soybean Crop Depending on Cropping System and Tillage System. Agriculture 2020, 10, 208. [Google Scholar] [CrossRef]
- De Castro, S.G.Q.; Dinardo-Miranda, L.L.; Fracasso, J.V.; Bordonal, R.O.; Menandro, L.; Franco, H.C.J.; Carvalho, J.L.N. Changes in Soil Pest Populations Caused by Sugarcane Straw Removal in Brazil. BioEnergy Res. 2019, 12, 878–887. [Google Scholar] [CrossRef]
- Zhang, J.; Hang, X.; Lamine, S.M.; Jiang, Y.; Afreh, D.; Qian, H.; Feng, X.; Zheng, C.; Deng, A.; Song, Z.; et al. Interactive effects of straw incorporation and tillage on crop yield and greenhouse gas emissions in double rice cropping system. Agric. Ecosyst. Environ. 2017, 250, 37–43. [Google Scholar] [CrossRef]
- Popin, G.V.; Santos, A.K.B.; Oliveira, T.D.P.; De Camargo, P.B.; Cerri, C.E.; Siqueira-Neto, M. Sugarcane straw management for bioenergy: Effects of global warming on greenhouse gas emissions and soil carbon storage. Mitig. Adapt. Strat. Glob. Chang. 2020, 25, 559–577. [Google Scholar] [CrossRef]
- Vasconcelos, A.L.S.; Cherubin, M.R.; Feigl, B.J.; Cerri, C.E.; Gmach, M.R.; Siqueira-Neto, M. Greenhouse gas emission responses to sugarcane straw removal. Biomass Bioenergy 2018, 113, 15–21. [Google Scholar] [CrossRef]
- Carneiro, A.M.; Moreira, E.A.; Bragagnolo, F.S.; Borges, M.S.; Pilon, A.C.; Rinaldo, D.; De Funari, C.S. Soya agricultural waste as a rich source of isoflavones. Food Res. Int. 2020, 130, 108949. [Google Scholar] [CrossRef]
- Chatterjee, C.; Gleddie, S.; Xiao, C.-W. Soybean Bioactive Peptides and Their Functional Properties. Nutrients 2018, 10, 1211. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-M.; Yoon, Y.; Yoon, H.; Park, H.-M.; Song, S.; Yeum, K.-J. Dietary Anthocyanins against Obesity and Inflammation. Nutrients 2017, 9, 1089. [Google Scholar] [CrossRef] [Green Version]
- Křížová, L.; Dadáková, K.; Kašparovská, J.; Kašparovský, T. Isoflavones. Molecules 2019, 24, 1076. [Google Scholar] [CrossRef] [Green Version]
- Messina, M. Soy and Health Update: Evaluation of the Clinical and Epidemiologic Literature. Nutrients 2016, 8, 754. [Google Scholar] [CrossRef] [Green Version]
- Landete, J.M.; Arques, J.L.; Medina, M.; Gaya, P.; Rivas, B.D.L.; Muñoz, R. Bioactivation of Phytoestrogens: Intestinal Bacteria and Health. Crit. Rev. Food Sci. Nutr. 2016, 56, 1826–1843. [Google Scholar] [CrossRef]
- Yamagata, K. Soy Isoflavones Inhibit Endothelial Cell Dysfunction and Prevent Cardiovascular Disease. J. Cardiovasc. Pharmacol. 2019, 74, 201–209. [Google Scholar] [CrossRef]
- Pabich, M.; Materska, M. Biological Effect of Soy Isoflavones in the Prevention of Civilization Diseases. Nutrients 2019, 11, 1660. [Google Scholar] [CrossRef] [Green Version]
- Thangavel, P.; Puga-Olguín, A.; Rodríguez-Landa, J.F.; Zepeda, R.C. Genistein as Potential Therapeutic Candidate for Menopausal Symptoms and Other Related Diseases. Molecules 2019, 24, 3892. [Google Scholar] [CrossRef] [Green Version]
- Taku, K.; Melby, M.K.; Nishi, N.; Omori, T.; Kurzer, M.S. Soy isoflavones for osteoporosis: An evidence-based approach. Maturitas 2011, 70, 333–338. [Google Scholar] [CrossRef]
- Moreira, E.A.; Pilon, A.C.; Andrade, L.E.; Lopes, N.P. New Perspectives on Chlorogenic Acid Accumulation in Harvested Leaf Tissue: Impact on Traditional Medicine Preparations. ACS Omega 2018, 3, 18380–18386. [Google Scholar] [CrossRef]
- Neilson, E.H.; Goodger, J.Q.; Woodrow, I.E.; Møller, B.L. Plant chemical defense: At what cost? Trends Plant Sci. 2013, 18, 250–258. [Google Scholar] [CrossRef]
- Gottlieb, O.R. Phytochemicals: Differentiation and function. Phytochemistry 1990, 29, 1715–1724. [Google Scholar] [CrossRef]
- Lu, Y.; Lam, H.-M.; Pi, E.; Zhan, Q.; Tsai, S.; Wang, C.; Kwan, Y.; Ngai, S. Comparative Metabolomics in Glycine Max and Glycine soja under Salt Stress To Reveal the Phenotypes of Their Offspring. J. Agric. Food Chem. 2013, 61, 8711–8721. [Google Scholar] [CrossRef]
- Yun, D.-Y.; Kang, Y.-G.; Yun, B.; Kim, E.-H.; Kim, M.; Park, J.S.; Lee, J.H.; Hong, Y.-S. Distinctive Metabolism of Flavonoid between Cultivated and Semiwild Soybean Unveiled through Metabolomics Approach. J. Agric. Food Chem. 2016, 64, 5773–5783. [Google Scholar] [CrossRef]
- Patent Database Search Results: TTL/“Soybean Cultivar” OR TTL/“Soybean Variety” in US Patent Collection. Available online: http://patft.uspto.gov/netacgi/nph-Parser?Sect1=PTO2&Sect2=HITOFF&p=1&u=%2Fnetahtml%2FPTO%2Fsearch-bool.html&r=0&f=S&l=50&TERM1=%22soybean+cultivar%22&FIELD1=TI&co1=OR&TERM2=%22soybean+variety%22&FIELD2=TI&d=PTXT (accessed on 10 May 2021).
