Discrimination of the Geographical Origin of Soybeans Using NMR-Based Metabolomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Soybean Samples
2.2. Chemicals and Reagents
2.3. Climate Data for Soybean Cultivation Regions in Korea and China
2.4. NMR Measurement and Peak Assignment
2.5. Data Processing and Statistical Analyses
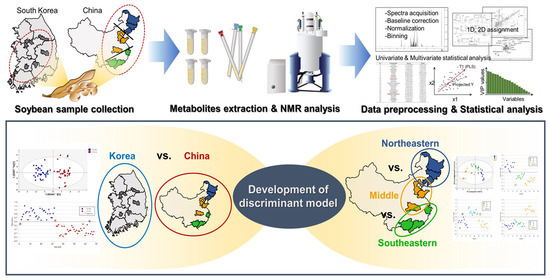
3. Results & Discussion
3.1. Size Measurement of Soybean Samples
3.2. Identification of Soybean Metabolites Using NMR Spectroscopy
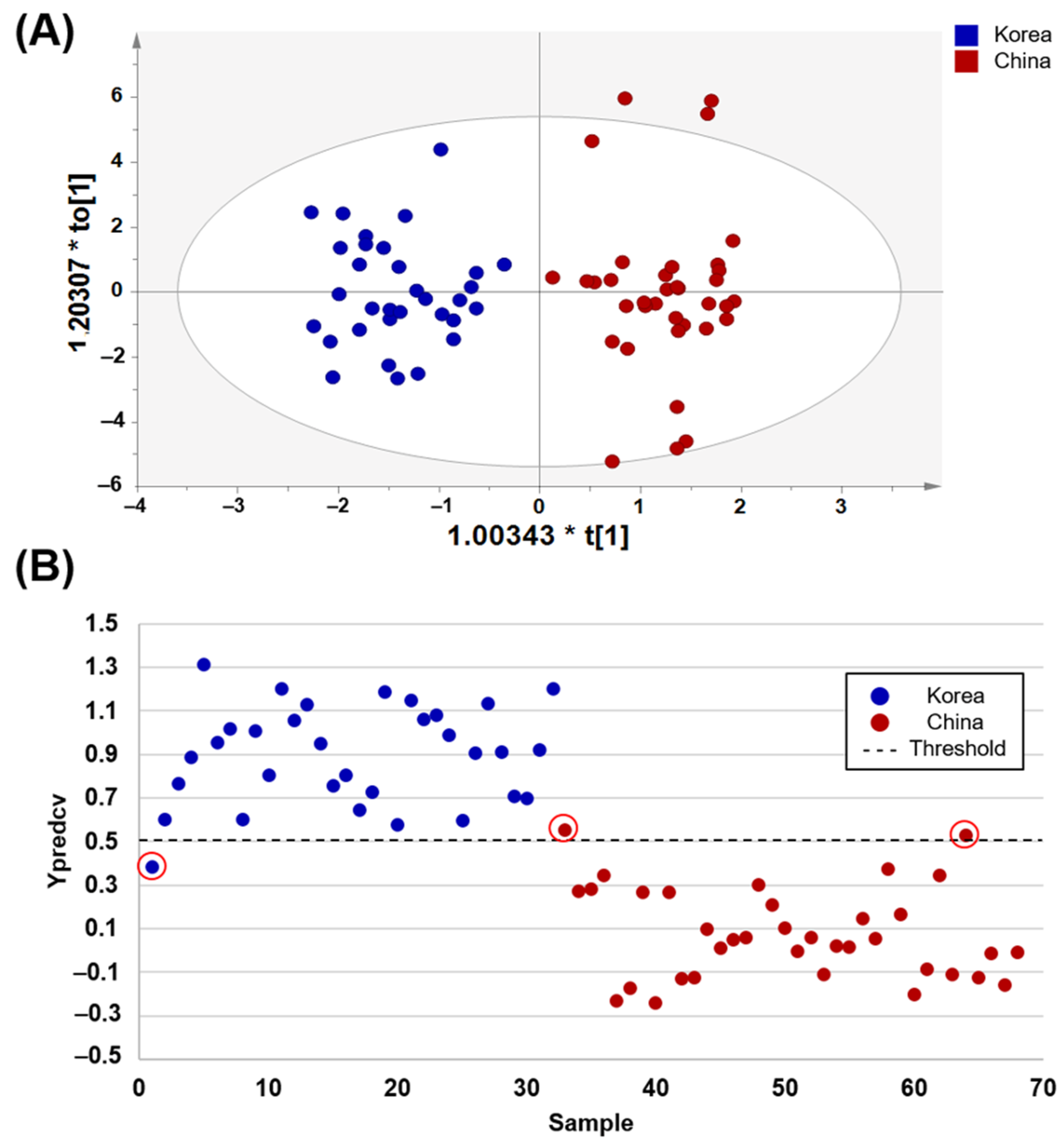
3.3. Discrimination and Prediction of Korean and Chinese Soybean Samples
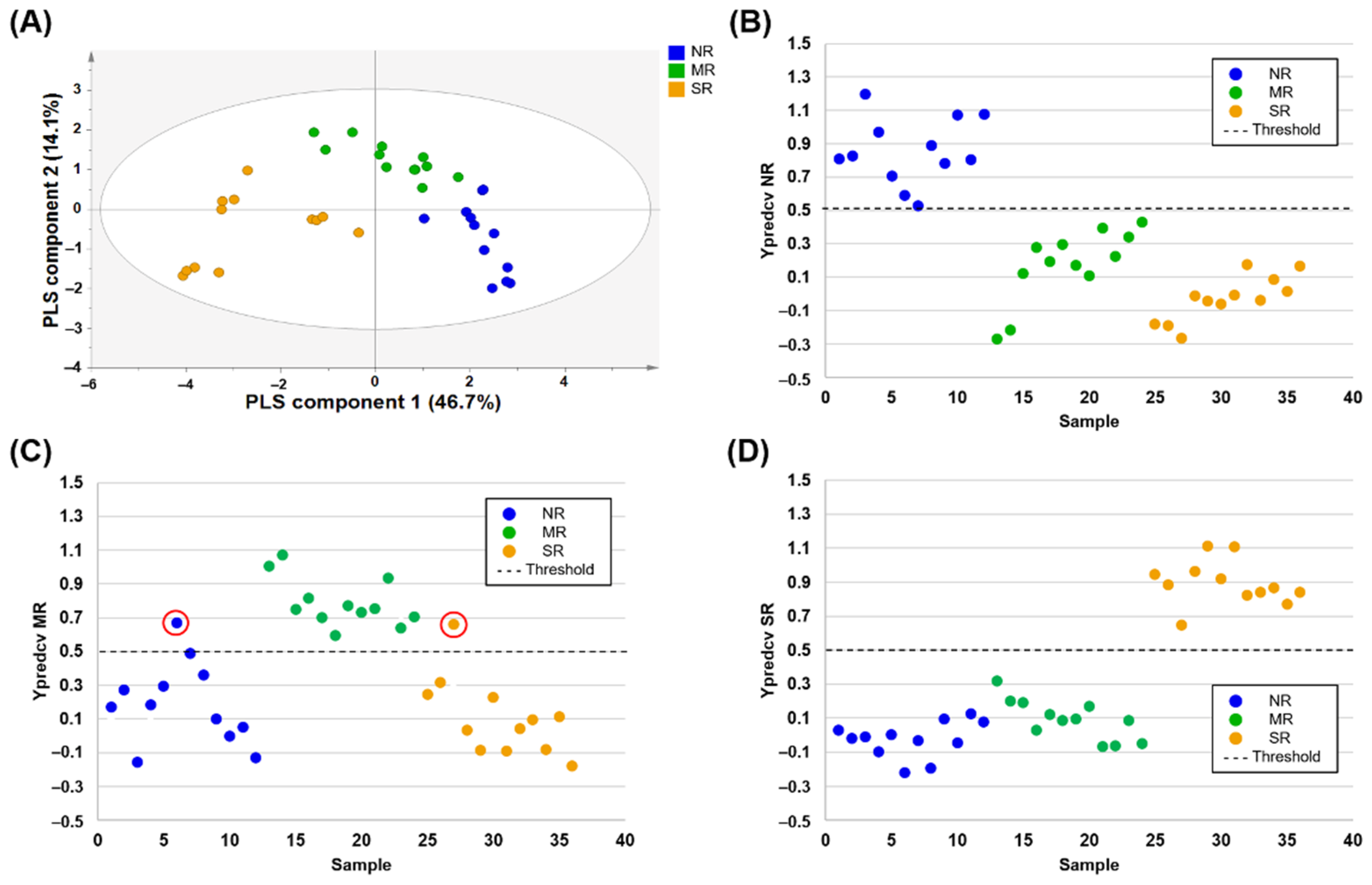
3.4. Discrimination and Prediction of Domestic Chinese Soybean Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.-H.; Lin, Y.-H.; Reid, D.E.; Gresshoff, P.M. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Jooyandeh, H. Soy products as healthy and functional foods. Middle East J. Sci. Res. 2011, 7, 71–80. [Google Scholar]
- Fournier, D.B.; Erdman, J.W., Jr.; Gordon, G.B. Soy, its components, and cancer prevention: A review of the in vitro, animal, and human data. Cancer Epidemiol. Biomark. Prev. 1998, 7, 1055–1065. [Google Scholar]
- Berdal, K.G.; Holst-Jensen, A. Roundup Ready® soybean event-specific real-time quantitative PCR assay and estimation of the practical detection and quantification limits in GMO analyses. Eur. Food Res. Technol. 2001, 213, 432–438. [Google Scholar] [CrossRef]
- Stevenson, S.E.; Woods, C.A.; Hong, B.; Kong, X.; Thelen, J.J.; Ladics, G.S. Environmental effects on allergen levels in commercially grown non-genetically modified soybeans: Assessing variation across North America. Front. Plant Sci. 2012, 3, 196. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Choung, M.G. Comparison of nutritional components in soybean varieties with different geographical origins. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 254–263. [Google Scholar] [CrossRef]
- Wu, H.J.; Deng, J.C.; Yang, C.Q.; Zhang, J.; Zhang, Q.; Wang, X.C.; Yang, F.; Yang, W.Y.; Liu, J. Metabolite profiling of isoflavones and anthocyanins in black soybean [Glycine max (L.) Merr.] seeds by HPLC-MS and geographical differentiation analysis in Southwest China. Anal. Methods 2017, 9, 792–802. [Google Scholar] [CrossRef]
