Rapid Identification of Adulteration in Edible Vegetable Oils Based on Low-Field Nuclear Magnetic Resonance Relaxation Fingerprints
Abstract
1. Introduction
2. Materials and Methods
2.1. Oil Sample Preparation
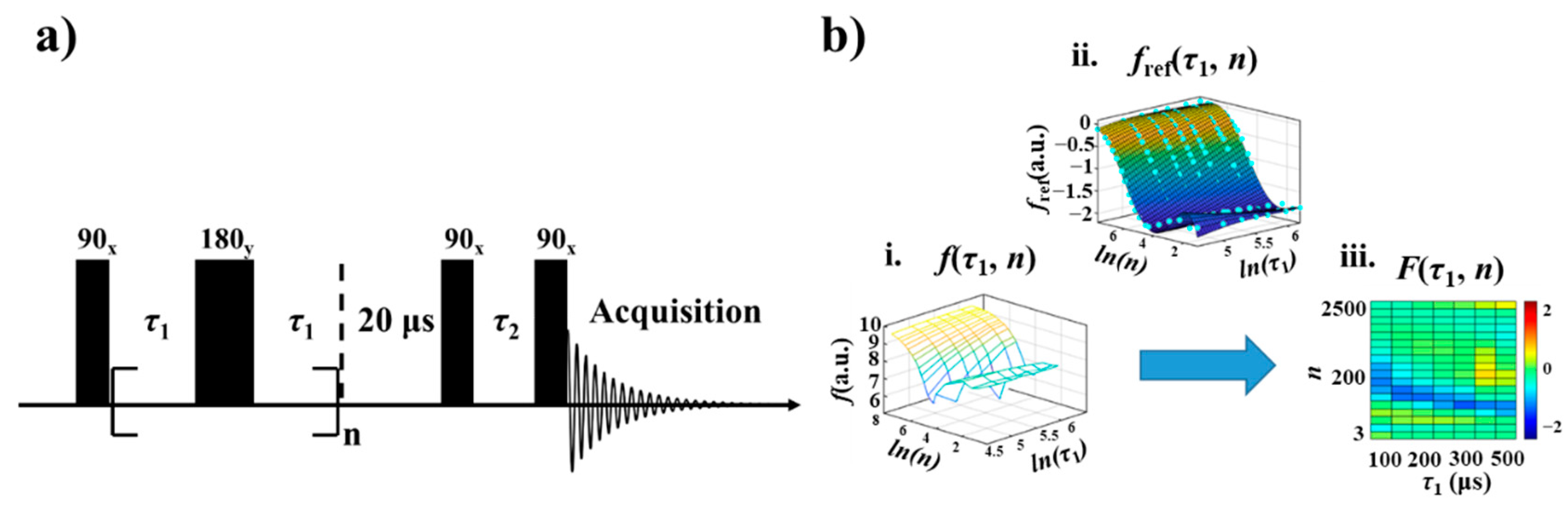
2.2. NMR Measurements
2.3. NMR Relaxation Fingerprints
2.4. Statistical Analysis
3. Results and Discussion
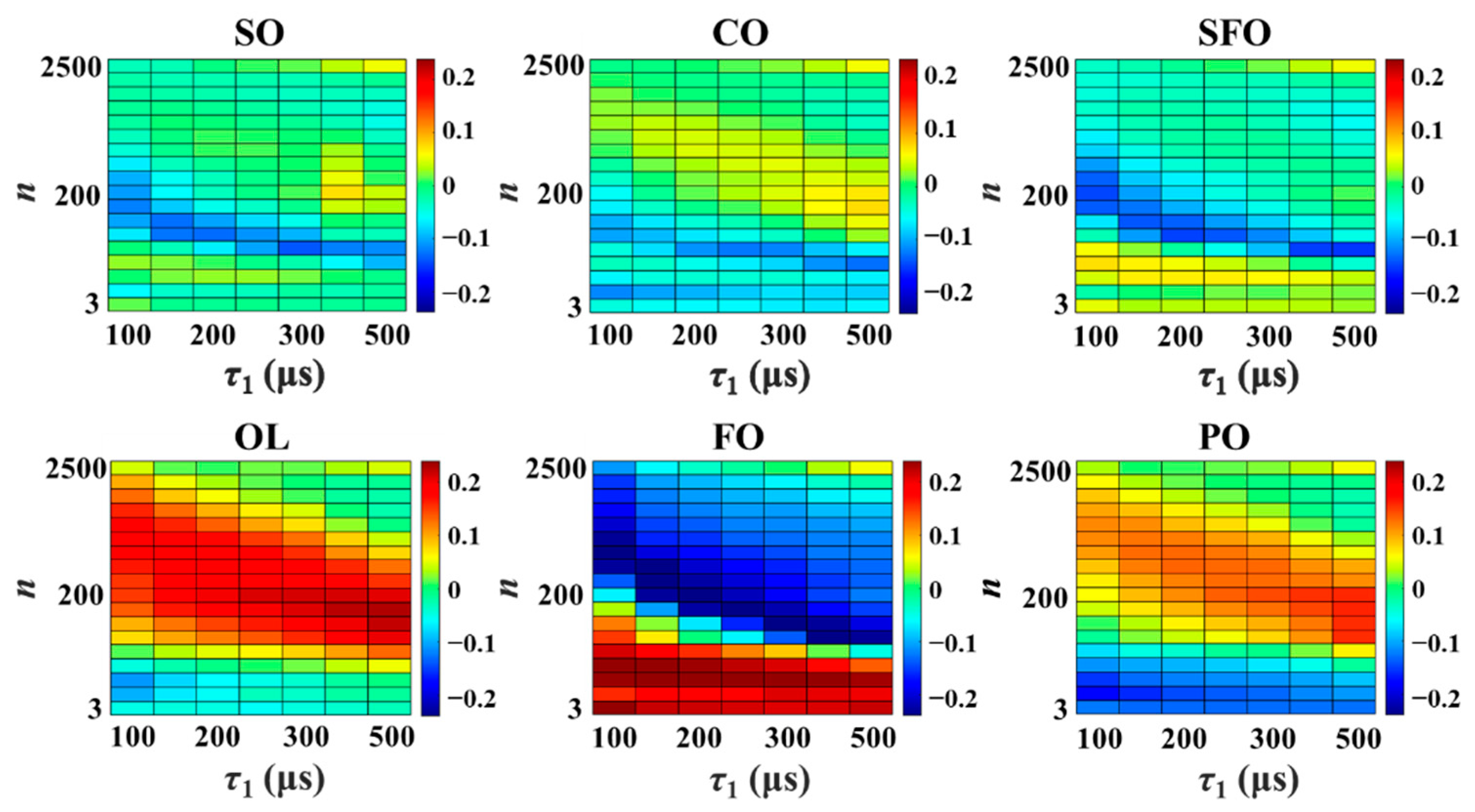
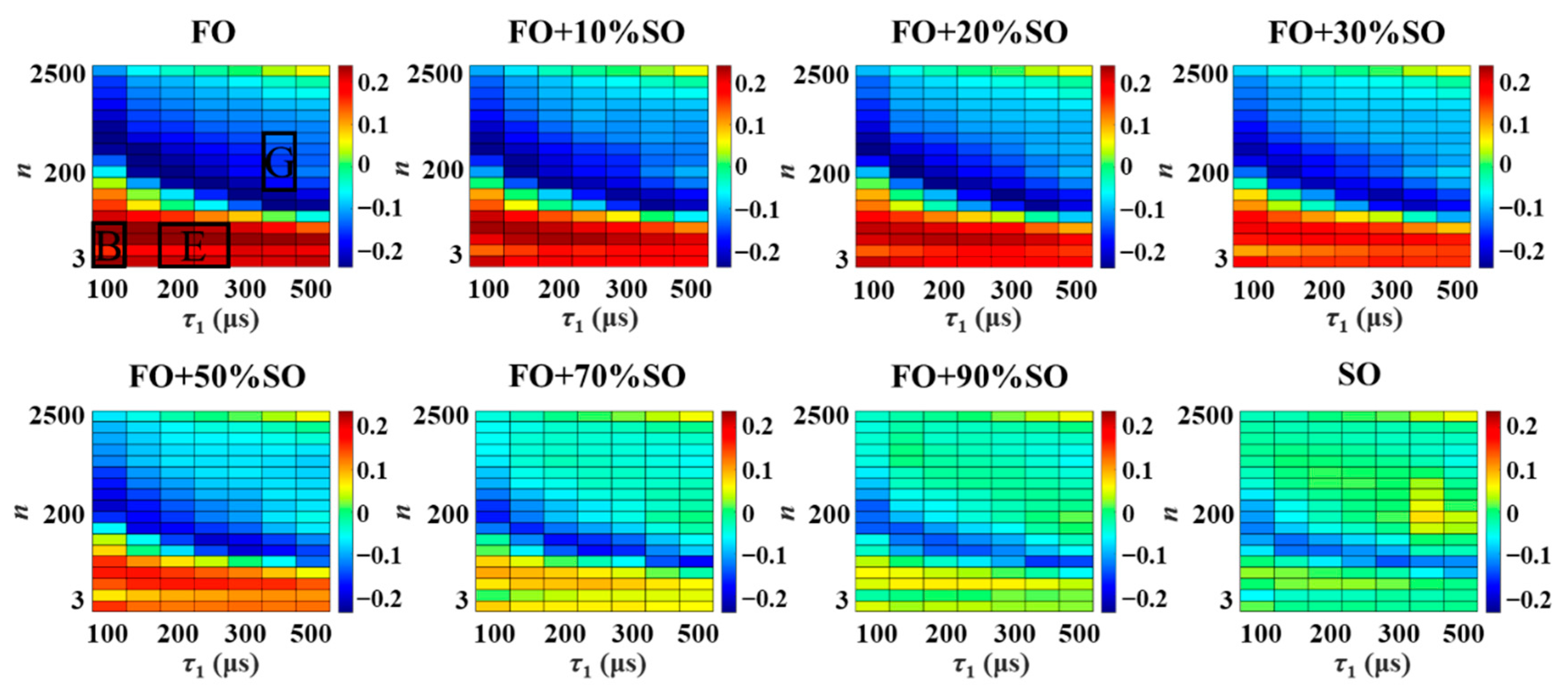
3.1. Relaxation Fingerprints
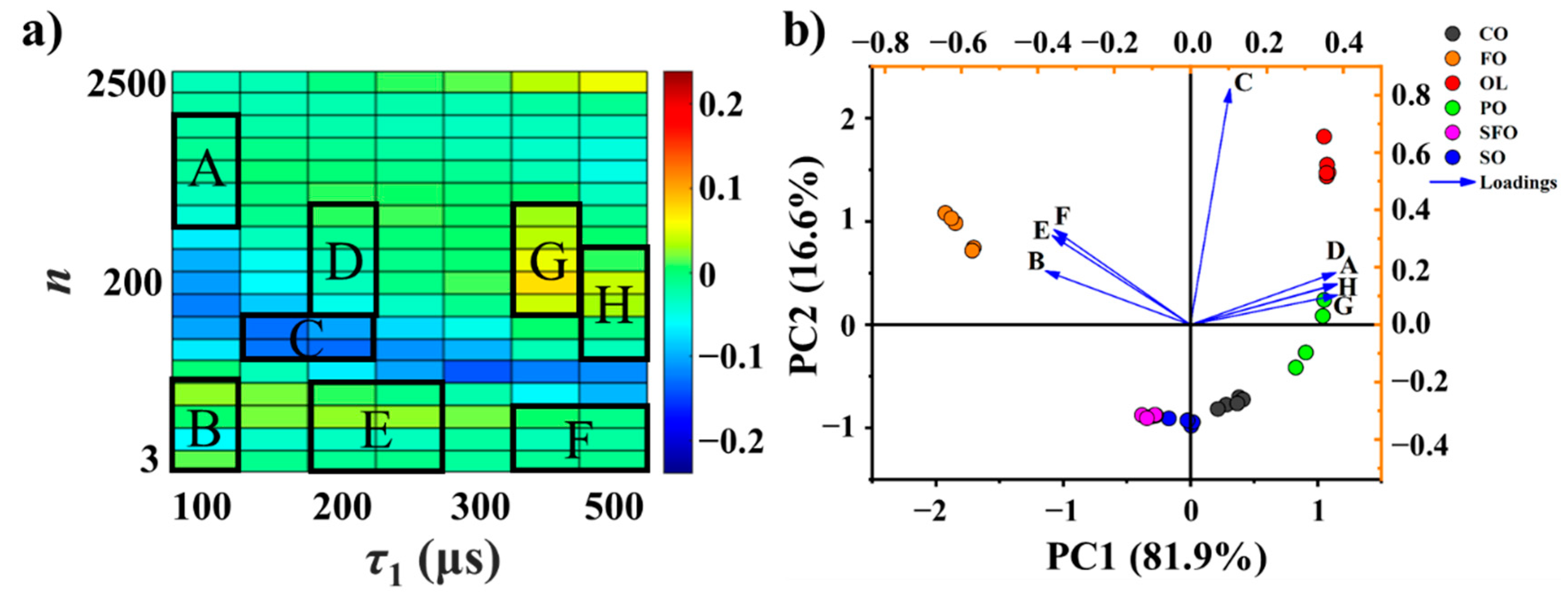
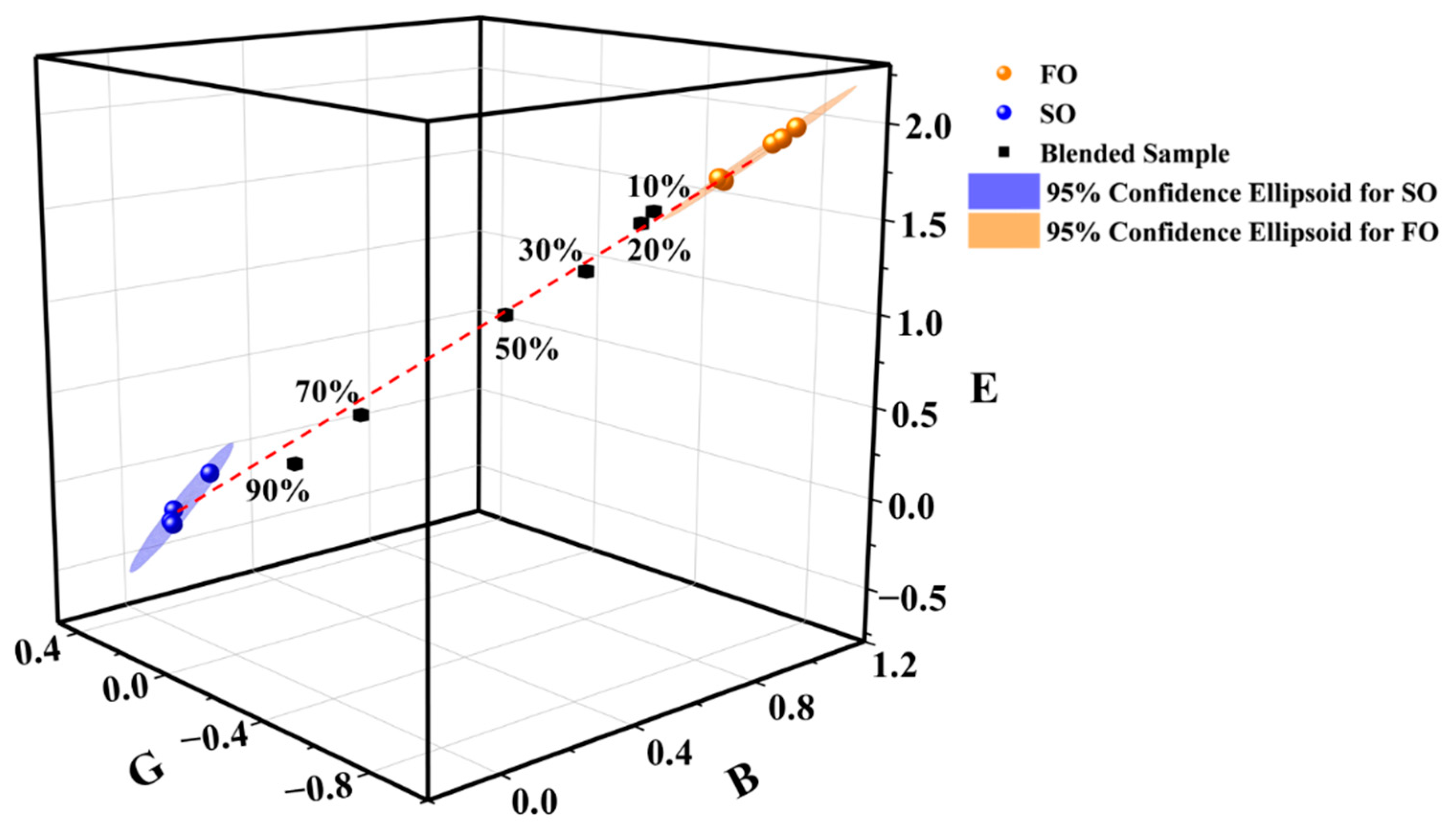
3.2. Characteristic Coordinate System
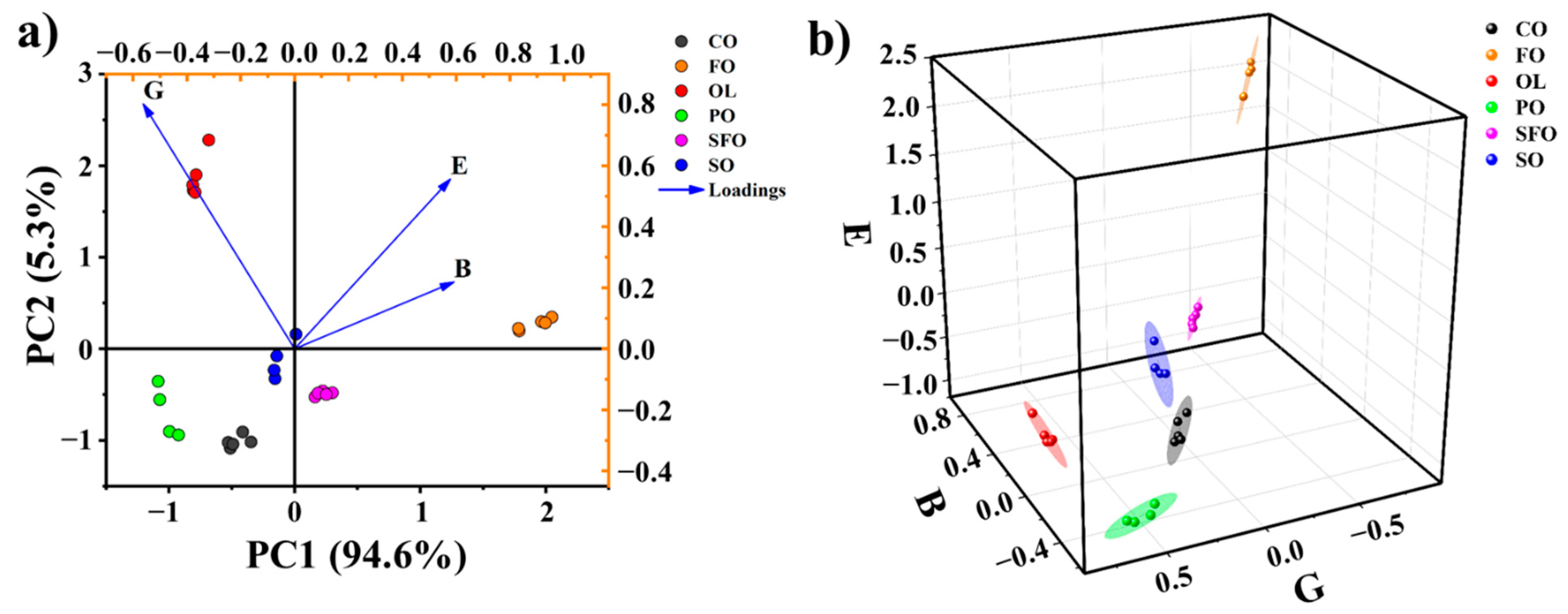
3.3. Adulteration Identification
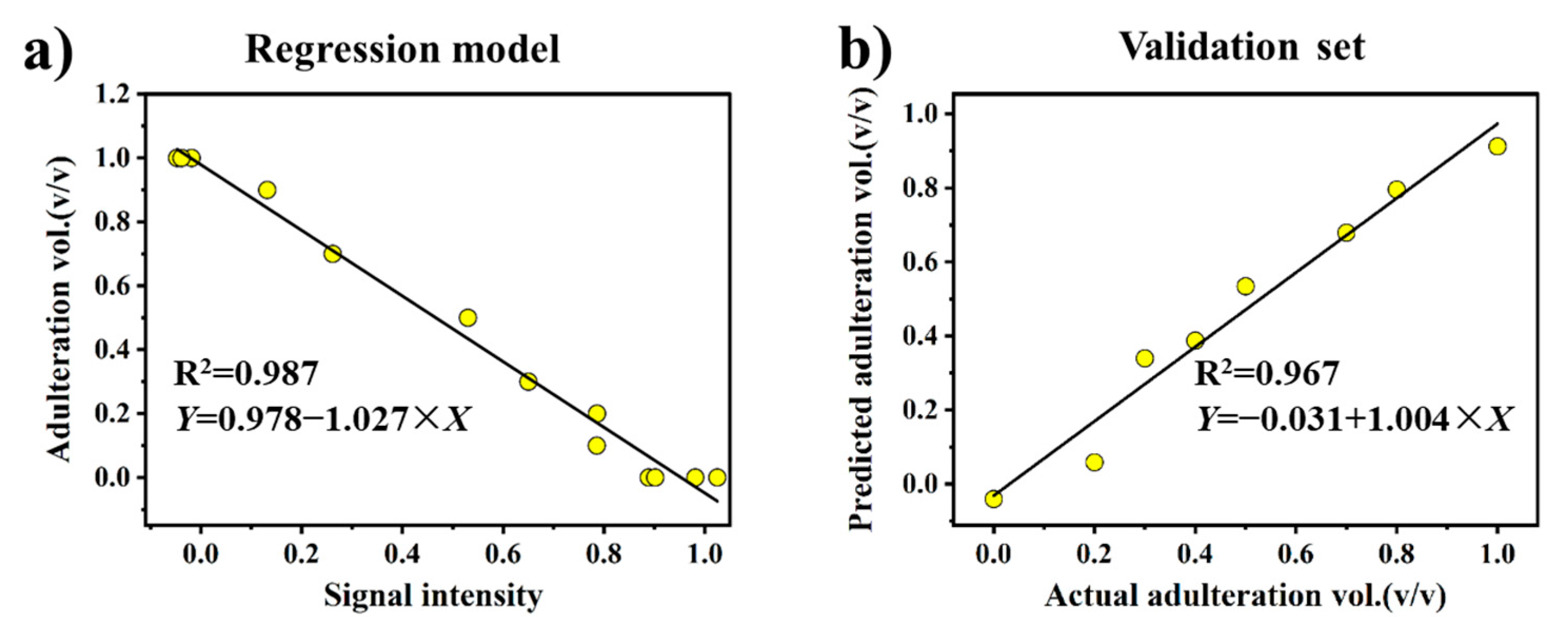
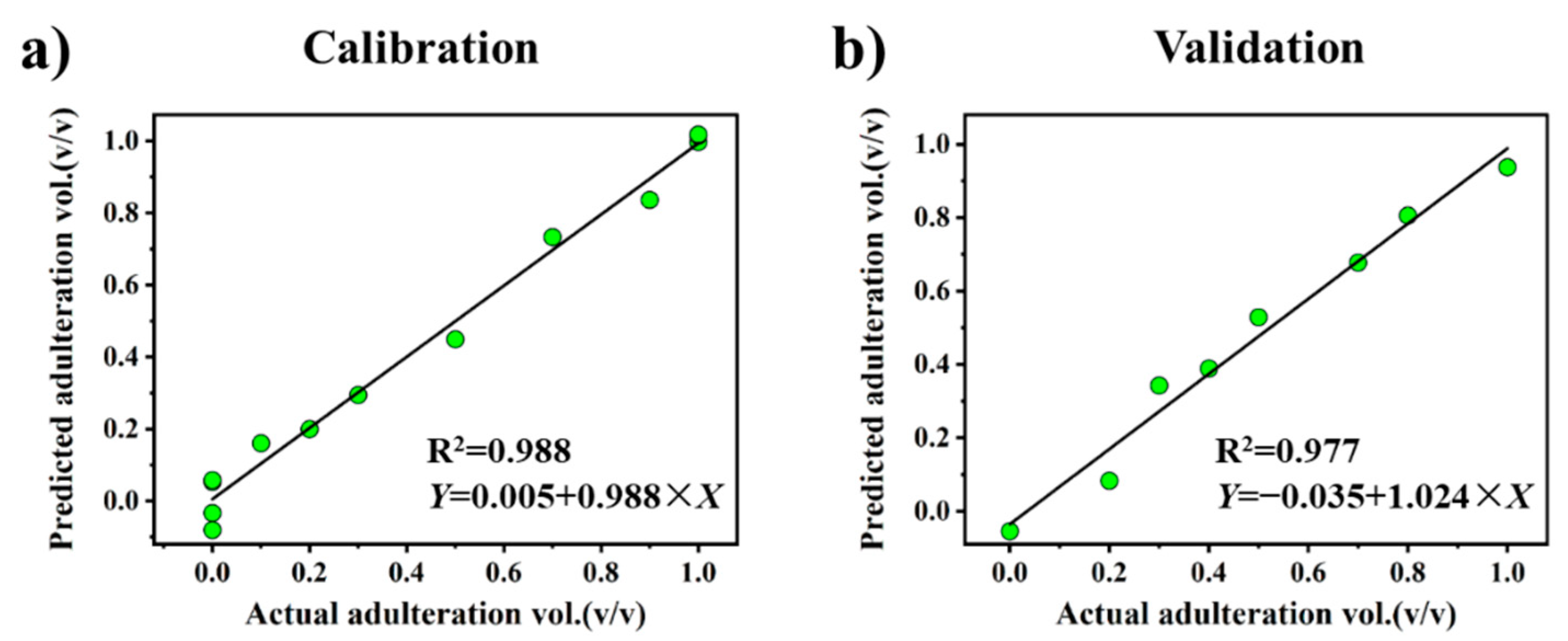
3.4. Quantitative Analysis of Oil Adulteration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niu, B.; Zhang, H.; Zhou, G.; Zhang, S.; Chen, Q. Safety risk assessment and early warning of chemical contamination in vegetable oil. Food Control 2021, 125, 107970. [Google Scholar] [CrossRef]
- Everstine, K.; Spink, J.; Kennedy, S. Economically motivated adulteration (EMA) of food: Common characteristics of EMA incidents. J. Food Prot. 2013, 76, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Hashempour-Baltork, F.; Torbati, M.; Azadmard-Damirchi, S.; Savage, G.P. Vegetable oil blending: A review of physicochemical, nutritional and health effects. Trends Food Ence Technol. 2016, 57, 52–58. [Google Scholar] [CrossRef]
