LC-QTOF-MS/MS Based Molecular Networking Approach for the Isolation of α-Glucosidase Inhibitors and Virucidal Agents from Coccinia grandis (L.) Voigt
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Materials and Extraction Method
2.3. UHPLC-ESI-QTOF-MS/MS Analysis
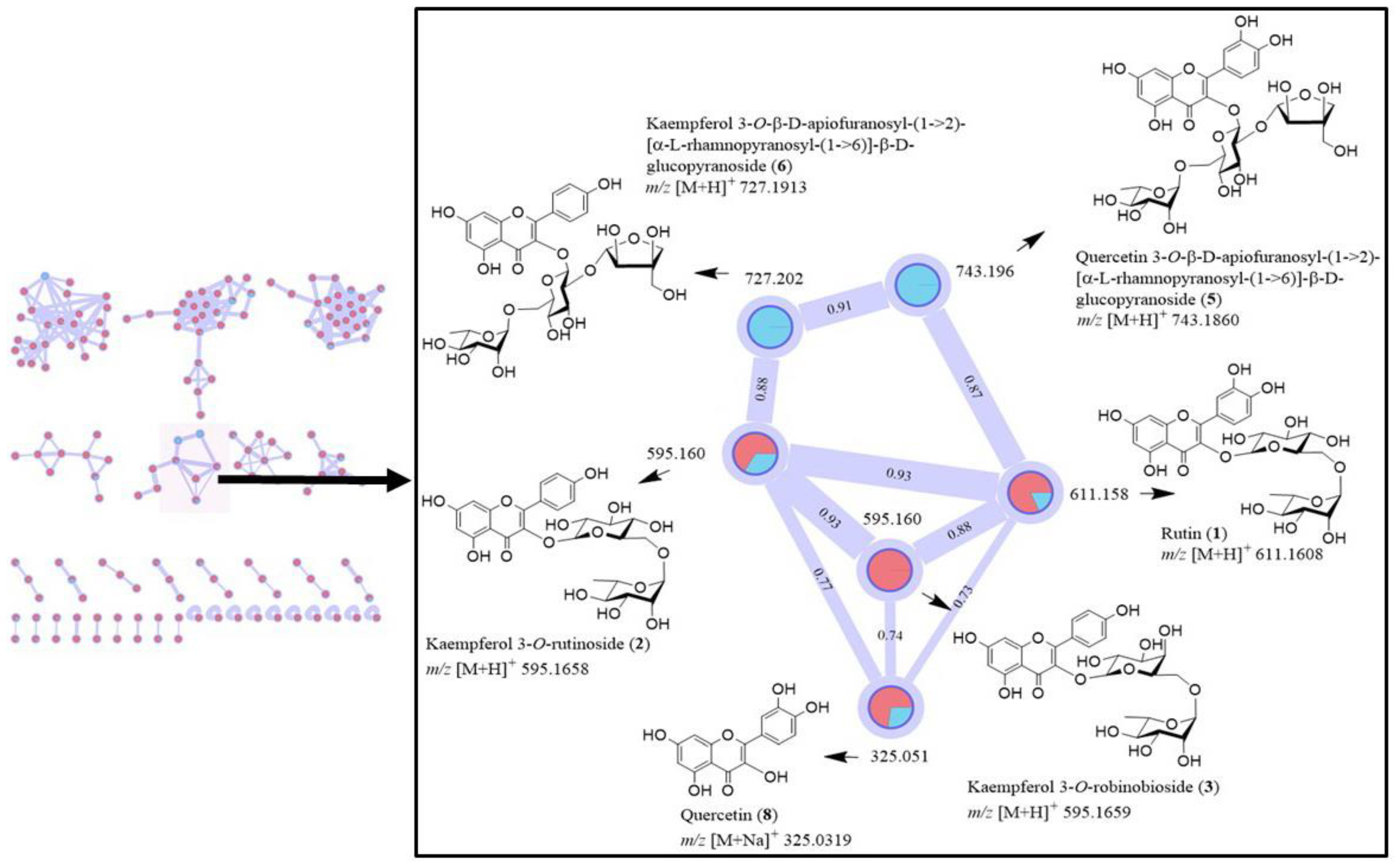
2.4. MS-Based Molecular Networking
2.5. NMR Experiments
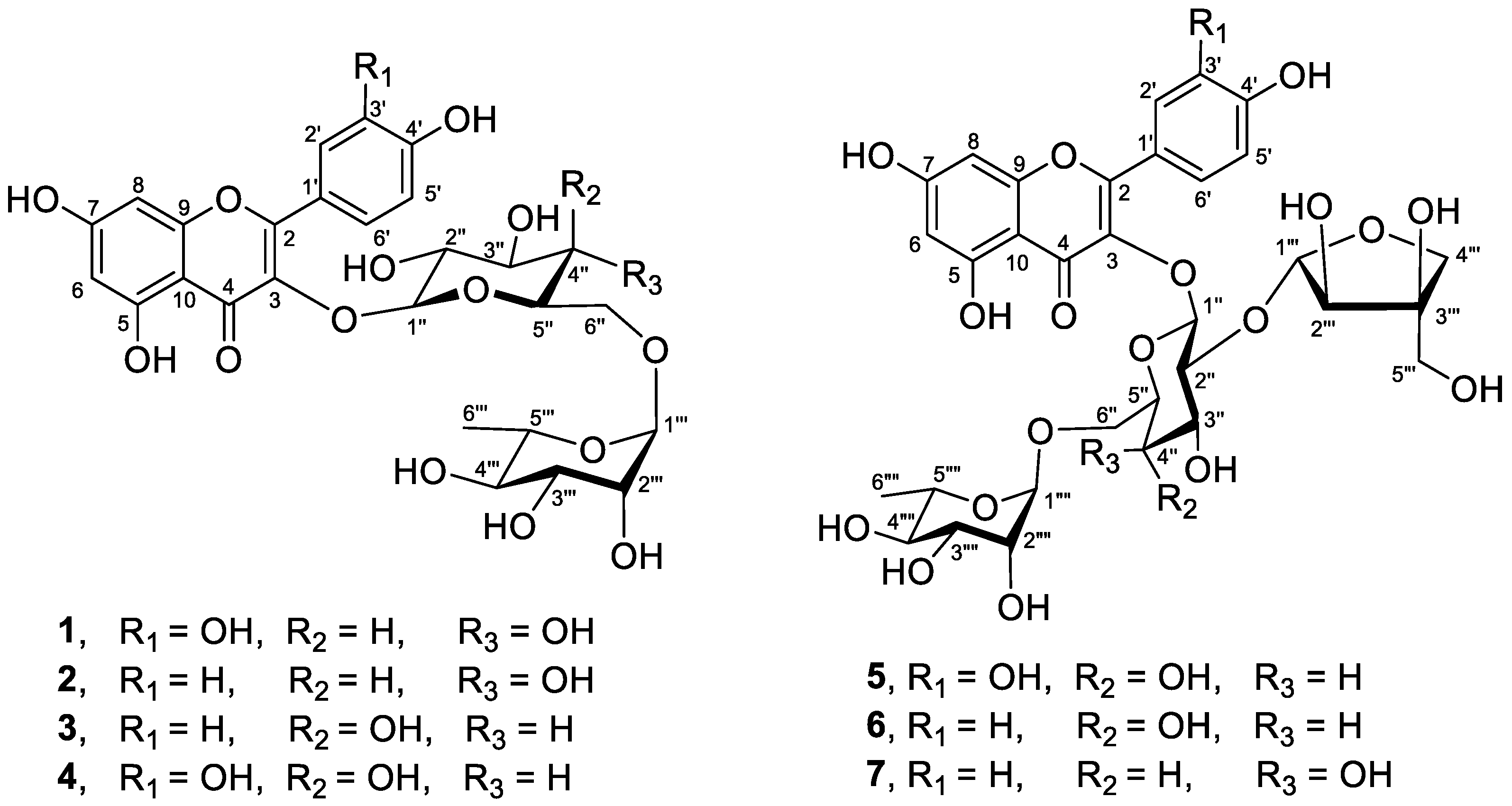
2.6. Isolation of Flavonoid Glycosides 1–7
2.7. In-Vitro α-Glucosidase Inhibitory Assay
2.8. Evaluation of Virucidal Activity
3. Results and Discussion
3.1. Profile of Compounds in a Crude Extract of C. Grandis Revealed by 1H NMR Spectrum and LC-MS/MS Analysis, and GNPS Molecular Networking of Flavonoid Glycosides
3.2. Isolation and Characterization of Flavonoid Glycosides
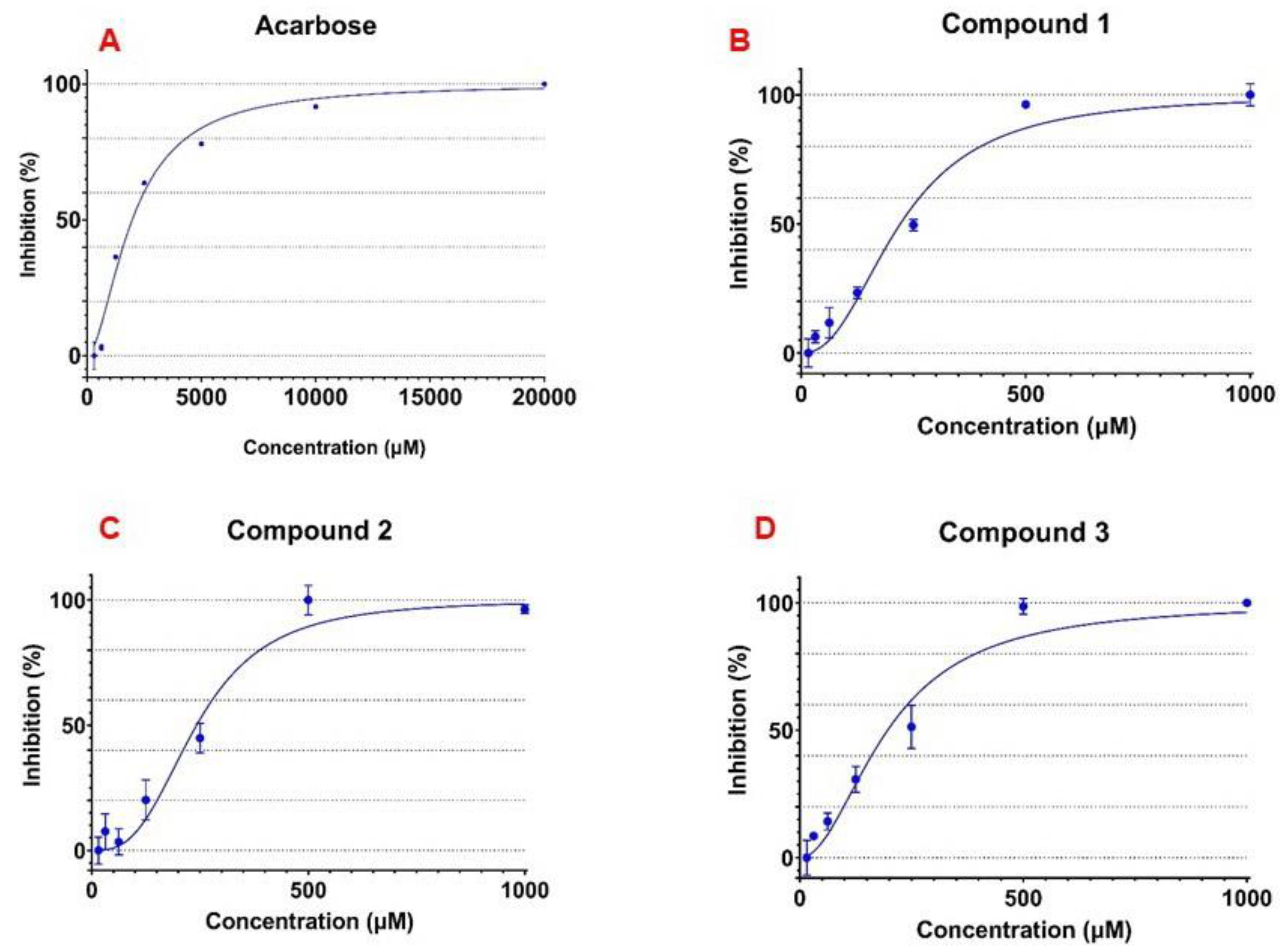
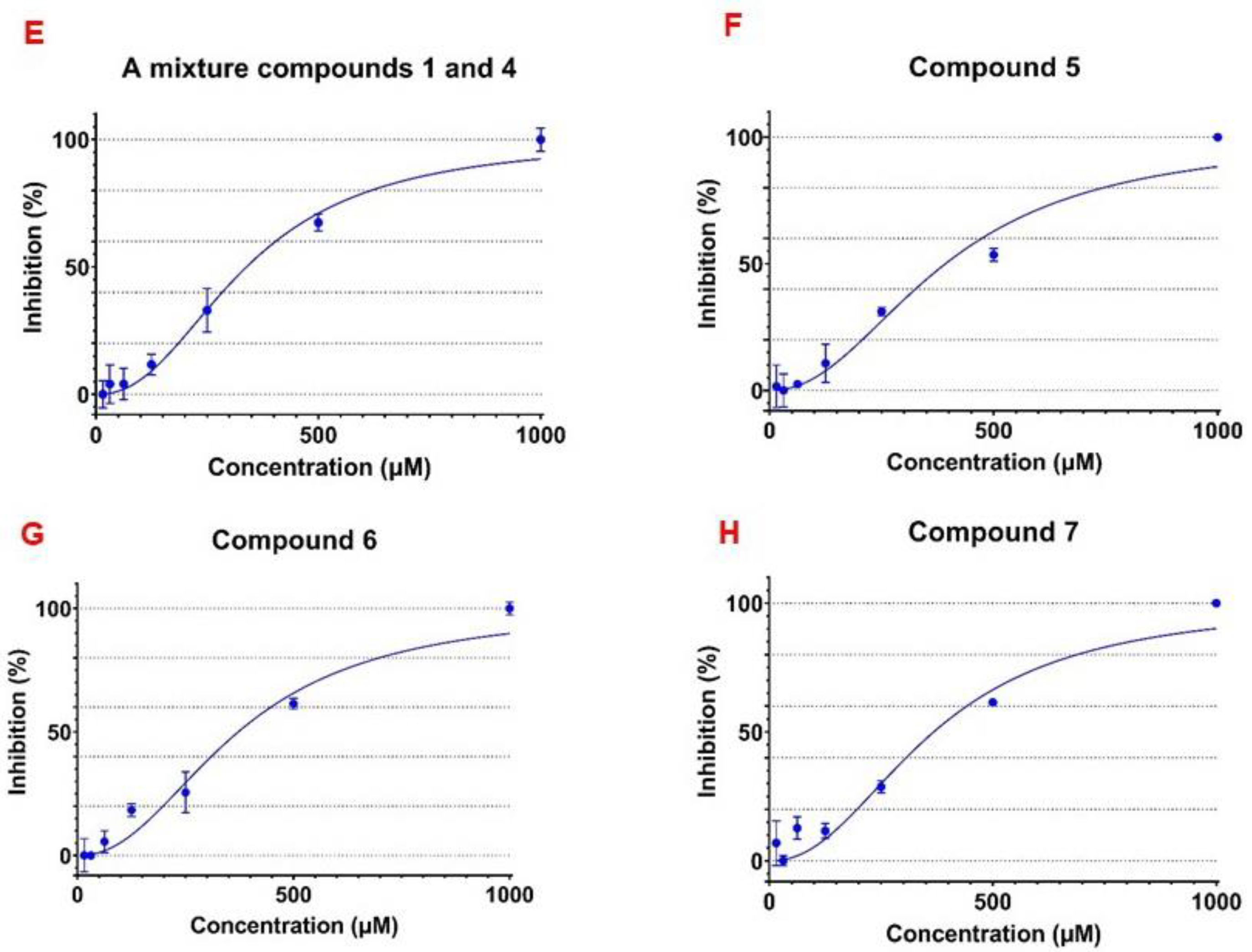
3.3. α-Glucosidase Inhibitory Activity of the Isolated Flavonoid Glycosides
3.4. Virucidal Activity of the Isolated Flavonoid Glycosides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wasana, K.G.P.; Attanayake, A.P.; Weerarathna, T.P.; Jayatilaka, K. Efficacy and safety of a herbal drug of Coccinia grandis (Linn.) Voigt in patients with type 2 diabetes mellitus: A double blind randomized placebo controlled clinical trial. Phytomedicine 2021, 81, 153431. [Google Scholar] [CrossRef]