- Lee, J.; Hwang, Y.-S.; Kim, S.T.; Yoon, W.-B.; Han, W.Y.; Kang, I.-K.; Choung, M.-G. Seed coat color and seed weight contribute differential responses of targeted metabolites in soybean seeds. Food Chem. 2017, 214, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Min, C.W.; Kim, S.W.; Wang, Y.; Agrawal, G.K.; Rakwal, R.; Kim, S.G.; Lee, B.W.; Ko, J.M.; Baek, I.Y.; et al. Comparative investigation of seed coats of brown- versus yellow-colored soybean seeds using an integrated proteomics and metabolomics approach. Proteomics 2015, 15, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-Q.; Zheng, L.; Wu, H.-J.; Zhu, Z.-K.; Zou, Y.-F.; Deng, J.-C.; Qin, W.-T.; Zhang, J.; Wang, X.-C.; Yang, W.-Y.; et al. Yellow- and green-cotyledon seeds of black soybean: Phytochemical and bioactive differences determine edibility and medical applications. Food Biosci. 2021, 39, 100842. [Google Scholar] [CrossRef]
- Yun, Y.J.; Lee, H.; Yoo, D.J.; Yang, J.Y.; Woo, S.-Y.; Seo, W.D.; Kim, Y.-C.; Lee, J.H. Molecular analysis of soyasaponin biosynthetic genes in two soybean (Glycine Max L. Merr.) cultivars. Plant Biotechnol. Rep. 2021, 15, 117–124. [Google Scholar] [CrossRef]
- García-Villalba, R.; León, C.; Dinelli, G.; Carretero, A.S.; Fernández-Gutiérrez, A.; Garcia-Cañas, V.; Cifuentes, A. Comparative metabolomic study of transgenic versus conventional soybean using capillary electrophoresis–time-of-flight mass spectrometry. J. Chromatogr. A 2008, 1195, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, G.G.; Skogerson, K.; MacIsaac, S.; Bickel, A.; Perez, T.; Li, X. Application of 1H NMR Profiling To Assess Seed Metabolomic Diversity. A Case Study on a Soybean Era Population. J. Agric. Food Chem. 2015, 63, 4690–4697. [Google Scholar] [CrossRef]
- Clarke, J.D.; Alexander, D.C.; Ward, D.P.; Ryals, J.A.; Mitchell, M.W.; Wulff, J.E.; Guo, L. Assessment of Genetically Modified Soybean in Relation to Natural Variation in the Soybean Seed Metabolome. Sci. Rep. 2013, 3, 3082. [Google Scholar] [CrossRef]
- de Campos, B.K.; Galazzi, R.M.; dos Santos, B.M.; Balbuena, T.S.; dos Santos, F.N.; Mokochinski, J.B.; Eberlin, M.N.; Arruda, M.A. Comparison of generational effect on proteins and metabolites in non-transgenic and transgenic soybean seeds through the insertion of the cp4-EPSPS gene assessed by omics-based platforms. Ecotoxicol. Environ. Saf. 2020, 202, 110918. [Google Scholar] [CrossRef]
- Rodrigues, J.M.; Coutinho, F.S.; dos Santos, D.S.; Vital, C.E.; Ramos, J.R.L.S.; Reis, P.B.; Oliveira, M.G.A.; Mehta, A.; Fontes, E.P.B.; Ramos, H.J.O. BiP-overexpressing soybean plants display accelerated hypersensitivity response (HR) affecting the SA-dependent sphingolipid and flavonoid pathways. Phytochemistry 2021, 185, 112704. [Google Scholar] [CrossRef]
- Kang, W.-S.; Chen, L.-J.; Wang, Y.-Y.; Zhu, X.-F.; Liu, X.-Y.; Fan, H.-Y.; Duan, Y.-X. Bacillus simplex treatment promotes soybean defence against soybean cyst nematodes: A metabolomics study using GC-MS. PLoS ONE 2020, 15, e0237194. [Google Scholar] [CrossRef]
- Nakata, R.; Yano, M.; Hiraga, S.; Teraishi, M.; Okumoto, Y.; Mori, N.; Kaga, A. Molecular Basis Underlying Common Cutworm Resistance of the Primitive Soybean Landrace Peking. Front. Genet. 2020, 11, 11. [Google Scholar] [CrossRef]
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 2019, 18, 1384–1395. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Wang, X.; Li, Q.; Yu, L.; Song, Q.; Gai, J.; Yang, S. Metabolomics Studies on Cytoplasmic Male Sterility during Flower Bud Development in Soybean. Int. J. Mol. Sci. 2019, 20, 2869. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, A.; Westrick, N.M.; Jain, S.; Piotrowski, J.S.; Ranjan, M.; Kessens, R.; Stiegman, L.; Grau, C.R.; Conley, S.; Smith, D.L.; et al. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 2019, 17, 1567–1581. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, C.S.; Lião, L.M.; Alcantara, G.B. Metabolic response of soybean plants to Sclerotinia sclerotiorum infection. Phytochemistry 2019, 167, 112099. [Google Scholar] [CrossRef]
- Cui, J.-Q.; Sun, H.-B.; Sun, M.-B.; Liang, R.-T.; Jie, W.-G.; Cai, B.-Y. Effects of Funneliformis mosseae on Root Metabolites and Rhizosphere Soil Properties to Continuously-Cropped Soybean in the Potted-Experiments. Int. J. Mol. Sci. 2018, 19, 2160. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Zhou, Y.; Li, X.; Zhao, J.; Guo, N.; Xing, H. Metabolomics Analysis of Soybean Hypocotyls in Response to Phytophthora sojae Infection. Front. Plant Sci. 2018, 9, 1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, W.; Zhu, X.; Wang, Y.; Chen, L.; Duan, Y. Transcriptomic and metabolomic analyses reveal that bacteria promote plant defense during infection of soybean cyst nematode in soybean. BMC Plant Biol. 2018, 18, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copley, T.R.; Aliferis, K.A.; Kliebenstein, D.J.; Jabaji, S.H. An integrated RNAseq-1H NMR metabolomics approach to understand soybean primary metabolism regulation in response to Rhizoctonia foliar blight disease. BMC Plant Biol. 2017, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lardi, M.; Murset, V.; Fischer, H.-M.; Mesa, S.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures. Int. J. Mol. Sci. 2016, 17, 815. [Google Scholar] [CrossRef] [Green Version]