- Jabeur, H.; Drira, M.; Rebai, A.; Bouaziz, M. Putative markers of adulteration of higher-grade olive oil with less expensive pomace olive oil by GC Combined with Chemometrics. J. Agric. Food Chem. 2017, 65, 5375–5383. [Google Scholar] [CrossRef] [PubMed]
- Celik, S.E.; Asfoor, A.; Senol, O.; Apak, R. Screening method for argan oil adulteration with vegetable oils: An online HPLC assay with postcolumn detection utilizing chemometric multidata analysis. J. Agric. Food Chem. 2019, 67, 8279–8289. [Google Scholar] [CrossRef]
- Zhang, L.; Li, P.; Sun, X.; Wang, X.; Xu, B.; Wang, X.; Ma, F.; Zhang, Q.; Ding, X. Classification and adulteration detection of vegetable oils based on fatty acid profiles. J. Agric. Food Chem. 2014, 62, 8745–8751. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, L.; Wang, D.; Jiang, J.; Harrington, P.D.; Mao, J.; Zhang, Q.; Li, P. Detection of flaxseed oil multiple adulteration by near-infrared spectroscopy and nonlinear one class partial least squares discriminant analysis. LWT 2020, 125, 109247. [Google Scholar] [CrossRef]
- Rohman, A.; Man, Y.B. The use of Fourier transform mid infrared (FT-MIR) spectroscopy for detection and quantification of adulteration in virgin coconut oil—ScienceDirect. Food Chem. 2011, 129, 583–588. [Google Scholar] [CrossRef]
- Zou, M.Q.; Zhang, X.F.; Qi, X.H.; Ma, H.L.; Dong, Y.; Liu, C.W.; Guo, X.; Wang, H. Rapid authentication of olive oil adulteration by Raman spectrometry. J. Agric. Food Chem. 2009, 57, 6001–6006. [Google Scholar] [CrossRef]