- Qin, G.N.; Verstegen, M.W.A.; Van der Poel, A.F.B. Effect of temperature and time during steam treatment on the protein quality of full-fat soybeans from different origins. J. Sci. Food Agric. 1998, 77, 393–398. [Google Scholar] [CrossRef]
- Longobardi, F.; Ventrella, A.; Napoli, C.; Humpfer, E.; Schütz, B.; Schäfer, H.; Kontominas, M.G.; Sacco, A. Classification of olive oils according to geographical origin by using 1H NMR fingerprinting combined with multivariate analysis. Food Chem. 2012, 130, 177–183. [Google Scholar] [CrossRef]
- Wu, Z.; Zhao, Y.; Zhang, J.; Wang, Y. Quality assessment of Gentiana rigescens from different geographical origins using FT-IR spectroscopy combined with HPLC. Molecules 2017, 22, 1238. [Google Scholar] [CrossRef]
- Yudthavorasit, S.; Wongravee, K.; Leepipatpiboon, N. Characteristic fingerprint based on gingerol derivative analysis for discrimination of ginger (Zingiber officinale) according to geographical origin using HPLC-DAD combined with chemometrics. Food Chem. 2014, 158, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Guo, J.; Xia, Q.; Zhao, G.; Zhou, H.; Xie, F. Metabolic profiling of Chinese tobacco leaf of different geographical origins by GC-MS. J. Agric. Food Chem. 2013, 61, 2597–2605. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, Y.; Hu, C.; Zhao, C.; Zhang, J.; Li, L.; Zeng, J.; Peng, X.; Lu, X.; Xu, G. Metabolic profiling with gas chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry reveals the carbon-nitrogen status of tobacco leaves across different planting areas. J. Proteome Res. 2016, 15, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Consonni, R.; Cagliani, L.R. Geographical characterization of polyfloral and acacia honeys by nuclear magnetic resonance and chemometrics. J. Agric. Food Chem. 2008, 56, 6873–6880. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.E.; Gaudillere, J.P.; Van Leeuwen, C.; Hilbert, G.; Maucourt, M.; Deborde, C.; Moing, A.; Rolin, D. 1H NMR metabolite fingerprints of grape berry: Comparison of vintage and soil effects in Bordeaux grapevine growing areas. Anal. Chim. Acta 2006, 563, 346–352. [Google Scholar] [CrossRef]