- Abeysekara, N.S.; Swaminathan, S.; Desai, N.; Guo, L.; Bhattacharyya, M.K. The plant immunity inducer pipecolic acid accumulates in the xylem sap and leaves of soybean seedlings following Fusarium virguliforme infection. Plant Sci. 2016, 243, 105–114. [Google Scholar] [CrossRef]
- Scandiani, M.M.; Luque, A.G.; Razori, M.V.; Casalini, L.C.; Aoki, T.; O’Donnell, K.; Cervigni, G.D.L.; Spampinato, C.P. Metabolic profiles of soybean roots during early stages of Fusarium tucumaniae infection. J. Exp. Bot. 2014, 66, 391–402. [Google Scholar] [CrossRef] [Green Version]
- Sato, D.; Akashi, H.; Sugimoto, M.; Tomita, M.; Soga, T. Metabolomic profiling of the response of susceptible and resistant soybean strains to foxglove aphid, Aulacorthum solani Kaltenbach. J. Chromatogr. B 2013, 925, 95–103. [Google Scholar] [CrossRef]
- Brechenmacher, L.; Lei, Z.; Libault, M.; Findley, S.; Sugawara, M.; Sadowsky, M.J.; Sumner, L.W.; Stacey, G. Soybean Metabolites Regulated in Root Hairs in Response to the Symbiotic Bacterium Bradyrhizobium japonicum. Plant Physiol. 2010, 153, 1808–1822. [Google Scholar] [CrossRef] [Green Version]
- Silva, E.; Perez Da Graça, J.; Porto, C.; Martin Do Prado, R.; Nunes, E.; Corrêa Marcelino-Guimarães, F.; Conrado Meyer, M.; Jorge Pilau, E. Untargeted Metabolomics Analysis by UHPLC-MS/MS of Soybean Plant in a Compatible Response to Phakopsora Pachyrhizi Infection. Metabolites 2021, 11, 179. [Google Scholar] [CrossRef]
- Zanzarin, D.M.; Hernandes, C.P.; Leme, L.M.; Silva, E.; Porto, C.; Prado, R.M.D.; Meyer, M.C.; Favoreto, L.; Nunes, E.; Pilau, E.J. Metabolomics of soybean green stem and foliar retention (GSFR) disease using mass spectrometry and molecular networking. Rapid Commun. Mass Spectrom. 2019, 34, e8655. [Google Scholar] [CrossRef]
- Silva, E.; Da Graça, J.P.; Porto, C.; Prado, R.M.D.; Hoffmann-Campo, C.B.; Meyer, M.C.; Nunes, E.; Pilau, E.J. Unraveling Asian Soybean Rust metabolomics using mass spectrometry and Molecular Networking approach. Sci. Rep. 2020, 10, 138. [Google Scholar] [CrossRef]
- Lee, J.; Hwang, Y.S.; Chang, W.S.; Moon, J.K.; Choung, M.G. Seed Maturity Differentially Mediates Metabolic Responses in Black Soybean. Food Chem. 2013, 141, 2052–2059. [Google Scholar] [CrossRef]
- Collakova, E.; Aghamirzaie, D.; Fang, Y.; Klumas, C.; Tabataba, F.; Kakumanu, A.; Myers, E.; Heath, L.S.; Grene, R. Metabolic and Transcriptional Reprogramming in Developing Soybean (Glycine Max) Embryos. Metabolites 2013, 3, 347–372. [Google Scholar] [CrossRef]
- Li, L.; Hur, M.; Lee, J.Y.; Zhou, W.; Song, Z.; Ransom, N.; Demirkale, C.Y.; Nettleton, D.; Westgate, M.; Arendsee, Z.; et al. A Systems Biology Approach toward Understanding Seed Composition in Soybean. BMC Genom. 2015, 16, S9. [Google Scholar] [CrossRef] [Green Version]
- Gu, E.J.; Kim, D.W.; Jang, G.J.; Song, S.H.; Lee, J.I.; Lee, S.B.; Kim, B.M.; Cho, Y.; Lee, H.J.; Kim, H.J. Mass-Based Metabolomic Analysis of Soybean Sprouts during Germination. Food Chem. 2017, 217, 311–319. [Google Scholar] [CrossRef]
- Song, H.H.; Ryu, H.W.; Lee, K.J.; Jeong, I.Y.; Kim, D.S.; Oh, S.R. Metabolomics Investigation of Flavonoid Synthesis in Soybean Leaves Depending on the Growth Stage. Metabolomics 2014, 10, 833–841. [Google Scholar] [CrossRef]
- Makino, Y.; Nishizaka, A.; Yoshimura, M.; Sotome, I.; Kawai, K.; Akihiro, T. Influence of Low O2 and High CO2 Environment on Changes in Metabolite Concentrations in Harvested Vegetable Soybeans. Food Chem. 2020, 317, 126380. [Google Scholar] [CrossRef]
- Pi, E.; Zhu, C.; Fan, W.; Huang, Y.; Qu, L.; Li, Y.; Zhao, Q.; Ding, F.; Qiu, L.; Wang, H.; et al. Quantitative Phosphoproteomic and Metabolomic Analyses Reveal GmMYB173 Optimizes Flavonoid Metabolism in Soybean under Salt Stress. Mol. Cell. Proteom. 2018, 17, 1209–1224. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Rushton, P.J.; Rohila, J.S. Metabolomic Profiling of Soybeans (Glycine Max L.) Reveals the Importance of Sugar and Nitrogen Metabolism under Drought and Heat Stress. Plants 2017, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Yu, H.; Yu, Q.; Jin, X.; Cao, L.; Wang, M.; Wang, M.; Ren, C.; Zhang, Y. Physiological and UPLC-MS/MS Widely Targeted Metabolites Mechanisms of Alleviation of Drought Stress-Induced Soybean Growth Inhibition by Melatonin. Ind. Crop. Prod. 2021, 163, 113323. [Google Scholar] [CrossRef]
- Gupta, R.; Min, C.W.; Kramer, K.; Agrawal, G.K.; Rakwal, R.; Park, K.H.; Wang, Y.; Finkemeier, I.; Kim, S.T. A Multi-Omics Analysis of Glycine Max Leaves Reveals Alteration in Flavonoid and Isoflavonoid Metabolism Upon Ethylene and Abscisic Acid Treatment. Proteomics 2018, 18, 1700366. [Google Scholar] [CrossRef]
