Exploration of Spanish Olive Oil Quality with a Miniaturized Low-Cost Fluorescence Sensor and Machine Learning Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Olive Oil Samples
2.2. Miniaturized Low-Cost Fluorescence Sensor
2.3. Dataset Preparation and Description
2.4. Machine Learning Classifiers
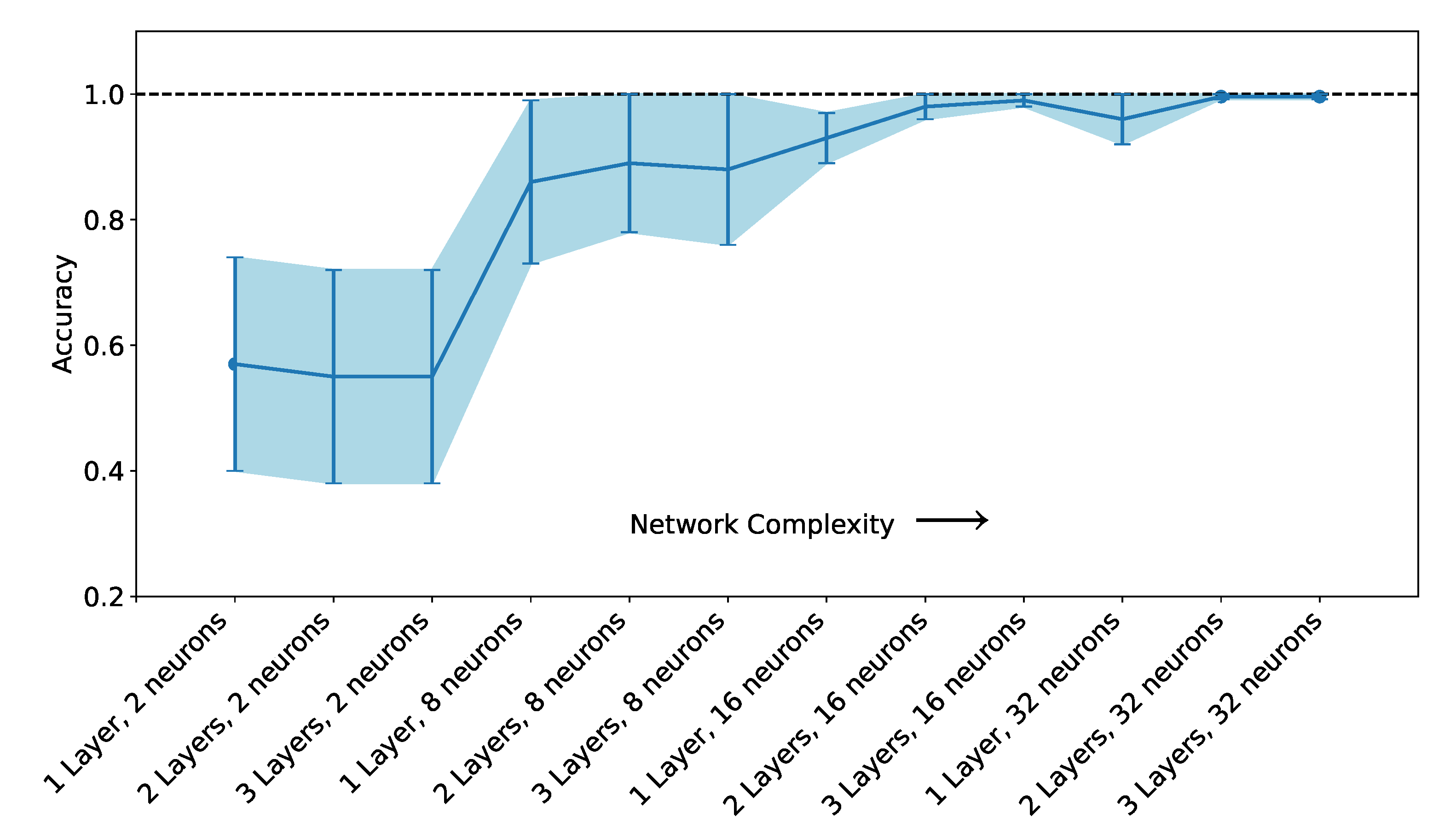
2.5. Artificial Neural Network-Based Classifiers
2.6. External Validation of Models
3. Results and Discussion
3.1. Spectral Response of the Olive Oils
3.2. Classification with Machine Learning Methods
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EVOO | Extra Virgin Olive Oil |
| VOO | Virgin Olive Oil |
| LOO | Lampante Olive Oil |
| EEC | European Economic Community |
| EEM | Excitation Emission Matrix |
| PCA | Principal Component Analysis |
| PLS | Partial Least Square Regression |
| PLS-DA | Partial Least Square Discriminant Analysis |
| LDA | Linear Discriminant Analysis |
| KNN | K-Nearest Neighbors |
| RF | Random Forest |
| ANN | Artificial Neural Network |
| NNM | Neural Network Model |
| UV | Ultraviolet |
| LED | Light Emitting Diode |
| CCD | Charge-Coupled Device |
| SVM | Support Vector Machine |
| NB | Naïve Bayes |
| DT | Decision Tree |
| MLR | Multinomial Logistic Regression |
| ReLU | Rectified Linear Unit |
| Adam | Adaptive Moment Estimation |
| HCA | Hyerarchical Cluster Analysis |
References
- Georgiou, C.A.; Danezis, G.P. Food Authentication: Management, Analysis and Regulation; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Regulation, H. Commission Regulation (EEC) No. 2568/91 of 11 July 1991 on the characteristics of olive oil and olive-residue oil and on the relevant methods of analysis Official Journal L 248, 5 September 1991. Offic. JL 1991, 248, 1–83. [Google Scholar]
- Kongbonga, Y.G.M.; Ghalila, H.; Onana, M.B.; Majdi, Y.; Lakhdar, Z.B.; Mezlini, H.; Sevestre-Ghalila, S. Characterization of vegetable oils by fluorescence spectroscopy. Food Nutr. Sci. 2011, 2, 692–699. [Google Scholar] [CrossRef]