- Lia, F.; Castellano, A.M.; Zammit-Mangion, M.; Farrugia, C. Application of fluorescence spectroscopy and chemometric models for the detection of vegetable oil adulterants in Maltese virgin olive oils. J. Food Sci. Technol. 2018, 55, 2143–2151. [Google Scholar] [CrossRef]
- Roy, M.; Yadav, B.K. Electronic nose for detection of food adulteration: A review. J. Food Sci. Technol. Mysore 2021. [Google Scholar] [CrossRef]
- Zhang, L.; Shuai, Q.; Li, P.; Zhang, Q.; Ma, F.; Zhang, W.; Ding, X. Ion mobility spectrometry fingerprints: A rapid detection technology for adulteration of sesame oil. Food Chem. 2016, 192, 60–66. [Google Scholar] [CrossRef]
- Torrecilla, J.S.; Rojo, E.; Dominguez, J.C.; Rodríguez, F. A novel method to quantify the adulteration of extra virgin olive oil with low-grade olive oils by UV-vis. J. Agric. Food Chem. 2010, 58, 1679–1684. [Google Scholar] [CrossRef] [PubMed]
- Hatzakis, E. Nuclear magnetic resonance (NMR) spectroscopy in food science: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef] [PubMed]
- Truzzi, E.; Marchetti, L.; Benvenuti, S.; Ferroni, A.; Bertelli, D. Novel strategy for the recognition of adulterant vegetable oils in essential oils commonly used in food industries by applying 13C NMR spectroscopy. J. Agric. Food Chem. 2021, 69, 8276–8286. [Google Scholar] [CrossRef]
- Karkoula, E.; Skantzari, A.; Melliou, E. Direct measurement of oleocanthal and oleacein levels in olive oil by quantitative ~1H NMR. Establishment of a new index for the characterization of extra virgin olive oils. J. Agric. Food Chem. 2012, 60, 11696–11703. [Google Scholar] [CrossRef] [PubMed]