- Cheng, J.; Yuan, C.; Graham, T.L. Potential Defense-Related Prenylated Isoflavones in Lactofen-Induced Soybean. Phytochemistry 2011, 72, 875–881. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Yu, Y.; Li, X.; Tan, H. Integrated proteomics, metabolomics and physiological analyses for dissecting the toxic effects of halosulfuron-methyl on soybean seedlings (Glycine Max merr.). Plant Physiol. Biochem. 2020, 157, 303–315. [Google Scholar] [CrossRef]
- Zhong, Z.; Kobayashi, T.; Zhu, W.; Imai, H.; Zhao, R.; Ohno, T.; Rehman, S.U.; Uemura, M.; Tian, J.; Komatsu, S. Plant-derived smoke enhances plant growth through ornithine-synthesis pathway and ubiquitin-proteasome pathway in soybean. J. Proteom. 2020, 221, 103781. [Google Scholar] [CrossRef]
- Ban, Y.J.; Song, Y.H.; Kim, J.Y.; Baiseitova, A.; Lee, K.W.; Kim, K.D.; Park, K.H. Comparative investigation on metabolites changes in soybean leaves by ethylene and activation of collagen synthesis. Ind. Crop. Prod. 2020, 154, 112743. [Google Scholar] [CrossRef]
- Yilmaz, A.; Rudolph, H.L.; Hurst, J.J.; Wood, T.D. High-Throughput Metabolic Profiling of Soybean Leaves by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal. Chem. 2015, 88, 1188–1194. [Google Scholar] [CrossRef] [Green Version]
- Yun, D.-Y.; Kang, Y.-G.; Kim, E.-H.; Kim, M.; Park, N.-H.; Choi, H.-T.; Go, G.H.; Lee, J.H.; Park, J.S.; Hong, Y.-S. Metabolomics approach for understanding geographical dependence of soybean leaf metabolome. Food Res. Int. 2018, 106, 842–852. [Google Scholar] [CrossRef]
- Feng, Z.; Ding, C.; Li, W.; Wang, D.; Cui, D. Applications of metabolomics in the research of soybean plant under abiotic stress. Food Chem. 2020, 310, 125914. [Google Scholar] [CrossRef]
- Seo, W.D.; Kang, J.E.; Choi, S.-W.; Lee, K.-S.; Lee, M.-J.; Park, K.-D.; Lee, J.H. Comparison of nutritional components (isoflavone, protein, oil, and fatty acid) and antioxidant properties at the growth stage of different parts of soybean [Glycine Max (L.) Merrill]. Food Sci. Biotechnol. 2017, 26, 339–347. [Google Scholar] [CrossRef]
- Jiang, Z.-F.; Liu, D.-D.; Wang, T.-Q.; Liang, X.-L.; Cui, Y.-H.; Liu, Z.-H.; Li, W.-B. Concentration difference of auxin involved in stem development in soybean. J. Integr. Agric. 2020, 19, 953–964. [Google Scholar] [CrossRef]
- Hu, B.-Y.; Yang, C.-Q.; Iqbal, N.; Deng, J.-C.; Zhang, J.; Yang, W.-Y.; Liu, J. Development and validation of a GC–MS method for soybean organ-specific metabolomics. Plant Prod. Sci. 2018, 21, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Dresler, S.; Wójciak-Kosior, M.; Sowa, I.; Strzemski, M.; Sawicki, J.; Kováčik, J.; Blicharski, T. Effect of Long-Term Strontium Exposure on the Content of Phytoestrogens and Allantoin in Soybean. Int. J. Mol. Sci. 2018, 19, 3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, K.-H.; Kim, D.Y.; Kim, H.J.; Pack, I.-S.; Chung, Y.S.; Kim, S.Y.; Kim, C.-G. Global metabolite profiling based on GC–MS and LC–MS/MS analyses in ABF3-overexpressing soybean with enhanced drought tolerance. Appl. Biol. Chem. 2019, 62, 15. [Google Scholar] [CrossRef] [Green Version]
- Coutinho, I.D.; Henning, L.M.M.; Döpp, S.A.; Nepomuceno, A.; Moraes, L.A.C.; Marcolino-Gomes, J.; Richter, C.; Schwalbe, H.; Colnago, L.A. Identification of primary and secondary metabolites and transcriptome profile of soybean tissues during different stages of hypoxia. Data Brief 2018, 21, 1089–1100. [Google Scholar] [CrossRef]
- Silva, F.A.C.; Carrão-Panizzi, M.C.; Blassioli-Moraes, M.C.; Panizzi, A.R. Influence of Volatile and Nonvolatile Secondary Metabolites From Soybean Pods on Feeding and on Oviposition Behavior of Euschistus heros (Hemiptera: Heteroptera: Pentatomidae). Environ. Entomol. 2013, 42, 1375–1382. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.-C.; Yang, C.-Q.; Zhang, J.; Zhang, Q.; Yang, F.; Yang, W.-Y.; Liu, J. Organ-Specific Differential NMR-Based Metabonomic Analysis of Soybean [Glycine Max (L.) Merr.] Fruit Reveals the Metabolic Shifts and Potential Protection Mechanisms Involved in Field Mold Infection. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xu, J.; Wang, X.; Fu, H.; Zhao, M.; Wang, H.; Shi, L. Photosynthetic characteristics and metabolic analyses of two soybean genotypes revealed adaptive strategies to low-nitrogen stress. J. Plant Physiol. 2018, 229, 132–141. [Google Scholar] [CrossRef]
- Dillon, F.M.; Chludil, H.D.; Mithöfer, A.; Zavala, J.A. Solar UVB-inducible ethylene alone induced isoflavonoids in pods of field-grown soybean, an important defense against stink bugs. Environ. Exp. Bot. 2020, 178, 104167. [Google Scholar] [CrossRef]
- Tsuno, Y.; Fujimatsu, T.; Endo, K.; Sugiyama, A.; Yazaki, K. Soyasaponins: A New Class of Root Exudates in Soybean (Glycine Max). Plant Cell Physiol. 2018, 59, 366–375. [Google Scholar] [CrossRef] [Green Version]
- ChemAxon-Software Solutions and Services for Chemistry & Biology. Available online: https://chemaxon.com/products/marvin (accessed on 8 April 2021).