- Lin, H.; Rao, J.; Shi, J.; Hu, C.; Cheng, F.; Wilson, Z.A.; Zhang, D.; Quan, S. Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. J. Integr. Plant Biol. 2014, 56, 826–836. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Skogerson, K.; Macisaac, S.; Bickel, A.; Perez, T.; Li, X. Application of 1H NMR profiling to assess seed metabolomic diversity. A case study on a soybean era population. J. Agric. Food Chem. 2015, 63, 4690–4697. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, F.; Innamorato, V.; Di Gioia, A.; Ventrella, A.; Lippolis, V.; Logrieco, A.F.; Catucci, L.; Agostiano, A. Geographical origin discrimination of lentils (Lens culinaris Medik.) using 1H NMR fingerprinting and multivariate statistical analyses. Food Chem. 2017, 237, 743–748. [Google Scholar] [CrossRef]
- Mannino, G.; Di Stefano, V.; Lauria, A.; Pitonzo, R.; Gentile, C. Vaccinium macrocarpon (Cranberry)-based dietary supplements: Variation in mass uniformity, proanthocyanidin dosage and anthocyanin profile demonstrates quality control standard needed. Nutrients 2020, 12, 992. [Google Scholar] [CrossRef]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2009, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.W.; Kim, S.H.; Park, S.J.; Hyun, S.H.; Lee, S.Y.; Auh, J.H.; Lee, H.J.; Cho, S.M.; Kim, J.H.; Choi, H.K. Effect of Korean black raspberry (Rubus coreanus Miquel) fruit administration on DNA damage levels in smokers and screening biomarker investigation using 1H-NMR-based metabolic profiling. Food Res. Int. 2013, 54, 1255–1262. [Google Scholar] [CrossRef]
- Rubingh, C.M.; Bijlsma, S.; Derks, E.P.; Bobeldijk, I.; Verheij, E.R.; Kochhar, S.; Smilde, A.K. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics 2006, 2, 53–61. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zeng, N.; Wang, N. Sensitivity, Specificity, Accuracy, Associated Confidence Interval and Roc Analysis with Practical Sas Implementations; NESUG Proceeding of Health Care and Life Sciences: Baltimore, MD, USA, 2010. [Google Scholar]
- Mattioni, N.M.; Schuch, L.O.B.; Villela, F.A.; Mertz, L.M.; Peske, S.T. Soybean seed size and quality as a function of soil compaction. Seed Sci. Technol. 2012, 40, 333–343. [Google Scholar] [CrossRef]