- Miaw, C.; Santos, P.M.; Silva, A.; Gozzi, A.; Souza, S. Comparison of Different multivariate classification methods for the detection of adulterations in grape nectars by using low-field nuclear magnetic resonance. Food Anal. Methods 2020, 13, 108–118. [Google Scholar] [CrossRef]
- Akkaya, S.; Ozel, B.; Oztop, M.H.; Yanik, D.K.; Gogus, F. Physical characterization of high methoxyl pectin and sunflower oil wax emulsions: A low-field 1H NMR relaxometry study. J. Food Sci. 2020, 86, 120–128. [Google Scholar] [CrossRef]
- Rudszuck, T.; Förster, E.; Nirschl, H.; Guthausen, G. Low-field NMR for quality control on oils. Magn. Reson. Chem. 2019, 57, 777–793. [Google Scholar] [CrossRef]
- Hansen, C.L.; Thybo, A.K.; Bertram, H.C.; Viereck, N.; Engelsen, S.B. Determination of dry matter content in potato tubers by low-field nuclear magnetic resonance (LF-NMR). J. Agric. Food Chem. 2010, 58, 10300–10304. [Google Scholar] [CrossRef]
- Ancora, D.; Milavec, J.; Gradiek, A.; Cifelli, M.; Domenici, V. Sensitivity of proton NMR relaxation and proton NMR diffusion measurements to olive oil adulterations with vegetable oils. J. Agric. Food Chem. 2021. [Google Scholar] [CrossRef]
- Wang, R.; Liu, K.; Wang, X.; Tan, M. Detection of Sesame Oil Adulteration Using Low-Field Nuclear Magnetic Resonance and Chemometrics. Int. J. Food Eng. 2019, 15, 1–8. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, X.; Chen, L. Rapid detection of peanut oil adulteration using low-field nuclear magnetic resonance and chemometrics. Food Chem. 2017, 216, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, G.; Hou, X.; Nie, S. A rapid screening approach for authentication of olive oil and classification of binary blends of olive oils using low-field nuclear magnetic resonance spectra and support vector machine. Food Anal. Methods 2020, 13, 1894–1905. [Google Scholar] [CrossRef]
- Hou, X.; Wang, G.; Wang, X.; Ge, X.; Nie, S. Convolutional neural network based approach for classification of edible oils using low-field nuclear magnetic resonance. J. Food Compos. Anal. 2020, 92, 103566. [Google Scholar] [CrossRef]
- Hou, X.; Wang, G.; Wang, X.; Ge, X.; Fan, Y.; Jiang, R.; Nie, S. Rapid screening for hazelnut oil and high-oleic sunflower oil in extra virgin olive oil using low-field nuclear magnetic resonance relaxometry and machine learning. J. Sci. Food Agric. 2020, 101, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Pan, K.; Yu, Z.; Xiao, R.H. Pattern recognition based on machine learning identifies oil adulteration and edible oil mixtures. Nat. Commun. 2020, 11, 5353. [Google Scholar] [CrossRef] [PubMed]
- Dan, M.; Gulani, V.; Seiberlich, N.; Liu, K.; Sunshine, J.L.; Duerk, J.L.; Griswold, M.A. Magnetic resonance fingerprinting. Nature 2013, 495, 187–192. [Google Scholar]
- Hong, T.; Han, D.; Kim, M.O.; Kim, D.H. RF slice profile effects in magnetic resonance fingerprinting. Magn. Reson. Imaging 2017, 41, 73–79. [Google Scholar] [CrossRef]
- Hamilton, J.I.; Yun, J.; Yong, C.; Dan, M.; Lo, W.; Griswold, M.; Seiberlich, N. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn. Reson. Med. 2017, 77, 1446–1458. [Google Scholar] [CrossRef] [PubMed]