- Sikorska, E.; Khmelinskii, I.; Sikorski, M. Analysis of olive oils by fluorescence spectroscopy: Methods and applications. In Olive Oil-Constituents, Quality, Health Properties and Bioconversions; IntechOpen: Rijeka, Croatia, 2012; pp. 63–88. [Google Scholar]
- Skoog, D.A.; Holler, F.J.; Crouch, S.R. Principles of Instrumental Analysis; Cengage Learning: Boston, MA, USA, 2017. [Google Scholar]
- Guzmán, E.; Baeten, V.; Pierna, J.A.F.; García-Mesa, J.A. Evaluation of the overall quality of olive oil using fluorescence spectroscopy. Food Chem. 2015, 173, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Merás, I.D.; Manzano, J.D.; Rodríguez, D.A.; de la Peña, A.M. Detection and quantification of extra virgin olive oil adulteration by means of autofluorescence excitation-emission profiles combined with multi-way classification. Talanta 2018, 178, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Guimet, F.; Boqué, R.; Ferré, J. Cluster analysis applied to the exploratory analysis of commercial Spanish olive oils by means of excitation- emission fluorescence spectroscopy. J. Agric. Food Chem. 2004, 52, 6673–6679. [Google Scholar] [CrossRef]
- Poulli, K.I.; Mousdis, G.A.; Georgiou, C.A. Classification of edible and lampante virgin olive oil based on synchronous fluorescence and total luminescence spectroscopy. Anal. Chim. Acta 2005, 542, 151–156. [Google Scholar] [CrossRef]
- Sayago, A.; Morales, M.; Aparicio, R. Detection of hazelnut oil in virgin olive oil by a spectrofluorimetric method. Eur. Food Res. Technol. 2004, 218, 480–483. [Google Scholar] [CrossRef]
- Poulli, K.I.; Mousdis, G.A.; Georgiou, C.A. Rapid synchronous fluorescence method for virgin olive oil adulteration assessment. Food Chem. 2007, 105, 369–375. [Google Scholar] [CrossRef]
- Ali, H.; Saleem, M.; Anser, M.R.; Khan, S.; Ullah, R.; Bilal, M. Validation of fluorescence spectroscopy to detect adulteration of edible oil in extra virgin olive oil (EVOO) by applying chemometrics. Appl. Spectrosc. 2018, 72, 1371–1379. [Google Scholar] [CrossRef]