- Al-Madhagy, S.A.; Mostafa, N.M.; Youssef, F.S.; Awad, G.E.A.; Eldahshan, O.A.; Singab, A.N.B. Metabolic profiling of a polyphenolic-rich fraction of Coccinia grandis leaves using LC-ESI-MS/MS and in vivo validation of its antimicrobial and wound healing activities. Food Funct. 2019, 10, 6267–6275. [Google Scholar] [CrossRef]
- Lim, T.K. Coccinia grandis. In Coccinia grandis Edible Medicinal and Non-Medicinal Plants; Springer: Dordrecht, The Netherlands, 2012; Volume 2, pp. 191–200. [Google Scholar]
- Albrahim, T.; Alnasser, M.M.; Al-Anazi, M.R.; Alkahtani, M.D.; Alkahtani, S.; Al-Qahtani, A.A. Potential anti-inflammatory and anti-apoptotic effect of Coccinia grandis plant extract in LPS stimulated-THP-1 cells. Environ. Sci. Pollut. Res. 2020, 27, 21892–21904. [Google Scholar] [CrossRef]
- Banerjee, A.; Das, D.; Paul, R.; Roy, S.; Das, U.; Saha, S.; Dey, S.; Adhikary, A.; Mukherjee, S.; Maji, B.K. Mechanistic study of attenuation of monosodium glutamate mixed high lipid diet induced systemic damage in rats by Coccinia grandis. Sci. Rep. 2020, 10, 15443. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, A.; Paik, D.; Pramanik, P.K.; Chakraborti, T. Serine protease inhibitors rich Coccinia grandis (L.) Voigt leaf extract induces protective immune responses in murine visceral leishmaniasis. Biomed. Pharmacother. 2019, 111, 224–235. [Google Scholar] [CrossRef]
- Banerjee, A.; Mukherjee, S.; Maji, B.K. Efficacy of Coccinia grandis against monosodium glutamate induced hepato-cardiac anomalies by inhibiting NF-kB and caspase 3 mediated signalling in rat model. Hum. Exp. Toxicol. 2021, 40, 1825–1851. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, G.; Muthukumar, T.; Tirichurapalli Sivagnanam, U. In vivo efficiency of the collagen coated nanofibrous scaffold and their effect on growth factors and pro-inflammatory cytokines in wound healing. Eur. J. Pharmacol. 2017, 814, 45–55. [Google Scholar] [CrossRef]
- Namchaiw, P.; Jaisin, Y.; Niwaspragrit, C.; Malaniyom, K.; Auvuchanon, A.; Ratanachamnong, P. The leaf extract of Coccinia grandis (L.) Voigt accelerated in vitro wound healing by reducing oxidative stress injury. Oxid. Med. Cell. Longev. 2021, 2021, 3963510. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Choo, C.; Watawana, M.I.; Jayawardena, N.; Waisundara, V.Y. An appraisal of eighteen commonly consumed edible plants as functional food based on their antioxidant and starch hydrolase inhibitory activities. J. Sci. Food Agric. 2015, 95, 2956–2964. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Gupta, P.; Rawat, P.; Puri, A.; Bhatia, G.; Maurya, R. Antidyslipidemic activity of polyprenol from Coccinia grandis in high-fat diet-fed hamster model. Phytomedicine 2007, 14, 792–798. [Google Scholar] [CrossRef]
- Tupe, R.S.; Sankhe, N.M.; Shaikh, S.A.; Kemse, N.G.; Khaire, A.A.; Phatak, D.V.; Parikh, J.U. Nutraceutical properties of dietary plants extracts: Prevention of diabetic nephropathy through inhibition of glycation and toxicity to erythrocytes and HEK293 cells. Pharm. Biol. 2015, 53, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Bunkrongcheap, R.; Hutadilok-Towatana, N.; Noipha, K.; Wattanapiromsakul, C.; Inafuku, M.; Oku, H. Ivy