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Chemin. 2016, 8, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Salama, A.A.A.; Allam, R.M. Promising Targets of Chrysin and Daidzein in Colorectal Cancer: Amphiregulin, CXCL1, and MMP-9. Eur. J. Pharmacol. 2021, 892, 173763. [Google Scholar] [CrossRef]
- He, Y.; Huang, M.; Tang, C.; Yue, Y.; Liu, X.; Zheng, Z.; Dong, H.; Liu, D. Dietary Daidzein Inhibits Hepatitis C Virus Replication by Decreasing MicroRNA-122 Levels. Virus Res. 2021, 298, 198404. [Google Scholar] [CrossRef]
- Yang, M.H.; Jung, S.H.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Sethi, G.; Ahn, K.S. Attenuation of STAT3 Signaling Cascade by Daidzin Can Enhance the Apoptotic Potential of Bortezomib against Multiple Myeloma. Biomolecules 2020, 10, 23. [Google Scholar] [CrossRef] [Green Version]
- Kazmi, Z.; Zeeshan, S.; Khan, A.; Malik, S.; Shehzad, A.; Seo, E.K.; Khan, S. Anti-Epileptic Activity of Daidzin in PTZ-Induced Mice Model by Targeting Oxidative Stress and BDNF/VEGF Signaling. NeuroToxicology 2020, 79, 150–163. [Google Scholar] [CrossRef]
- Xie, X.; Cong, L.; Liu, S.; Xiang, L.; Fu, X. Genistein Alleviates Chronic Vascular Inflammatory Response via the MiR-21/NF-ΚB P65 Axis in Lipopolysaccharide-Treated Mice. Mol. Med. Rep. 2021, 23, 192. [Google Scholar] [CrossRef]
- Zang, Y.Q.; Feng, Y.Y.; Luo, Y.H.; Zhai, Y.Q.; Ju, X.Y.; Feng, Y.C.; Wang, J.R.; Yu, C.Q.; Jin, C.H. Glycitein Induces Reactive Oxygen Species-Dependent Apoptosis and G0/G1 Cell Cycle Arrest through the MAPK/STAT3/NF-ΚB Pathway in Human Gastric Cancer Cells. Drug Dev. Res. 2019, 80, 573–584. [Google Scholar] [CrossRef]
- Hwang, S.T.; Yang, M.H.; Baek, S.H.; Um, J.Y.; Ahn, K.S. Genistin Attenuates Cellular Growth and Promotes Apoptotic Cell Death Breast Cancer Cells through Modulation of ERalpha Signaling Pathway. Life Sci. 2020, 263, 118594. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, B.; Yu, H.; Zhao, W.; Song, X.; Liu, Y.; Wang, K.; Peacher, N.; Zhao, X.; Zhang, H.T. Biochanin A Attenuates Ovariectomy-Induced Cognition Deficit via Antioxidant Effects in Female Rats. Front. Pharmacol. 2021, 12, 171. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Y.; Liang, H.; Liu, X. Coumestrol Mitigates Retinal Cell Inflammation, Apoptosis, and Oxidative Stress in a Rat Model of Diabetic Retinopathy via Activation of SIRT1. Aging 2021, 13, 5342–5357. [Google Scholar] [CrossRef]
- Yamamoto, T.; Sakamoto, C.; Tachiwana, H.; Kumabe, M.; Matsui, T.; Yamashita, T.; Shinagawa, M.; Ochiai, K.; Saitoh, N.; Nakao, M. Endocrine Therapy-Resistant Breast Cancer Model Cells Are Inhibited by Soybean Glyceollin I through Eleanor Non-Coding RNA. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mansoori, M.N.; Raghuvanshi, A.; Shukla, P.; Awasthi, P.; Trivedi, R.; Goel, A.; Singh, D. Medicarpin Prevents Arthritis in Post-Menopausal Conditions by Arresting the Expansion of TH17 Cells and pro-Inflammatory Cytokines. Int. Immunopharmacol. 2020, 82, 106299. [Google Scholar] [CrossRef]
- Luo, L.; Zhou, J.; Zhao, H.; Fan, M.; Gao, W. The Anti-Inflammatory Effects of Formononetin and Ononin on Lipopolysaccharide-Induced Zebrafish Models Based on Lipidomics and Targeted Transcriptomics. Metabolomics 2019, 15, 153. [Google Scholar] [CrossRef]
- Tsai, W.; Nakamura, Y.; Akasaka, T.; Katakura, Y.; Tanaka, Y.; Shirouchi, B.; Jiang, Z.; Yuan, X.; Sato, M. Soyasaponin Ameliorates Obesity and Reduces Hepatic Triacylglycerol Accumulation by Suppressing Lipogenesis in High-fat Diet-fed Mice. J. Food Sci. 2021. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, E.J.; Kim, H.S.; Choi, C.W.; Choi, S.W.; Kim, S.L.; Seo, W.D.; Do, S.H. Soyasaponin Ab Alleviates Postmenopausal Obesity through Browning of White Adipose Tissue. J. Funct. Foods 2019, 57, 453–464. [Google Scholar] [CrossRef]
- Liu, X.; Chen, K.; Zhu, L.; Liu, H.; Ma, T.; Xu, Q.; Xie, T. Soyasaponin Ab Protects against Oxidative Stress in HepG2 Cells via Nrf2/HO-1/NQO1 Signaling Pathways. J. Funct. Foods 2018, 45, 110–117. [Google Scholar] [CrossRef]
- Wang, F.; Gong, S.; Wang, T.; Li, L.; Luo, H.; Wang, J.; Huang, C.; Zhou, H.; Chen, G.; Liu, Z.; et al. Soyasaponin II Protects against Acute Liver Failure through Diminishing YB-1 Phosphorylation and Nlrp3-Inflammasome Priming in Mice. Theranostics 2020, 10, 2714–2726. [Google Scholar] [CrossRef]