- Hernández-Sánchez, N.; Lleó, L.; Ammari, F.; Cuadrado, T.R.; Roger, J.M. Fast fluorescence spectroscopy methodology to monitor the evolution of extra virgin olive oils under illumination. Food Bioprocess Technol. 2017, 10, 949–961. [Google Scholar] [CrossRef]
- Mishra, P.; Lleó, L.; Cuadrado, T.; Ruiz-Altisent, M.; Hernández-Sánchez, N. Monitoring oxidation changes in commercial extra virgin olive oils with fluorescence spectroscopy-based prototype. Eur. Food Res. Technol. 2018, 244, 565–575. [Google Scholar] [CrossRef]
- Baltazar, P.; Hernández-Sánchez, N.; Diezma, B.; Lleó, L. Development of rapid extra virgin olive oil quality assessment procedures based on spectroscopic techniques. Agronomy 2020, 10, 41. [Google Scholar] [CrossRef]
- Lobo-Prieto, A.; Tena, N.; Aparicio-Ruiz, R.; García-González, D.L.; Sikorska, E. Monitoring Virgin Olive Oil Shelf-Life by Fluorescence Spectroscopy and Sensory Characteristics: A Multidimensional Study Carried Out under Simulated Market Conditions. Foods 2020, 9, 1846. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, N.; Le Dréau, Y.; Ollivier, D.; Artaud, J.; Pinatel, C.; Kister, J. Origin of French virgin olive oil registered designation of origins predicted by chemometric analysis of synchronous excitation- emission fluorescence spectra. J. Agric. Food Chem. 2005, 53, 9361–9368. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Carvelo, A.M.; Lozano, V.A.; Olivieri, A.C. Comparative chemometric analysis of fluorescence and near infrared spectroscopies for authenticity confirmation and geographical origin of Argentinean extra virgin olive oils. Food Control 2019, 96, 22–28. [Google Scholar] [CrossRef]
- Al Riza, D.F.; Kondo, N.; Rotich, V.K.; Perone, C.; Giametta, F. Cultivar and geographical origin authentication of Italian extra virgin olive oil using front-face fluorescence spectroscopy and chemometrics. Food Control 2021, 121, 107604. [Google Scholar] [CrossRef]
- Michelucci, U. Applied Deep Learning—A Case-Based Approach to Understanding Deep Neural Networks; APRESS Media, LLC: New York, NY, USA, 2018. [Google Scholar]