gourd (Coccinia grandis L. Voigt) root suppresses adipocyte differentiation in 3T3-L1 cells. Lipids Health Dis. 2014, 13, 88. [Google Scholar] [CrossRef] [Green Version]
- Kondhare, D.; Lade, H. Phytochemical profile, aldose reductase inhibitory, and antioxidant activities of Indian traditional medicinal Coccinia grandis (L.) fruit extract. 3 Biotech 2017, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, V.; Haselgrübler, R.; Lanzerstorfer, P.; Plochberger, B.; Borgmann, D.; Jacak, J.; Winkler, S.M.; Schröder, K.; Höglinger, O.; Weghuber, J. Biomolecular characterization of putative antidiabetic herbal extracts. PLoS ONE 2016, 11, e0148109. [Google Scholar] [CrossRef]
- Meenatchi, P.; Purushothaman, A.; Maneemegalai, S. Antioxidant, antiglycation and insulinotrophic properties of Coccinia grandis (L.) in vitro: Possible role in prevention of diabetic complications. J. Tradit. Complement. Med. 2017, 7, 54–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chanda, J.; Mukherjee, P.K.; Kar, A.; Maitra, P.K.; Singha, S.; Halder, P.K.; Gajbhiye, R.; Vishnuvardh, R. LC–QTOF–MS-based metabolite profiling and evaluation of α-glucosidase inhibitory kinetics of Coccinia grandis fruit. Biomed. Chromatogr. 2020, 34, e4950. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- GNPS: Global Natural Products Molecular Networking. Available online: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp (accessed on 15 August 2021).