- Carr, H.Y.; Purcell, E.M. Effect of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments. Phys. Rev. 1954, 94, 630–638. [Google Scholar] [CrossRef]
- Meiboom, S. Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 1958, 29, 688–691. [Google Scholar] [CrossRef]
- Kowalewski, J.; Levy, G.C.; Johnson, L.; Palmer, L. A three-parameter non-linear procedure for fitting inversion-recovery measurements of spin-lattice relaxation times. J. Magn. Reson. 1977, 26, 533–536. [Google Scholar] [CrossRef]
- Vermeir, L.; Declerck, A.; To, C.M.; Kerkaert, B.; Meeren, P. Water and oil signal assignment in low-moisture mozzarella as determined by time-domain NMR T 2 relaxometry. Magn. Reson. Chem. 2019, 57, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.L.; Rovnyak, D.; Strein, T.G. Identification of edible oils by principal component analysis of 1H NMR spectra. J. Chem. Educ. 2017, 94, 1377–1382. [Google Scholar] [CrossRef]
- Hsg, A.; Xl, B.; Mdp, C.; Ksl, C.; Fs, C.; Ina, C.; Scwa, B. A rapid method for the detection of extra virgin olive oil adulteration using UHPLC-CAD profiling of triacylglycerols and PCA. Food Control 2020, 107, 106773. [Google Scholar]
- Jović, O.; Smolić, T.; Primožič, I.; Hrenar, T. Spectroscopic and chemometric analysis of binary and ternary edible oil mixtures: Qualitative and quantitative study. Anal. Chem. 2016, 88, 4516–4524. [Google Scholar] [CrossRef]