- Sikorska, E.; Khmelinskii, I.; Sikorski, M. Vibrational and electronic spectroscopy and chemometrics in analysis of edible oils. In Methods in Food Analysis; Cruz, R.M.S., Khmelinskii, I., Vieira, M., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 201–234. [Google Scholar]
- Zaroual, H.; Chénè, C.; El Hadrami, E.M.; Karoui, R. Application of new emerging techniques in combination with classical methods for the determination of the quality and authenticity of olive oil: A review. Crit. Rev. Food Sci. Nutr. 2021, 1–24. [Google Scholar] [CrossRef]
- Meenu, M.; Cai, Q.; Xu, B. A critical review on analytical techniques to detect adulteration of extra virgin olive oil. Trends Food Sci. Technol. 2019, 91, 391–408. [Google Scholar] [CrossRef]
- Gonzalez-Fernandez, I.; Iglesias-Otero, M.; Esteki, M.; Moldes, O.; Mejuto, J.; Simal-Gandara, J. A critical review on the use of artificial neural networks in olive oil production, characterization and authentication. Crit. Rev. Food Sci. Nutr. 2019, 59, 1913–1926. [Google Scholar] [CrossRef] [PubMed]
- Iqdiam, B.M.; Welt, B.A.; Goodrich-Schneider, R.; Sims, C.A.; Baker IV, G.L.; Marshall, M.R. Influence of headspace oxygen on quality and shelf life of extra virgin olive oil during storage. Food Packag. Shelf Life 2020, 23, 100433. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Ferreiro-González, M.; Barbero, G.F.; Álvarez, J.A.; Ruiz, A.; Palma, M.; Ayuso, J. Authentication of virgin olive oil by a novel curve resolution approach combined with visible spectroscopy. Food Chem. 2017, 220, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Torreblanca-Zanca, A.; Aroca-Santos, R.; Lastra-Mejias, M.; Izquierdo, M.; Cancilla, J.C.; Torrecilla, J.S. Laser diode induced excitation of PDO extra virgin olive oils for cognitive authentication and fraud detection. Sens. Actuators B Chem. 2019, 280, 1–9. [Google Scholar] [CrossRef]
- Borello, E.; Domenici, V. Determination of pigments in virgin and extra-virgin olive oils: A comparison between two near UV-Vis spectroscopic techniques. Foods 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Huang, J.Z. An Introduction to Statistical Learning: With Applications in R By Gareth James, Trevor Hastie, Robert Tibshirani, Daniela Witten; Springer: New York, NY, USA, 2014. [Google Scholar]