- Liu, S.; Yu, Z.; Zhu, H.; Zhang, W.; Chen, Y. In vitro α-glucosidase inhibitory activity of isolated fractions from water extract of Qingzhuan dark tea. BMC Complement. Altern. Med. 2016, 16, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Society for Testing and Materials (ASTM). Standard Practice to Assess Virucidal Activity of Chemicals Intended for Disinfection of Inanimate, Nonporous Environmental Surfaces; ASTM E1053–20; ASTM: West Conshohocken, PA, USA, 2020; Available online: https://www.astm.org/Standards/E1053.htm (accessed on 15 August 2021).
- Dellanno, C.; Vega, Q.; Boesenberg, D. The antiviral action of common household disinfectants and antiseptics against murine hepatitis virus, a potential surrogate for SARS coronavirus. Am. J. Infect. Control 2009, 37, 649–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, A.; Kaleem, M.; Ahmed, Z.; Shafiq, H. Therapeutic potential of flavonoids and their mechanism of action against microbial and viral infection-A review. Food Res. Int. 2015, 77, 221–235. [Google Scholar] [CrossRef]
- Şöhretoğlu, D.; Sari, S. Flavonoids as alpha-glucosidase inhibitors: Mechanistic approaches merged with enzyme kinetics and molecular modelling. Phytochem. Rev. 2020, 19, 1081–1092. [Google Scholar] [CrossRef]
- Barber, E.; Houghton, M.J.; Williamson, G. Flavonoids as human intestinal α-glucosidase inhibitors. Foods 2021, 10, 1939. [Google Scholar] [CrossRef]
- Xu, Q.-D.; Yu, Z.-L.; Zeng, W.-C. Structural and functional modifications of myofibrillar protein by natural phenolic compounds and their application in pork meatball. Food Res. Int. 2021, 148, 110593. [Google Scholar] [CrossRef]
- Cedeño, H.; Espinosa, S.; Andrade, J.M.; Cartuche, L.; Malagón, O. Novel flavonoid glycosides of quercetin from leaves and flowers of Gaiadendron punctatum G.Don. (Violeta de Campo), used by the Saraguro community in Southern Ecuador, inhibit α-glucosidase enzyme. Molecules 2019, 24, 4267. [Google Scholar] [CrossRef] [Green Version]
- Ganbaatar, C.; Gruner, M.; Mishig, D.; Duger, R.; Schmidt, A.W.; Knölker, H.-J. Flavonoid glycosides from the aerial parts of Polygonatum odoratum (Mill.) Druce growing in Mongolia. Open Nat. Prod. J. 2015, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Hou, W.-C.; Lin, R.-D.; Lee, T.-H.; Huang, Y.-H.; Hsu, F.-L.; Lee, M.-H. The phenolic constituents and free radical scavenging activities of Gynura formosana Kiamnra. J. Sci. Food Agric. 2005, 85, 615–621. [Google Scholar] [CrossRef]
- Hassan, A.R.; Amer, K.F.; El-Toumy, S.A.; Nielsen, J.; Christensen, S.B. A new flavonol glycoside and other flavonoids from the aerial parts of Taverniera aegyptiaca. Nat. Prod. Res. 2019, 33, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Piccinelli, A.L.; Veneziano, A.; Passi, S.J.; Simone, F.D.; Rastrelli, L. Flavonol glycosides from whole cottonseed by-product. Food Chem. 2007, 100, 344–349. [Google Scholar] [CrossRef]
- Zhu, N.; Sheng, S.; Li, D.; Lavoie, E.J.; Karwe, M.V.; Rosen, R.T.; Ho, C.T. Antioxidative flavonoid glycosides from quinoa seeds (Chenopodium quinoa Willd). J. Food Lipids 2001, 8, 37–44. [Google Scholar] [CrossRef]
- Li, Y.F.; Yang, M.; Yuan, L.; Zhao, Y.M.; Luan, X.H.; Luo, Z.P. Antidepressant effect of quercetin 3-O-apiosyl (l→2)-[rhamnosyl(1 →6)]-glucoside in mice. Chin. J. Pharmacol. Toxicol. 2000, 14, 125–127. [Google Scholar]
- Zhang, L.M.; Zhang, Y.Z.; Liu, Y.Q.; Gong, Z.H.; Zhao, Y.M.; Li, Y.F. CTN-986, a compound extracted from cottonseeds, increases cell proliferation in hippocampus in vivo and in cultured neural progenitor cells in vitro. Eur. J. Pharmacol. 2009, 607, 110–113. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, Y.; Zhao, L.; Zhang, W.; Zhang, A.; Xu, B. Simultaneous quantification of CTN986 and its deglycosylation products in rat serum using liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 1701–1708. [Google Scholar] [CrossRef]