- Brown, E.A.; Caviness, C.E.; Brown, D.A. Response of selected soybean cultivars to soil moisture deficit. Agron. J. 1985, 77, 274–278. [Google Scholar] [CrossRef]
- Gutierrez-Boem, F.; Thomas, G.W. Phosphorus nutrition and water deficits in field-grown soybeans. Plant Soil 1999, 207, 87–96. [Google Scholar] [CrossRef]
- Moles, A.T.; Westoby, M. Seedling survival and seed size: A synthesis of the literature. J. Ecol. 2004, 92, 372–383. [Google Scholar] [CrossRef]
- Lee, G.A.; Crawford, G.W.; Liu, L.; Sasaki, Y.; Chen, X. Archaeological soybean (Glycine max) in east Asia: Does size matter? PLoS ONE. 2011, 6, e26720. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.M.; Klein, M.S.; Hochrein, J.; Oefner, P.J.; Spang, R.; Gronwald, W. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics 2012, 8, 146–160. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.; Cloareo, O.; Holmes, E.; Nicholson, J.K.; Lindon, J.C. Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Anal. Chem. 2006, 78, 2262–2267. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Johansson, E.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Part I: Basic Principles and Applications. In Multi-and Megavariate Data Analysis, 2nd ed.; Umetrics Academy: Stockholm, Sweden, 2006; pp. 39–101. [Google Scholar]
- Li, B.; Tang, J.; Yang, Q.; Cui, X.; Li, S.; Chen, S. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci. Rep. 2016, 6, 38881. [Google Scholar] [CrossRef]
- Weljie, A.M.; Newton, J.; Mercier, P.; Carlson, E.; Slupsky, C.M. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006, 78, 4430–4442. [Google Scholar] [CrossRef]
- Beleggia, R.; Platani, C.; Nigro, F.; de Vita, P.; Cattivelli, L.; Papa, R. Effect of genotype, environment and genotype-by-environment interaction on metabolite profiling in durum wheat (Triticum durum Desf.) grain. J. Cereal Sci. 2013, 57, 183–192. [Google Scholar] [CrossRef]
- Cohen, H.; Shir, O.M.; Yu, Y.; Hou, W.; Sun, S.; Han, T.; Amir, R. Genetic background and environmental conditions drive metabolic variation in wild type and transgenic soybean (Glycine max) seeds. Plant Cell Environ. 2016, 39, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.S.; Zhao, L.M.; Liu, B.; Wang, Z.W.; Jin, Z.O.; Sun, H. The genetic diversity of cultivated soybean grown in China. Theor. Appl. Genet. 2004, 108, 931–936. [Google Scholar] [CrossRef]