- Platt, J.C. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In Advances in Large Margin Classifiers; MIT Press: Cambridge, MA, USA, 1999; pp. 61–74. [Google Scholar]
- Rish, I. An empirical study of the naive Bayes classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, DC, USA, 4–10 August 2001; Volume 3, pp. 41–46. [Google Scholar]
- Islam, M.J.; Wu, Q.J.; Ahmadi, M.; Sid-Ahmed, M.A. Investigating the performance of naive-bayes classifiers and k-nearest neighbor classifiers. In Proceedings of the 2007 International Conference on Convergence Information Technology (ICCIT 2007), Gwangju, Korea, 21–23 November 2007; pp. 1541–1546. [Google Scholar]
- Berrar, D. Bayes’ theorem and naive Bayes classifier. In Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics; Elsevier Science Publisher: Amsterdam, The Netherlands, 2018; pp. 403–412. [Google Scholar]
- Bewick, V.; Cheek, L.; Ball, J. Statistics review 14: Logistic regression. Crit. Care 2005, 9, 1–7. [Google Scholar] [CrossRef]
- Lemeshow, S.; Hosmer, D.W., Jr. A review of goodness of fit statistics for use in the development of logistic regression models. Am. J. Epidemiol. 1982, 115, 92–106. [Google Scholar] [CrossRef]
- Van Houwelingen, J.; Le Cessie, S. Logistic regression, a review. Stat. Neerl. 1988, 42, 215–232. [Google Scholar] [CrossRef]
- Martinez, A.M.; Kak, A.C. Pca versus lda. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 228–233. [Google Scholar] [CrossRef]
- Tominaga, Y. Comparative study of class data analysis with PCA-LDA, SIMCA, PLS, ANNs, and k-NN. Chemom. Intell. Lab. Syst. 1999, 49, 105–115. [Google Scholar] [CrossRef]
- Rokach, L.; Maimon, O. Top-down induction of decision trees classifiers—A survey. IEEE Trans. Syst. Man Cybern. Part C Appl. Rev. 2005, 35, 476–487. [Google Scholar] [CrossRef]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man, Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef]