- Guo, J.; Meng, F.; Ren, Z.; Zhao, Y. Pharmacokinetics and tissue distribution of a water-soluble flavonol triglycoside, CTN986, in mice. Planta Med. 2008, 74, 228–232. [Google Scholar] [CrossRef]
- Pičmanová, M.; Møller, B.L. Apiose: One of nature’s witty games. Glycobiology 2016, 26, 430–442. [Google Scholar] [CrossRef] [Green Version]
- Eixelsberger, T.; Horvat, D.; Gutmann, A.; Weber, H.; Nidetzky, B. Isotope probing of the UDP-apiose/UDP-xylose synthase reaction: Evidence of a mechanism via a coupled oxidation and aldol cleavage. Angew. Chem. Int. Ed. 2017, 56, 2503–2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Kang, S.; Rhee, Y.H. De novo synthesis of furanose sugars: Catalytic asymmetric synthesis of apiose and apiose-containing oligosaccharides. Angew. Chem. Int. Ed. 2016, 55, 9733–9737. [Google Scholar] [CrossRef]
- Hong, H.-C.; Li, S.-L.; Zhang, X.-Q.; Ye, W.-C.; Zhang, Q.-W. Flavonoids with α-glucosidase inhibitory activities and their contents in the leaves of Morus atropurpurea. Chin. Med. 2013, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Bose, M.; Kamra, M.; Mullick, R.; Bhattacharya, S.; Das, S.; Karande, A.A. Identification of a flavonoid isolated from plum (Prunus domestica) as a potent inhibitor of Hepatitis C virus entry. Sci. Rep. 2017, 7, 3965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Sarmah, S.; Lyndem, S.; Singha Roy, A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2021, 39, 3347–3357. [Google Scholar] [CrossRef]
- Dwivedi, V.D.; Bharadwaj, S.; Afroz, S.; Khan, N.; Ansari, M.A.; Yadava, U.; Tripathi, R.C.; Tripathi, I.P.; Mishra, S.K.; Kang, S.G. Anti-dengue infectivity evaluation of bioflavonoid from Azadirachta indica by dengue virus serine protease inhibition. J. Biomol. Struct. Dyn. 2021, 39, 1417–1430. [Google Scholar] [CrossRef]
- Yarmolinsky, L.; Huleihel, M.; Zaccai, M.; Ben-Shabat, S. Potent antiviral flavone glycosides from Ficus benjamina leaves. Fitoterapia 2012, 83, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Sabrin, M.S.; Selenge, E.; Takeda, Y.; Batkhuu, J.; Ogawa, H.; Jamsransuren, D.; Suganuma, K.; Murata, T. Isolation and evaluation of virucidal activities of flavanone glycosides and rosmarinic acid derivatives from Dracocephalum spp. against feline calicivirus. Phytochemistry 2021, 191, 112896. [Google Scholar] [CrossRef] [PubMed]
- Di Cola, G.; Fantilli, A.C.; Pisano, M.B.; Ré, V.E. Foodborne transmission of hepatitis A and hepatitis E viruses: A literature review. Int. J. Food Microbiol. 2021, 338, 108986. [Google Scholar] [CrossRef]
- Sarowska, J.; Wojnicz, D.; Jama-Kmiecik, A.; Frej-Mądrzak, M.; Choroszy-Król, I. Antiviral potential of plants against noroviruses. Molecules 2021, 26, 4669. [Google Scholar] [CrossRef]
- Li, D.; Baert, L.; Uyttendaele, M. Inactivation of food-borne viruses using natural biochemical substances. Food Microbiol. 2013, 35, 1–9. [Google Scholar] [CrossRef] [PubMed]
| No. | RT (Min) | Compounds (Identification by Database or Isolation) | Molecular Formula | Mass | Adduct Ions | Observed m/z | Calculated m/z | Δ (ppm) | Fragment Ions (m/z) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.638 | Maltopentaose (M) | C30H52O26 | 828.2741 | (M − H)− | 827.2674 | 827.2674 | 0.71 | 683.2247, 533.1721, 471.0754, 387.1144, 341.1092, 299.0983, 191.0562, 162.8391, 133.0143 |
| 2 | 0.640 | L-Mannitol (H, M) | C6H14O6 | 182.079 | (M − H)− | 181.0717 | 181.0717 | 0.27 | 179.0563, 133.0142 |
| 3 | 0.648 | Gulonic acid (M) | C6H12O7 | 196.0582 | (M − H)− | 195.0508 | 195.0508 | 0.34 | 191.0562, 179.0563, 165.0404, 133.0142 |