- Martins, J.P.A.; Teófilo, R.F.; Ferreira, M.M.C. Computational performance and cross-validation error precision of five PLS algorithms using designed and real data sets. J. Chemom. 2010, 24, 320–332. [Google Scholar] [CrossRef]
- Orsavova, J.; Misurcova, L.; Ambrozova, J.V.; Vicha, R.; Mlcek, J. Fatty Acids Composition of Vegetable Oils and Its Contribution to Dietary Energy Intake and Dependence of Cardiovascular Mortality on Dietary Intake of Fatty Acids. Int. J. Mol. Sci. 2015, 16, 12871–12890. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Yoshida, M.; Maeda, I.; Miyashita, K. Proton NMR Relaxation Times of Polyunsaturated Fatty Acids in Chloroform Solutions and Aqueous Micelles. J. Oleo Sci. 2004, 53, 105–108. [Google Scholar] [CrossRef][Green Version]
- Kamal-Eldin, A. Minor components of fats and oils. In Bailey’s Industrial Oil and Fat Products; Wiley: Sussex, UK, 2005. [Google Scholar]
- Zhu, M.T.; Shi, T.; Chen, Y.; Luo, S.H.; Leng, T.; Wang, Y.L.; Guo, C.; Xie, M.Y. Prediction of fatty acid composition in camellia oil by 1H NMR combined with PLS regression—ScienceDirect. Food Chem. 2019, 279, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Saulo, H.; Escobar, H.B.; Leao, J. A new model selection criterion for partial least squares regression—ScienceDirect. Chemom. Intell. Lab. Syst. 2017, 169, 64–78. [Google Scholar] [CrossRef]
| Univariate | PLSR | ||||
|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 3 | |||
| Input variables | B | E | G | A–H | |
| Accuracy | RMSEP | 6.39% | 5.88% | 5.92% | 5.46% |
| RMSEC | 4.66% | 4.66% | 6.55% | 4.45% | |