- Lee, J.D.; Yu, J.K.; Hwang, Y.H.; Blake, S.; So, Y.S.; Lee, G.J.; Nguyen, H.T.; Shannon, J.G. Genetic diversity of wild soybean (Glycine soja Sieb. and Zucc.) accessions from South Korea and other countries. Crop Sci. 2008, 48, 606–616. [Google Scholar] [CrossRef]
- Lee, J.D.; Vuong, T.D.; Moon, H.; Yu, J.K.; Nelson, R.; Nguyen, H.T. Genetic diversity and population structure of Korean and Chinese soybean [Glycine max (L.) Merr.] accessions. Crop Sci. 2011, 51, 1080–1088. [Google Scholar] [CrossRef]
- Li, Z.; Nelson, R.L. Genetic diversity among soybean accessions from three countries measured by RAPDs. Crop Sci. 2001, 41, 1337–1347. [Google Scholar] [CrossRef]
- Han, O.K.; Abe, J.; Shimamoto, Y. Genetic diversity of soybean landraces in Korea. Korean J. Crop Sci. 1999, 44, 256–262. [Google Scholar]
- Li, Y.; Guan, R.; Liu, Z.; Ma, Y.; Wang, L.; Li, L.; Lin, F.; Luan, W.; Chen, P.; Yan, Z.; et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 2008, 117, 857–871. [Google Scholar] [CrossRef]
- Qin, X.; Feng, F.; Li, D.; Herbert, S.J.; Liao, Y.; Siddique, K.H.M. Changes in yield and agronomic traits of soybean cultivars released in China in the last 60 years. Crop Pasture Sci. 2017, 68, 973–984. [Google Scholar] [CrossRef]
- Xiong, D.; Zhao, T.; Gai, J. Genetic bases of improved soybean cultivars released from 1923 to 2005 in China—A historical review. Front. Agric. China 2010, 4, 383–393. [Google Scholar] [CrossRef]
- Das, A.; Rushton, P.J.; Rohila, J.S. Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants 2017, 6, 21. [Google Scholar] [CrossRef]
- Cox, M.S.; Gerard, P.D.; Wardlaw, M.C.; Abshire, M.J. Variability of selected soil properties and their relationships with soybean yield. Soil Sci. Soc. Am. J. 2003, 67, 1296–1302. [Google Scholar] [CrossRef]
- Bellaloui, N.; Hanks, J.E.; Fisher, D.K.; Mengistu, A. Soybean seed composition is influenced by within-field variability in soil nutrients. Crop Manag. 2009, 8, 1–12. [Google Scholar] [CrossRef]
- Gao, M.; Li, S. Relationship between soybean yield/quality and soil quality in a major soybean producing area based on a 2D-QSAR model. AIP Conf. Proc. 2017, 1839, 020071. [Google Scholar] [CrossRef]