- Omar, A.; Kalra, R.S.; Putri, J.; Elwakeel, A.; Kaul, S.C.; Wadhwa, R. Soyasapogenol-A Targets CARF and Results in Suppression of Tumor Growth and Metastasis in P53 Compromised Cancer Cells. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Jandacek, R.J. Linoleic Acid: A Nutritional Quandary. Healthcare 2017, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Nouri, Z.; Fakhri, S.; El-Senduny, F.F.; Sanadgol, N.; Abd-Elghani, G.E.; Farzaei, M.H.; Chen, J.T. On the Neuroprotective Effects of Naringenin: Pharmacological Targets, Signaling Pathways, Molecular Mechanisms, and Clinical Perspective. Biomolecules 2019, 9, 690. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Aslam Gondal, T.; Atif, M.; Shahbaz, M.; Batool Qaisarani, T.; Hanif Mughal, M.; Salehi, B.; Martorell, M.; Sharifi-Rad, J. Apigenin as an Anticancer Agent. Phytother. Res. 2020, 34, 1812–1828. [Google Scholar] [CrossRef]
- Guo, Z.; Cao, W.; Zhao, S.; Han, Z.; Han, B. Protection against 1-Methyl-4-Phenyl Pyridinium-Induced Neurotoxicity in Human Neuroblastoma SH-SY5Y Cells by Soyasaponin i by the Activation of the Phosphoinositide 3-Kinase/AKT/GSK3β Pathway. NeuroReport 2016, 27, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Fouzder, C.; Mukhuty, A.; Mukherjee, S.; Malick, C.; Kundu, R. Trigonelline Inhibits Nrf2 via EGFR Signalling Pathway and Augments Efficacy of Cisplatin and Etoposide in NSCLC Cells. Toxicol. Vitr. 2021, 70, 105038. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Mukherjee, S.; Yun, J.W. Trigonelline Induces Browning in 3T3-L1 White Adipocytes. Phytother. Res. 2021, 35, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Farid, M.M.; Yang, X.; Kuboyama, T.; Tohda, C. Trigonelline Recovers Memory Function in Alzheimer’s Disease Model Mice: Evidence of Brain Penetration and Target Molecule. Sci. Rep. 2020, 10, 16424. [Google Scholar] [CrossRef]
- Guo, X.; Wang, L.; Xu, M.; Bai, J.; Shen, J.; Yu, B.; Liu, Y.; Sun, H.; Hao, Y.; Geng, D. Shikimic Acid Prevents Cartilage Matrix Destruction in Human Chondrocytes. Int. Immunopharmacol. 2018, 63, 155–160. [Google Scholar] [CrossRef]
- Ertugrul, B.; Iplik, E.S.; Cakmakoglu, B. In Vitro Inhibitory Effect of Succinic Acid on T-Cell Acute Lymphoblastic Leukemia Cell Lines. Arch. Med. Res. 2021, 52, 270–276. [Google Scholar] [CrossRef]
- Kim, H.S.; Yoo, H.J.; Lee, K.M.; Song, H.E.; Kim, S.J.; Lee, J.O.; Hwang, J.J.; Song, J.W. Stearic Acid Attenuates Profibrotic Signalling in Idiopathic Pulmonary Fibrosis. Respirology 2021, 26, 255–263. [Google Scholar] [CrossRef]
- Liu, C.; Weir, D.; Busse, P.; Yang, N.; Zhou, Z.; Emala, C.; Li, X.M. The Flavonoid 7,4′-Dihydroxyflavone Inhibits MUC5AC Gene Expression, Production, and Secretion via Regulation of NF-ΚB, STAT6, and HDAC2. Phytother. Res. 2015, 29, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Guo, S.; Jiang, K.; Wang, Y.; Yang, M.; Guo, M. Glycitin Alleviates Lipopolysaccharide-Induced Acute Lung Injury via Inhibiting NF-ΚB and MAPKs Pathway Activation in Mice. Int. Immunopharmacol. 2019, 75, 105749. [Google Scholar] [CrossRef]
- Baek, S.; Kim, J.; Moon, B.S.; Park, S.M.; Jung, D.E.; Kang, S.Y.; Lee, S.J.; Oh, S.J.; Kwon, S.H.; Nam, M.H.; et al. Camphene Attenuates Skeletal Muscle Atrophy by Regulating Oxidative Stress and Lipid Metabolism in Rats. Nutrients 2020, 12, 3731. [Google Scholar] [CrossRef]
- Khoshnazar, M.; Parvardeh, S.; Bigdeli, M.R. Alpha-Pinene Exerts Neuroprotective Effects via Anti-Inflammatory and Anti-Apoptotic Mechanisms in a Rat Model of Focal Cerebral Ischemia-Reperfusion. J. Stroke Cerebrovasc. Dis. 2020, 29, 104977. [Google Scholar] [CrossRef]
- Saeedi-Boroujeni, A.; Mahmoudian-Sani, M.R. Anti-Inflammatory Potential of Quercetin in COVID-19 Treatment. J. Inflamm. 2021, 18, 1–9. [Google Scholar] [CrossRef]
- Bharath, B.; Perinbam, K.; Devanesan, S.; AlSalhi, M.S.; Saravanan, M. Evaluation of the Anticancer Potential of Hexadecanoic Acid from Brown Algae Turbinaria Ornata on HT–29 Colon Cancer Cells. J. Mol. Struct. 2021, 1235, 130229. [Google Scholar] [CrossRef]
- Liu, H.-M.; Li, H.-Y. Application and Conversion of Soybean Hulls. In Soybean-The Basis of Yield, Biomass and Productivity; InTech: London, UK, 2017. [Google Scholar]
- Cabezudo, I.; Meini, M.R.; di Ponte, C.C.; Melnichuk, N.; Boschetti, C.E.; Romanini, D. Soybean (Glycine Max) Hull Valorization through the Extraction of Polyphenols by Green Alternative Methods. Food Chem. 2021, 338, 128131. [Google Scholar] [CrossRef]
- de Oliveira Silva, F.; Perrone, D. Characterization and Stability of Bioactive Compounds from Soybean Meal. LWT-Food Sci. Technol. 2015, 63, 992–1000. [Google Scholar] [CrossRef]