| Calibration | Slope | −1.027 | −0.515 | 1.021 | 0.988 |
| Intercept | 0.978 | 0.949 | 0.829 | 0.005 | |
| R2 | 0.987 | 0.987 | 0.974 | 0.988 | |
| Validation | Slope | 1.004 | 0.990 | 1.027 | 1.024 |
| Intercept | −0.031 | −0.030 | −0.056 | −0.035 | |
| R2 | 0.967 | 0.976 | 0.984 | 0.977 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.-M.; Xin, J.-X.; Sun, S.-S.; Li, Y.; Wei, D.-X.; Zhu, J.; Wang, X.-L.; Wang, J.; Yao, Y.-F. Rapid Identification of Adulteration in Edible Vegetable Oils Based on Low-Field Nuclear Magnetic Resonance Relaxation Fingerprints. Foods 2021, 10, 3068. https://doi.org/10.3390/foods10123068
Huang Z-M, Xin J-X, Sun S-S, Li Y, Wei D-X, Zhu J, Wang X-L, Wang J, Yao Y-F. Rapid Identification of Adulteration in Edible Vegetable Oils Based on Low-Field Nuclear Magnetic Resonance Relaxation Fingerprints. Foods. 2021; 10(12):3068. https://doi.org/10.3390/foods10123068
Chicago/Turabian StyleHuang, Zhi-Ming, Jia-Xiang Xin, Shan-Shan Sun, Yi Li, Da-Xiu Wei, Jing Zhu, Xue-Lu Wang, Jiachen Wang, and Ye-Feng Yao. 2021. "Rapid Identification of Adulteration in Edible Vegetable Oils Based on Low-Field Nuclear Magnetic Resonance Relaxation Fingerprints" Foods 10, no. 12: 3068. https://doi.org/10.3390/foods10123068
APA StyleHuang, Z.-M., Xin, J.-X., Sun, S.-S., Li, Y., Wei, D.-X., Zhu, J., Wang, X.-L., Wang, J., & Yao, Y.-F. (2021). Rapid Identification of Adulteration in Edible Vegetable Oils Based on Low-Field Nuclear Magnetic Resonance Relaxation Fingerprints. Foods, 10(12), 3068. https://doi.org/10.3390/foods10123068