- Wijewardana, C.; Reddy, K.R.; Bellaloui, N. Soybean seed physiology, quality, and chemical composition under soil moisture stress. Food Chem. 2019, 278, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Zulak, K.G.; Weljie, A.M.; Vogel, H.J.; Facchini, P.J. Quantitative 1H NMR metabolomics reveals extensive metabolic reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures. BMC Plant Biol. 2008, 22, 8. [Google Scholar] [CrossRef]
- Thonusin, C.; IglayReger, H.B.; Soni, T.; Rothberg, A.E.; Burant, C.F.; Evans, C.R. Evaluation of intensity drift correction strategies using MetaboDrift, a normalization tool for multi-batch metabolomics data. J. Chromatogr. A 2017, 1523, 265–274. [Google Scholar] [CrossRef]
- van der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Dauwe, R.; Holliday, J.A.; Aitken, S.N.; Mansfield, S.D. Metabolic dynamics during autumn cold acclimation within and among populations of Sitka spruce (Picea sitchensis). New Phytol. 2012, 194, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, I.D.; Henning, L.M.M.; Döpp, S.A.; Nepomuceno, A.; Moraes, L.A.C.; Marcolino-Gomes, J.; Richter, C.; Schwalbe, H.; Colnago, L.A. Flooded soybean metabolomic analysis reveals important primary and secondary metabolites involved in the hypoxia stress response and tolerance. Environ. Exp. Bot. 2018, 153, 176–187. [Google Scholar] [CrossRef]
- Lee, B.-J.; Zhou, Y.; Lee, J.S.; Shin, B.K.; Seo, J.-A.; Lee, D.; Kim, Y.S.; Choi, H.K. Discrimination and prediction of the origin of Chinese and Korean soybeans using Fourier transform infrared spectrometry (FT-IR) with multivariate statistical analysis. PLoS ONE 2018, 13, e0196315. [Google Scholar] [CrossRef] [PubMed]
| No. | Compounds | Chemical Shift | Multiplicity; J Value | Assignment | Assignment Method |
|---|---|---|---|---|---|
| 1 | 2-hydroxyisobutyrate | 1.34 | s | H-3, H-4 | One-dimensional proton NMR (1D)/heteronuclear single quantum correlation (HSQC) |
| 2 | 2-oxoglutarate | 2.44 | t; J = 6.92 | H-5 | 1D/correlation spectroscopy (COSY)/HSQC |
| 2.99 | t; J = 6.92 | H-4 | |||
| 3 | Acetate | 1.91 | s | H-2 | 1D/HSQC |
| 4 | Acetoacetate | 2.28 | s | H-4 | 1D/HSQC |
| 5 | Alanine | 1.47 | d; J = 7.19 | H-3 | 1D/COSY/HSQC |