- Song, Y.Y.; Ying, L. Decision tree methods: Applications for classification and prediction. Shanghai Arch. Psychiatry 2015, 27, 130. [Google Scholar]
- Swain, P.H.; Hauska, H. The decision tree classifier: Design and potential. IEEE Trans. Geosci. Electron. 1977, 15, 142–147. [Google Scholar] [CrossRef]
- Ho, T.K. Random decision forests. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 1, pp. 278–282. [Google Scholar]
- Ho, T.K. The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 832–844. [Google Scholar]
- Amit, Y.; Geman, D. Shape Quantization and Recognition with Randomized Trees. Neural Comput. 1997, 9, 1545–1588. [Google Scholar] [CrossRef]
- Hodges, J.L. Discriminatory Analysis; Number 11; USAF School of Aviation Medicine: Dayton, OH, USA, 1950. [Google Scholar]
- Altman, N.S. An Introduction to Kernel and Nearest-Neighbor Nonparametric Regression. Am. Stat. 1992, 46, 175–185. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.A. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Michelucci, U.; Venturini, F. Estimating Neural Network’s Performance with Bootstrap: A Tutorial. Mach. Learn. Knowl. Extr. 2021, 3, 357–373. [Google Scholar] [CrossRef]
- Galeano Díaz, T.; Durán Merás, I.; Correa, C.A.; Roldán, B.; Rodríguez Cáceres, M.I. Simultaneous fluorometric determination of chlorophylls a and b and pheophytins a and b in olive oil by partial least-squares calibration. J. Agric. Food Chem. 2003, 51, 6934–6940. [Google Scholar] [CrossRef] [PubMed]
- El Orche, A.; Bouatia, M.; Mbarki, M. Rapid analytical method to characterize the freshness of olive oils using fluorescence spectroscopy and chemometric algorithms. J. Anal. Methods Chem. 2020, 2020, 8860161. [Google Scholar] [CrossRef] [PubMed]
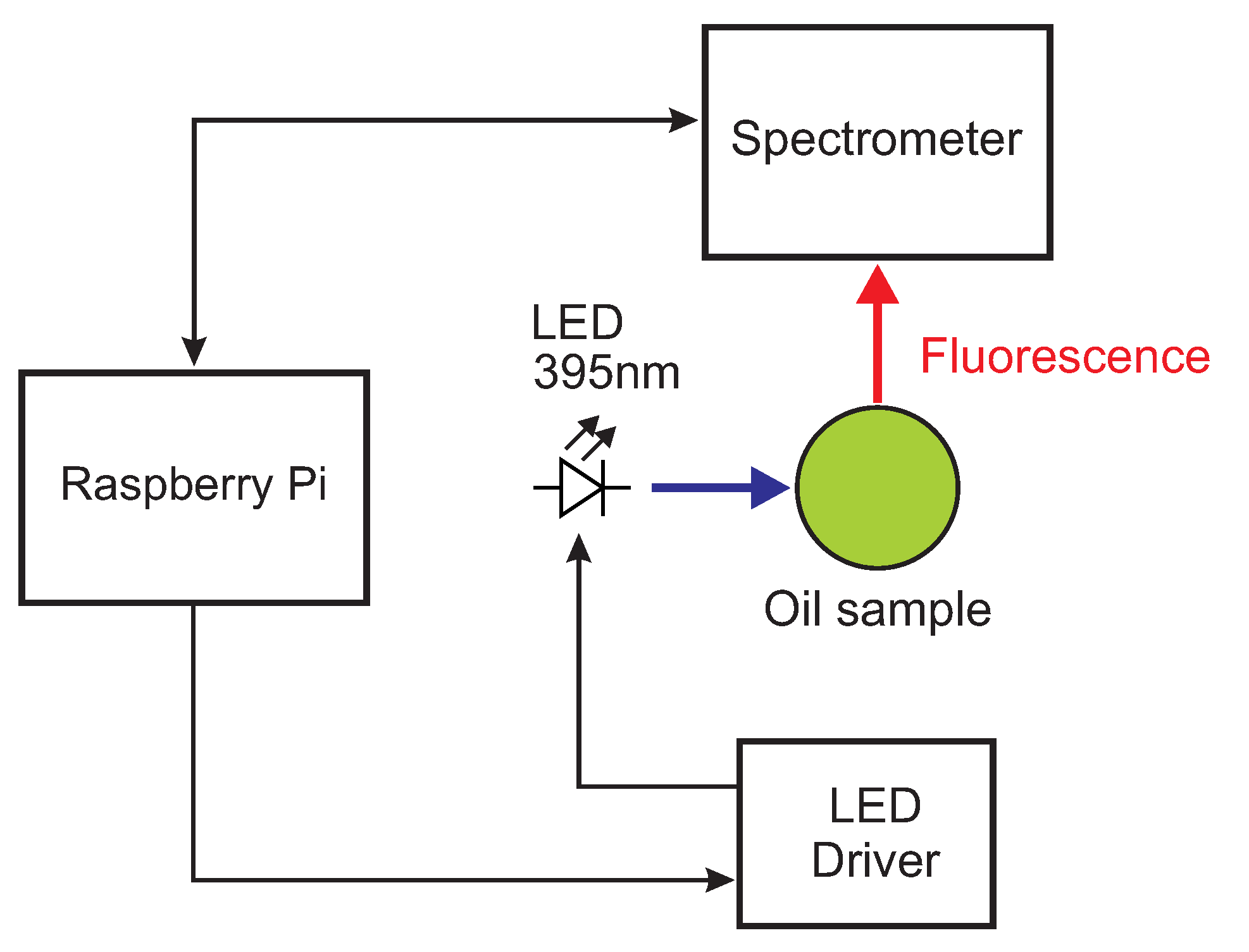

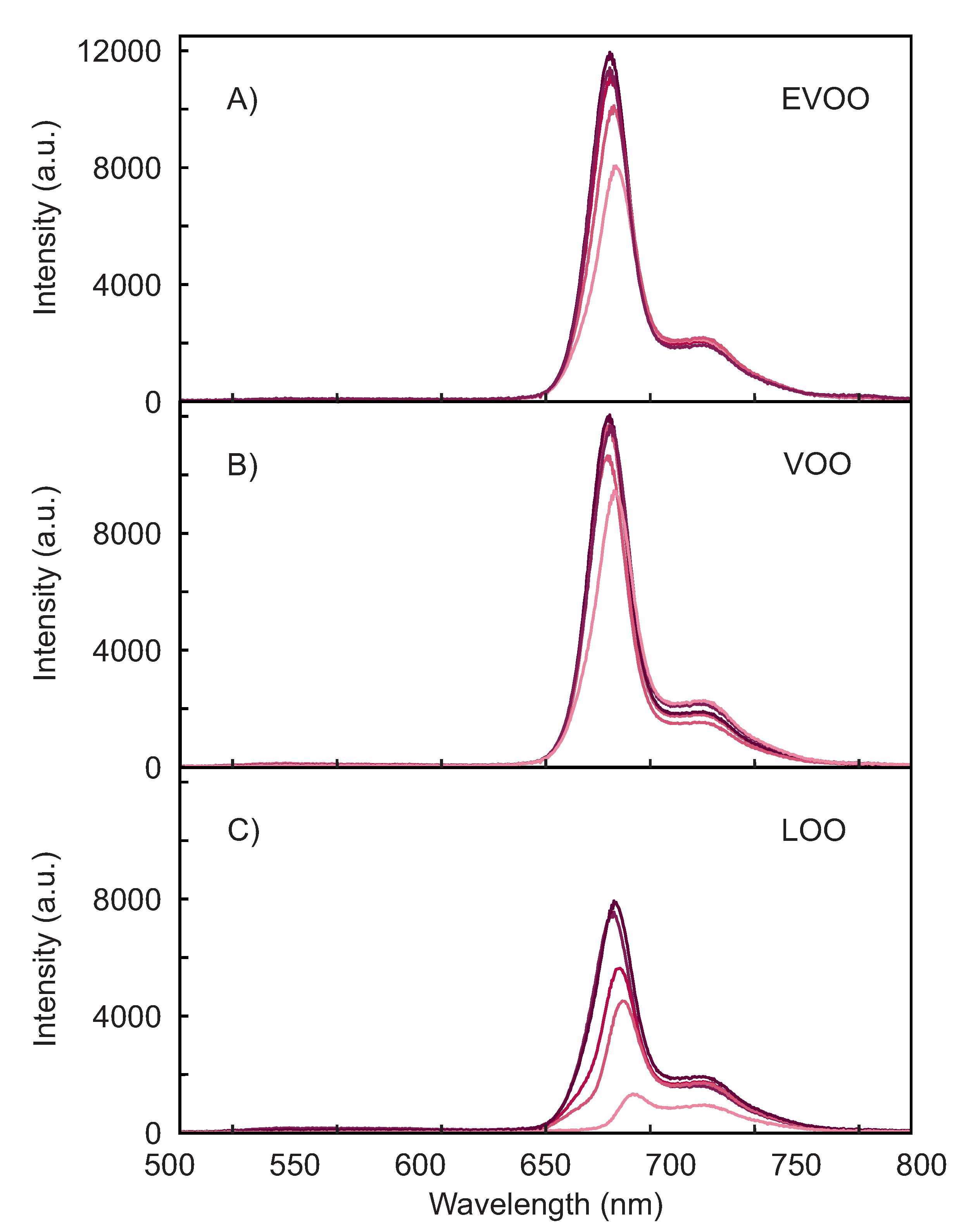
| Quality | Number of Samples |
|---|---|
| EVOO | 12 |
| VOO | 8 |
| LOO | 7 |
| Algorithm | Implementation Details | References |
|---|---|---|
| SVM | Regularization parameter , kernel = radial basis function | [31,32,33] |