- Alvarez, M.V.; Cabred, S.; Ramirez, C.L.; Fanovich, M.A. Valorization of an Agroindustrial Soybean Residue by Supercritical Fluid Extraction of Phytochemical Compounds. J. Supercrit. Fluids 2019, 143, 90–96. [Google Scholar] [CrossRef]
- Freitas, C.S.; da Silva, G.A.; Perrone, D.; Vericimo, M.A.; Dos S. Baião, D.; Pereira, P.R.; Paschoalin, V.M.F.; del Aguila, E.M. Recovery of Antimicrobials and Bioaccessible Isoflavones and Phenolics from Soybean (Glycine Max) Meal by Aqueous Extraction. Molecules 2019, 24, 74. [Google Scholar] [CrossRef] [Green Version]
- Nkurunziza, D.; Pendleton, P.; Chun, B.S. Optimization and Kinetics Modeling of Okara Isoflavones Extraction Using Subcritical Water. Food Chem. 2019, 295, 613–621. [Google Scholar] [CrossRef]
- Nile, S.H.; Nile, A.; Oh, J.W.; Kai, G. Soybean Processing Waste: Potential Antioxidant, Cytotoxic and Enzyme Inhibitory Activities. Food Biosci. 2020, 38, 100778. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, H.X.; Wu, Z.L.; Wang, Y.J.; Wang, L.J. Recovery of Isoflavone Aglycones from Soy Whey Wastewater Using Foam Fractionation and Acidic Hydrolysis. J. Agric. Food Chem. 2013, 61, 7366–7372. [Google Scholar] [CrossRef]
- Chua, J.Y.; Liu, S.Q. Soy Whey: More than Just Wastewater from Tofu and Soy Protein Isolate Industry. Trends Food Sci. Technol. 2019, 91, 24–32. [Google Scholar] [CrossRef]
- Davy, P.; Vuong, Q.V. Soy Milk By-Product: Its Composition and Utilisation. Food Rev. Int. 2020. [Google Scholar] [CrossRef]
- Melini, F.; Melini, V.; Luziatelli, F.; Ficca, A.G.; Ruzzi, M. Health-Promoting Components in Fermented Foods: An Up-to-Date Systematic Review. Nutrients 2019, 11, 1189. [Google Scholar] [CrossRef] [Green Version]
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented Foods: Definitions and Characteristics, Impact on the Gut Microbiota and Effects on Gastrointestinal Health and Disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health Benefits of Fermented Foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef]
- Tamang, J.P.; Shin, D.H.; Jung, S.J.; Chae, S.W. Functional Properties of Microorganisms in Fermented Foods. Front. Microbiol. 2016, 7, 578. [Google Scholar] [CrossRef] [Green Version]
- Adebo, O.A.; Oyeyinka, S.A.; Adebiyi, J.A.; Feng, X.; Wilkin, J.D.; Kewuyemi, Y.O.; Abrahams, A.M.; Tugizimana, F. Application of Gas Chromatography–Mass Spectrometry (GC-MS)-Based Metabolomics for the Study of Fermented Cereal and Legume Foods: A Review. Int. J. Food Sci. Technol. 2021, 56, 1514–1534. [Google Scholar] [CrossRef]
- Gupta, S.; Lee, J.J.L.; Chen, W.N. Analysis of Improved Nutritional Composition of Potential Functional Food (Okara) after Probiotic Solid-State Fermentation. J. Agric. Food Chem. 2018, 66, 5373–5381. [Google Scholar] [CrossRef]
- Chan, L.Y.; Takahashi, M.; Lim, P.J.; Aoyama, S.; Makino, S.; Ferdinandus, F.; Ng, S.Y.C.; Arai, S.; Fujita, H.; Tan, H.C.; et al. Eurotium Cristatum Fermented Okara as a Potential Food Ingredient to Combat Diabetes. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Mok, W.K.; Tan, Y.X.; Lee, J.; Kim, J.; Chen, W.N. A Metabolomic Approach to Understand the Solid-State Fermentation of Okara Using Bacillus Subtilis WX-17 for Enhanced Nutritional Profile. AMB Express 2019, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Mok, W.K.; Tan, Y.X.; Lyu, X.M.; Chen, W.N. Effects of Submerged Liquid Fermentation of Bacillus Subtilis WX-17 Using Okara as Sole Nutrient Source on the Composition of a Potential Probiotic Beverage. Food Sci. Nutr. 2020, 8, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chen, W.N. A Metabolomics Approach to Evaluate Post-Fermentation Enhancement of Daidzein and Genistein in a Green Okara Extract. J. Sci. Food Agric. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Chen, W.N. Characterization and in Vitro Bioactivity of Green Extract from Fermented Soybean Waste. ACS Omega 2019, 4, 21675–21683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghanem, K.Z.; Mahran, M.Z.; Ramadan, M.M.; Ghanem, H.Z.; Fadel, M.; Mahmoud, M.H. A Comparative Study on Flavour Components and Therapeutic Properties of Unfermented and Fermented Defatted Soybean Meal Extract. Sci. Rep. 2020, 10, 5998. [Google Scholar] [CrossRef]
- Azi, F.; Tu, C.; Meng, L.; Zhiyu, L.; Cherinet, M.T.; Ahmadullah, Z.; Dong, M. Metabolite Dynamics and Phytochemistry of a Soy Whey-Based Beverage Bio-Transformed by Water Kefir Consortium. Food Chem. 2021, 342, 128225. [Google Scholar] [CrossRef]