| 4 | 0.683 | Shikimic acid (H, M) | C7H10O5 | 174.0532 | (M − H)− | 173.0458 | 173.0458 | −0.36 | 191.0562, 133.0142 |
| 5 | 0.687 | D-malic acid (H, M) | C4H6O5 | 134.0215 | (M − H)− | 133.0142 | 133.0142 | 0.13 | 112.9856 |
| 6 | 0.992 | Succinic acid (H) | C4H6O4 | 118.0264 | (M − H)− | 117.0191 | 117.0191 | 2.06 | 112.9856 |
| 7 | 1.961 | Pantothenic acid (H, M) | C9H17NO5 | 219.1108 | (M + H)+ | 220.1183 | 220.1181 | −0.80 | 205.0863, 194.1177, 186.9566, 158.0030, 121.0509 |
| 8 | 2.402 | Damascenone (M) | C13H18O | 190.1357 | (M + H)+ | 191.1430 | 191.1430 | 0.20 | 158.0030, 141.9587, 121.0509 |
| 9 | 2.792 | Chlorogenic acid (H, M) | C16H18O9 | 354.0951 | (M + H)+ | 355.1022 | 355.1023 | 0.06 | 311.1854, 279.1593, 231.2068, 194.1177, 158.0031, 121.0509 |
| 10 | 3.430 | 3-Hydoxybenzoic acid (H, M) | C7H6O3 | 138.0318 | (M − H)− | 137.0245 | 137.0246 | −0.79 | 112.9856 |
| 11 | 4.503 | Chlorogenoquinone (M) | C16H16O9 | 352.0793 | (M − H)− | 351.0719 | 351.0720 | 0.35 | 191.0560 |
| 12 | 4.727 | Esculetin (H, M) | C9H6O4 | 178.0267 | (M − H)− | 177.0194 | 177.0194 | −0.69 | 147.0298 |
| 13 | 5.490 | Feruloylagmatine (M) | C15H22N4O3 | 306.1693 | (M + H)+ | 307.1766 | 307.1765 | −0.46 | 262.1801, 231.2067, 194.1178, 158.0030, 121.0509 |
| 14 | 5.756 | Kaempferol 3-(2G-xylosylrutinoside)-7-glucoside (M) | C38H48O24 | 888.2530 | (M − H)− | 887.2452 | 887.2455 | 0.59 | 435.2235 |
| 15 | 7.022 | Isopentyl gentiobioside (M) | C17H32O11 | 412.1938 | (M + HCOO)¯ | 457.1920 | 457.1919 | 1.6 | 427.1821 |
| 16 | 7.086 | Kaempferol 3-O-rutinoside (2) or nicotiflorin (M, S, isolated) | C27H30O15 | 594.1580 | (M + H)+ | 595.1641 | 595.1650 | 0.82 | 536.1648, 470.3695, 372.2498, 307.1764, 279.1595, 231.2067, 194.1177, 158.0030, 121.0509 |
| 17 | 8.867 | Quercetin 3-O-(β-D-apiofuranosyl(1→2)-α-rhamnopyranosyl(1→6)-β-D-glucopyranoside) (5) (M, isolated) | C32H38O20 | 742.1952 | (M − H)− | 741.1880 | 741.1880 | 0.65 | No fragment ions observed |
| 18 | 9.387 | Rutin (1) (H, M, S, isolated) | C27H30O16 | 610.1524 | (M + H)+ | 611.1578 | 611.1578 | 1.6 | 536.1649, 373.2333, 311.1854, 279.1596, 231.2066, 194.1177, 158.0030, 121.0509 |
| 19 | 9.388 | Herbacetin (M) | C15H10O7 | 302.0426 | (M + H)+ | 303.0501 | 303.0499 | 0.32 | 279.1596, 231.2066, 194.1177, 158.0030, 121.0509 |
| 20 | 9.660 | Kaempferol 3-O-(β-D-apiofuranosyl(1→2)-α-rhamnopyranosyl(1→6)-β-D-galactopyranoside (7) (M, isolated) | C32H38O19 | 726.1998 | (M − H)− | 725.1926 | 725.1926 | 1.22 | 609.1462 |
| 21 | 9.696 | Quercetin 3-O-glucoside (M) | C21H20O12 | 464.0957 | (M + H)+ | 465.1028 | 465.1028 | −0.57 | 453.3431, 393.2476, 357.2384, 279.1595, 231.2067, 194.1177, 158.0030, 121.0509 |
| 22 | 10.444 | Luteolin (M) | C15H10O6 | 286.0478 | (M + H)+ | 287.0550 | 287.0552 | −0.13 | 279.1595, 262.1801, 231.2067, 194.1178, 158.0030, 121.0509 |
| 23 | 10.800 | Citrusin B (M) | C27H36O13 | 568.2157 | (M − H)− | 567.2087 | 567.2090 | −0.26 | 146.9657 |
| 24 | 11.171 | Astragalin (M) | C21H20O11 | 448.1005 | (M + H)+ | 449.1080 | 449.1079 | 0.04 | 393.2466, 317.0603, 279.1594, 231.2066, 194.1177, 158.0031, 121.0509 |
| 25 | 16.837 | Jasmonic acid (H, M) | C12H18O3 | 210.1260 | (M + H)+ | 211.1324 | 211.1324 | −2.10 | 194.1177, 180.1361, 158.1540, 121.0509 |
| 26 | 17.190 | 2-Phenylpropionic acid (M) | C9H10O2 | 150.0680 | (M + H)+ | 151.0753 | 151.0753 | 0.53 | 130.1593, 124.0872, 121.0509, 118.0863 |
| 27 | 17.199 | Rishitin (M) | C14H22O2 | 222.1619 | (M − H)− | 221.1546 | 221.1546 | 0.52 | 132.9234 |
| 28 | 17.683 | 3,8-Dihydroxy-6-methoxy-7(11)-eremophilen-12,8-olide (M) | C16H24O5 | 296.1618 | (M − H)¯ | 295.1544 | 295.1547 | 1.97 | 194.0824, 132.9234 |
| 29 | 17.849 | Gingerglycolipid A (M) | C33H56O14 | 676.3665 | (M + HCOO)− | 721.3646 | 721.3646 | 0.77 | 339.1997, 311.1687, 132.9234 |