| NB | None | [32,34,35,36] |
| MLR | regularization penalty = l2, solver algorithm = Newton conjugate gradient | [32,37,38,39] |
| PCA + LDA | Number of components used with LDA: 2, 3, 4, 5, 10, 15, 20, 25, and 30 | [32,40,41] |
| DT | Split quality criterion used = Gini impurity | [32,42,43,44,45] |
| RF | Number of trees = 100, split quality criterion used = Gini impurity | [32,42,46,47,48] |
| k-NN | Numbers of neighbors | [32,49,50] |
| Algorithm | Average Accuracy | Standard Deviation |
|---|---|---|
| SVM | 0.51 | 0.07 |
| NB | 0.64 | 0.05 |
| MLR | 0.88 | 0.03 |
| PCA + LDA | 0.93 | 0.02 |
| (10 PCA Components) | ||
| DT | 0.99 | 0.01 |
| ANN | 0.99 | 0.04 |
| PCA + LDA | 0.999 | 0.006 |
| (30 PCA Components) | ||
| RF | 1.0 | 0.0 |
| k-NN | 1.0 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venturini, F.; Sperti, M.; Michelucci, U.; Herzig, I.; Baumgartner, M.; Caballero, J.P.; Jimenez, A.; Deriu, M.A. Exploration of Spanish Olive Oil Quality with a Miniaturized Low-Cost Fluorescence Sensor and Machine Learning Techniques. Foods 2021, 10, 1010. https://doi.org/10.3390/foods10051010
Venturini F, Sperti M, Michelucci U, Herzig I, Baumgartner M, Caballero JP, Jimenez A, Deriu MA. Exploration of Spanish Olive Oil Quality with a Miniaturized Low-Cost Fluorescence Sensor and Machine Learning Techniques. Foods. 2021; 10(5):1010. https://doi.org/10.3390/foods10051010
Chicago/Turabian StyleVenturini, Francesca, Michela Sperti, Umberto Michelucci, Ivo Herzig, Michael Baumgartner, Josep Palau Caballero, Arturo Jimenez, and Marco Agostino Deriu. 2021. "Exploration of Spanish Olive Oil Quality with a Miniaturized Low-Cost Fluorescence Sensor and Machine Learning Techniques" Foods 10, no. 5: 1010. https://doi.org/10.3390/foods10051010
APA StyleVenturini, F., Sperti, M., Michelucci, U., Herzig, I., Baumgartner, M., Caballero, J. P., Jimenez, A., & Deriu, M. A. (2021). Exploration of Spanish Olive Oil Quality with a Miniaturized Low-Cost Fluorescence Sensor and Machine Learning Techniques. Foods, 10(5), 1010. https://doi.org/10.3390/foods10051010