- Yang, J.; Wu, X.B.; Chen, H.L.; Sun-Waterhouse, D.; Zhong, H.B.; Cui, C. A Value-Added Approach to Improve the Nutritional Quality of Soybean Meal Byproduct: Enhancing Its Antioxidant Activity through Fermentation by Bacillus Amyloliquefaciens SWJS22. Food Chem. 2019, 272, 396–403. [Google Scholar] [CrossRef]
- Liang, S.; Jiang, W.; Song, Y.; Zhou, S.F. Improvement and Metabolomics-Based Analysis of d-Lactic Acid Production from Agro-Industrial Wastes by Lactobacillus Delbrueckii Submitted to Adaptive Laboratory Evolution. J. Agric. Food Chem. 2020, 68, 7660–7669. [Google Scholar] [CrossRef]
- Kupski, L.; Telles, A.C.; Gonçalves, L.M.; Nora, N.S.; Furlong, E.B. Recovery of Functional Compounds from Lignocellulosic Material: An Innovative Enzymatic Approach. Food Biosci. 2018, 22, 26–31. [Google Scholar] [CrossRef]
- Yang, S.O.; Kim, S.H.; Cho, S.; Lee, J.; Kim, Y.S.; Yun, S.S.; Choi, H.K. Classification of Fermented Soymilk during Fermentation by 1H NMR Coupled with Principal Component Analysis and Elucidation of Free-Radical Scavenging Activities. Biosci. Biotechnol. Biochem. 2009, 73, 1184–1188. [Google Scholar] [CrossRef]
- Simu, S.Y.; Castro-Aceituno, V.; Lee, S.; Ahn, S.; Lee, H.K.; Hoang, V.A.; Yang, D.C. Fermentation of Soybean Hull by Monascus Pilosus and Elucidation of Its Related Molecular Mechanism Involved in the Inhibition of Lipid Accumulation. An in Sílico and in Vitro Approach. J. Food Biochem. 2018, 42, e12442. [Google Scholar] [CrossRef]
| Name | Formula | B | L | P | R | References |
|---|---|---|---|---|---|---|
| 2′-hydroxydaidzein | C15H10O5 | X | [80] | |||
| 7,3′,4′-trihydroxyisoflavone | C15H10O5 | X | [79] | |||
| 7-O-methylluteone | C21H20O6 | X | [78] | |||
| acetyl daidzin | C22H22O9 | X | [104] | |||
| acetyl genistin | C23H22O11 | X | X | [94,104] | ||
| acetyl glycitin | C24H24O11 | X | [104] | |||
| afrormosin 7-O-glucoside | C23H24O10 | X | [80] | |||
| biochanin A | C16H12O5 | X | [80] | |||
| biochanin A 7-O-D-glucoside | C22H22O10 | X | [80] | |||
| biochanin A 7-O-glucoside-6′′-O-malonate | C25H24O13 | X | [80] | |||
| calycosin | C16H12O5 | X | [80] | |||
| coumestrol | C15H8O5 | X | X | [79,80,101] | ||
| daidzein | C15H10O4 | X | X | X | X | [38,78,79,80,94,98,101,103,104,106,107,108] |
| daidzin | C21H20O9 | X | X | X | X | [38,78,79,80,94,98,101,103,104,107,108] |
| formononetin | C16H12O4 | X | [80,102] | |||
| formononetin 7-O-glucoside | C22H22O9 | X | X | [79,80] | ||
| formononetin 7-O-glucoside-6′′-malonate | C25H24O12 | X | [78,80,94] | |||
| formononetin 7-O-glucoside-6-O-malonate | C25H24O12 | X | X | [78,79] | ||
| genistein | C15H10O5 | X | X | X | X | [38,79,94,98,104,108] |
| genistin | C21H20O10 | X | X | X | X | [38,78,79,94,101,104,107,108] |
| glyceollidin I/II | C20H20O5 | X | [80] | |||
| glyceollin I | C20H18O5 | X | [78,80] | |||
| glyceollin II | C20H18O5 | X | [78,80] | |||
| glyceollin III | C20H18O5 | X | [78,80] | |||
| glyceollin IV | C21H22O5 | X | [80] | |||
| glyceollin VI | C20H16O4 | X | [80] | |||
| glycitein | C16H12O5 | X | X | X | X | [38,80,98,104,108] |
| glycitein 7-O-glucoside | C22H22O10 | X | [80] | |||
| glycitin | C22H22O10 | X | X | X | X | [38,79,101,104,108] |
| isotrifoliol | C16H10O6 | X | [80] | |||
| malonyldaidzin | C24H22O12 | X | X | X | X | [38,78,79,80,94,101,103,104,107,108] |
| malonylgenistin | C24H22O13 | X | X | X | X | [78,79,80,94,101,104,107,108] |
| malonylglycitin | C25H24O13 | X | X | X | [80,94,104,108] | |
| medicarpin | C16H14O4 | X | [80] | |||
| neobavaisoflavone | C20H18O4 | X | X | [78,79] | ||
| phaseollin | C20H18O4 | X | [80] | |||
| pisatin | C17H14O6 | X | [80] | |||
| sojagol | C20H16O5 | X | [78,80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bragagnolo, F.S.; Funari, C.S.; Ibáñez, E.; Cifuentes, A. Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds. Foods 2021, 10, 1308. https://doi.org/10.3390/foods10061308
Bragagnolo FS, Funari CS, Ibáñez E, Cifuentes A. Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds. Foods. 2021; 10(6):1308. https://doi.org/10.3390/foods10061308
Chicago/Turabian StyleBragagnolo, Felipe Sanchez, Cristiano Soleo Funari, Elena Ibáñez, and Alejandro Cifuentes. 2021. "Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds" Foods 10, no. 6: 1308. https://doi.org/10.3390/foods10061308
APA StyleBragagnolo, F. S., Funari, C. S., Ibáñez, E., & Cifuentes, A. (2021). Metabolomics as a Tool to Study Underused Soy Parts: In Search of Bioactive Compounds. Foods, 10(6), 1308. https://doi.org/10.3390/foods10061308