| 30 | 18.296 | Gingerglycolipid B (M) | C33H58O14 | 678.3828 | (M + HCOO)− | 723.3810 | 723.3810 | −0.19 | 311.1687, 132.9234 |
| 31 | 18.563 | Isocurcumenol (M) | C15H22O2 | 234.1619 | (M − H)− | 233.1546 | 233.1546 | 0.52 | 132.9235 |
| 32 | 18.635 | Isokobusone (H, M) | C14H22O2 | 222.1620 | (M − H)− | 221.1546 | 221.1546 | 0.09 | 132.9235 |
| Compound | α-Glucosidase Inhibitory Activity, IC50 (µM) |
|---|---|
| 1 | 243.4 ± 5.29 |
| 2 | 235.8 ± 27.67 |
| 3 | 195.4 ± 18.13 |
| 4 (and 1), the ratio of 1.00:0.79 | 342.2 ± 28.72 |
| 5 | 431.0 ± 17.64 |
| 6 | 455.6 ± 26.00 |
| 7 | 432.5 ± 18.88 |
| Acarbose | 2023.3 ± 17.34 |
| Compound | Concentration of Compound (µg/mL) | Cytotoxicity (% Cell Viability) | Virucidal Activity Log Reduction |
|---|---|---|---|
| 1 | 125.0 | 77.2 ± 1.28 | 1.80 ± 0.105 |
| 62.5 | 83.8 ± 2.02 | 1.52 ± 0.045 | |
| 31.3 | 86.5 ± 2.16 | 1.34 ± 0.030 | |
| 15.6 | 94.7 ± 3.93 | 1.34 ± 0.044 | |
| 7.8 | 97.8 ± 0.31 | 1.32 ± 0.032 | |
| 3.9 | 99.3 ± 3.45 | 1.20 ± 0.025 | |
| 2 | 125.0 | 78.0 ± 1.75 | 1.95 ± 0.124 |
| 62.5 | 86.5 ± 3.82 | 1.82 ± 0.091 | |
| 31.3 | 88.5 ± 2.67 | 1.68 ± 0.046 | |
| 15.6 | 92.3 ± 1.95 | 1.66 ± 0.071 | |
| 7.8 | 96.3 ± 5.13 | 1.65 ± 0.084 | |
| 3.9 | 98.1 ± 1.34 | 1.66 ± 0.071 | |
| 3 | 125.0 | 73.3 ± 0.90 | 1.98 ± 0.088 |
| 62.5 | 83.6 ± 0.74 | 1.91 ± 0.062 | |
| 31.3 | 88.7 ± 0.21 | 1.95 ± 0.124 | |
| 15.6 | 89.7 ± 0.82 | 1.91 ± 0.062 | |
| 7.8 | 92.8 ± 4.84 | 1.66 ± 0.071 | |
| 3.9 | 95.6 ± 6.25 | 1.63 ± 0.069 | |
| A mixture of 4 and 1 with the ratio of 1.00:0.79 | 500.0 | 83.4 ± 1.63 | 2.41 ± 0.000 |
| 250.0 | 84.6 ± 2.08 | 2.01 ± 0.102 | |
| 125.0 | 89.9 ± 1.88 | 1.90 ± 0.142 | |
| 62.5 | 93.8 ± 1.83 | 1.84 ± 0.108 | |
| 31.3 | 94.7 ± 0.97 | 1.81 ± 0.091 | |
| 15.6 | 96.9 ± 0.54 | 1.87 ± 0.072 | |
| 5 | 500.0 | 82.9 ± 2.06 | 2.11 ± 0.198 |
| 250.0 | 83.7 ± 1.20 | 2.01 ± 0.239 | |
| 125.0 | 85.8 ± 2.87 | 1.97 ± 0.088 | |
| 62.5 | 90.0 ± 3.08 | 1.90 ± 0.062 | |
| 31.3 | 94.1 ± 1.85 | 1.84 ± 0.108 | |
| 15.6 | 95.8 ± 4.22 | 1.87 ± 0.072 | |
| 6 | 500.0 | 86.4 ± 2.06 | 1.97 ± 0.042 |
| 250.0 | 88.6 ± 0.43 | 1.93 ± 0.124 | |
| 125.0 | 89.5 ± 0.83 | 1.93 ± 0.124 | |
| 62.5 | 93.7 ± 4.87 | 1.93 ± 0.124 | |
| 31.3 | 95.7 ± 1.63 | 1.93 ± 0.124 | |
| 15.6 | 97.0 ± 2.99 | 1.93 ± 0.124 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Astiti, M.A.; Jittmittraphap, A.; Leaungwutiwong, P.; Chutiwitoonchai, N.; Pripdeevech, P.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. LC-QTOF-MS/MS Based Molecular Networking Approach for the Isolation of α-Glucosidase Inhibitors and Virucidal Agents from Coccinia grandis (L.) Voigt. Foods 2021, 10, 3041. https://doi.org/10.3390/foods10123041
Astiti MA, Jittmittraphap A, Leaungwutiwong P, Chutiwitoonchai N, Pripdeevech P, Mahidol C, Ruchirawat S, Kittakoop P. LC-QTOF-MS/MS Based Molecular Networking Approach for the Isolation of α-Glucosidase Inhibitors and Virucidal Agents from Coccinia grandis (L.) Voigt. Foods. 2021; 10(12):3041. https://doi.org/10.3390/foods10123041
Chicago/Turabian StyleAstiti, Maharani A., Akanitt Jittmittraphap, Pornsawan Leaungwutiwong, Nopporn Chutiwitoonchai, Patcharee Pripdeevech, Chulabhorn Mahidol, Somsak Ruchirawat, and Prasat Kittakoop. 2021. "LC-QTOF-MS/MS Based Molecular Networking Approach for the Isolation of α-Glucosidase Inhibitors and Virucidal Agents from Coccinia grandis (L.) Voigt" Foods 10, no. 12: 3041. https://doi.org/10.3390/foods10123041
APA StyleAstiti, M. A., Jittmittraphap, A., Leaungwutiwong, P., Chutiwitoonchai, N., Pripdeevech, P., Mahidol, C., Ruchirawat, S., & Kittakoop, P. (2021). LC-QTOF-MS/MS Based Molecular Networking Approach for the Isolation of α-Glucosidase Inhibitors and Virucidal Agents from Coccinia grandis (L.) Voigt. Foods, 10(12), 3041. https://doi.org/10.3390/foods10123041