| 3.78 | q; J = 7.19 | H-2 | |||
| 6 | Asparagine | 2.82–2.88 | m | H-2 | 1D/COSY/HSQC |
| 2.90–2.98 | m | H-2 | |||
| 4.01 | q; J = 4.26 | H-3 | |||
| 7 | Aspartate | 2.62 | dd; J = 8.7, 14.43 | H-2 | 1D/COSY/HSQC |
| 2.79 | dd; J = 3.78, 13.68 | H-2 | |||
| 3.91 | dd; J = 3.75, 4.92 | H-3 | |||
| 8 | Choline | 3.20 | s | H-3, H-4, H-5 | 1D/COSY/HSQC |
| 3.48–3.53 | m | H-2 | |||
| 4.03–4.09 | m | H-1 | |||
| 9 | Citrate | 2.54 | d; J = 15.36 | 2Ha, 4Ha | 1D/COSY/HSQC |
| 2.68 | d; J = 15.36 | 2Hb, 4Hb | |||
| 10 | Formate | 8.46 | s | H-1 | 1D |
| 11 | Fumarate | 6.52 | s | H-2, H-3 | 1D |
| 12 | Galactarate | 3.94 | s | H-3, H-4 | 1D/COSY/HSQC |
| 4.26 | s | H-2, H-5 | |||
| 13 | Glucose | 3.22 | dd; J = 1.44, 7.95 | H-2 | 1D/COSY/HSQC |
| 3.38–3.43 | m | H-4 | |||
| 3.48–3.54 | m | H-5 | |||
| 3.52 | dd; J = 3.7, 9.82 | H-2 | |||
| 3.72–3.78 | m | H-3, H-6 | |||
| 3.80–3.85 | m | H-5, H-6 | |||
| 4.62 | d, J = 7.92 | H-1 | |||
| 5.22 | d; J = 3.72 | H-1 | |||
| 14 | Glutamate | 2.00–2.08 | m | H-3 | 1D/COSY/HSQC |
| 2.10–2.18 | m | H-3 | |||
| 2.28–2.40 | m | H-4 | |||
| 3.75 | dd; J = 4.8, 2.4 | H-2 | |||
| 15 | Hypoxanthine | 8.17 | s | H-2 | 1D/COSY/HSQC |
| 8.20 | s | H-8 | |||
| 16 | Isoleucine | 0.93 | t; J = 7.15 | H-5 | 1D/COSY/HSQC |
| 1.00 | d; J = 7.15 | CH3 | |||
| 1.41–1.49 | m | H-4 | |||
| 1.92–2.01 | m | H-3 | |||
| 3.66 | d; J = 4.08 | H-2 | |||
| 17 | Leucine | 0.94 | t; J = 6.06 | H-5, CH3 | 1D/COSY/HSQC |
| 1.64–1.78 | m | H-3, H-4 | |||
| 3.69–3.76 | m | H-2 | |||
| 18 | Malonate | 3.13 | s | H-2 | 1D/HSQC |
| 19 | Oxypurinol | 8.27 | s | H-7 | 1D/HSQC |
| 20 | Raffinose/Stachyose | 3.52 | t; J = 4.5 | H-4′ | 1D/COSY/HSQC |
| 3.69 | br. s | H-6 | |||
| 3.95 | t; J = 6.36 | H-5″ | |||
| 4.95 | dd; J = 2.7, 4.1 | H-1″ | |||
| 5.41 | d; J = 4.5 | H-1 | |||
| 21 | Succinate | 2.42 | s | H-2, H-3 | 1D/HSQC |
| 22 | Sucrose | 3.55 | dd; J = 3.84, 6.12 | H-1′ | 1D/COSY/HSQC |
| 3.66 | s | H-1 | |||
| 3.75 | t; J = 9.05 | H-3 | |||
| 3.76–3.84 | m | H-6 | |||
| 4.04 | t; J = 9.05 | H-4′ | |||
| 5.39 | d; J = 3.84 | H-1 | |||
| 23 | Tartarate | 4.34 | s | H-2, H-3 | 1D/HSQC |
| 24 | Tryptophan | 7.20–7.24 | m | H-8 | 1D/COSY/HSQC |
| 7.18–7.28 | m | H-9 | |||
| 7.32 | s | H-2 | |||
| 7.71 | d; J = 7.92 | H-7 | |||
| 25 | Valine | 0.98 | d; J = 7.02 | CH3 | 1D/COSY/HSQC |
| 1.05 | d; J = 7.14 | H-4 | |||
| 2.20–2.32 | m | H-3 | |||
| 3.61 | d; J = 4.33 | H-2 |
| Group No. | Normalization Method | Scaling Method | Number of Component | R2Y | Q2Y | R2Y Intercept | Q2Y Intercept |
|---|---|---|---|---|---|---|---|
| 1 | Standard | UV | 4 | 0.844 | 0.762 | 0.218 | −0.430 |
| 2 | Par | 7 | 0.861 | 0.779 | 0.197 | −0.417 | |
| 3 | Total | UV | 5 | 0.882 | 0.783 | 0.254 | −0.487 |
| 4 | Par | 6 | 0.862 | 0.798 | 0.165 | −0.385 |
| Class | Sensitivity % | Specificity % | Accuracy % | |
|---|---|---|---|---|
| Korea vs. China | 96.9 | 94.4 | 95.6 | |
| China | NR vs. MR&SR | 100.0 | 100.0 | 100.0 |
| MR vs. NR&SR | 100.0 | 91.7 | 94.4 | |
| SR vs. NR&MR | 100.0 | 100.0 | 100.0 | |
| Group No. | Normalization Method | Scaling Method | Number of Component | R2Y | Q2Y | R2Y Intercept | Q2Y Intercept |
|---|---|---|---|---|---|---|---|
| 1 | Standard | UV | 6 | 0.898 | 0.651 | 0.348 | −0.821 |
| 2 | Par | 2 | 0.492 | 0.348 | 0.082 | −0.189 | |
| 3 | Total | UV | 3 | 0.731 | 0.622 | 0.197 | −0.345 |
| 4 | Par | 4 | 0.713 | 0.566 | 0.142 | −0.468 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Kim, S.-Y.; Lee, J.-S.; Shin, B.-K.; Seo, J.-A.; Kim, Y.-S.; Lee, D.-Y.; Choi, H.-K. Discrimination of the Geographical Origin of Soybeans Using NMR-Based Metabolomics. Foods 2021, 10, 435. https://doi.org/10.3390/foods10020435
Zhou Y, Kim S-Y, Lee J-S, Shin B-K, Seo J-A, Kim Y-S, Lee D-Y, Choi H-K. Discrimination of the Geographical Origin of Soybeans Using NMR-Based Metabolomics. Foods. 2021; 10(2):435. https://doi.org/10.3390/foods10020435
Chicago/Turabian StyleZhou, Yaoyao, Seok-Young Kim, Jae-Soung Lee, Byeung-Kon Shin, Jeong-Ah Seo, Young-Suk Kim, Do-Yup Lee, and Hyung-Kyoon Choi. 2021. "Discrimination of the Geographical Origin of Soybeans Using NMR-Based Metabolomics" Foods 10, no. 2: 435. https://doi.org/10.3390/foods10020435
APA StyleZhou, Y., Kim, S.-Y., Lee, J.-S., Shin, B.-K., Seo, J.-A., Kim, Y.-S., Lee, D.-Y., & Choi, H.-K. (2021). Discrimination of the Geographical Origin of Soybeans Using NMR-Based Metabolomics. Foods, 10(2), 435. https://doi.org/10.3390/foods10020435