Bioactivity of Essential Oils for Mitigation of Listeria monocytogenes Isolated from Fresh Retail Chicken Meat
Abstract
1. Introduction
- Investigate the prevalence of L. monocytogenes in fresh retail chicken meat;
- Detect the antibiogram profile of the isolated L. monocytogenes;
- Evaluate the biofilm-forming ability (BFA), virulence, and resistance genes;
- Investigate the activity of different EOs as antimicrobial agents against multidrug-resistant L. monocytogenes;
- Study the effect of EOs on bacterial cell viability and integrity loss, as indicated by increased electrical conductivity, ion leakage, and salt tolerance capacity loss.
2. Materials and Methods
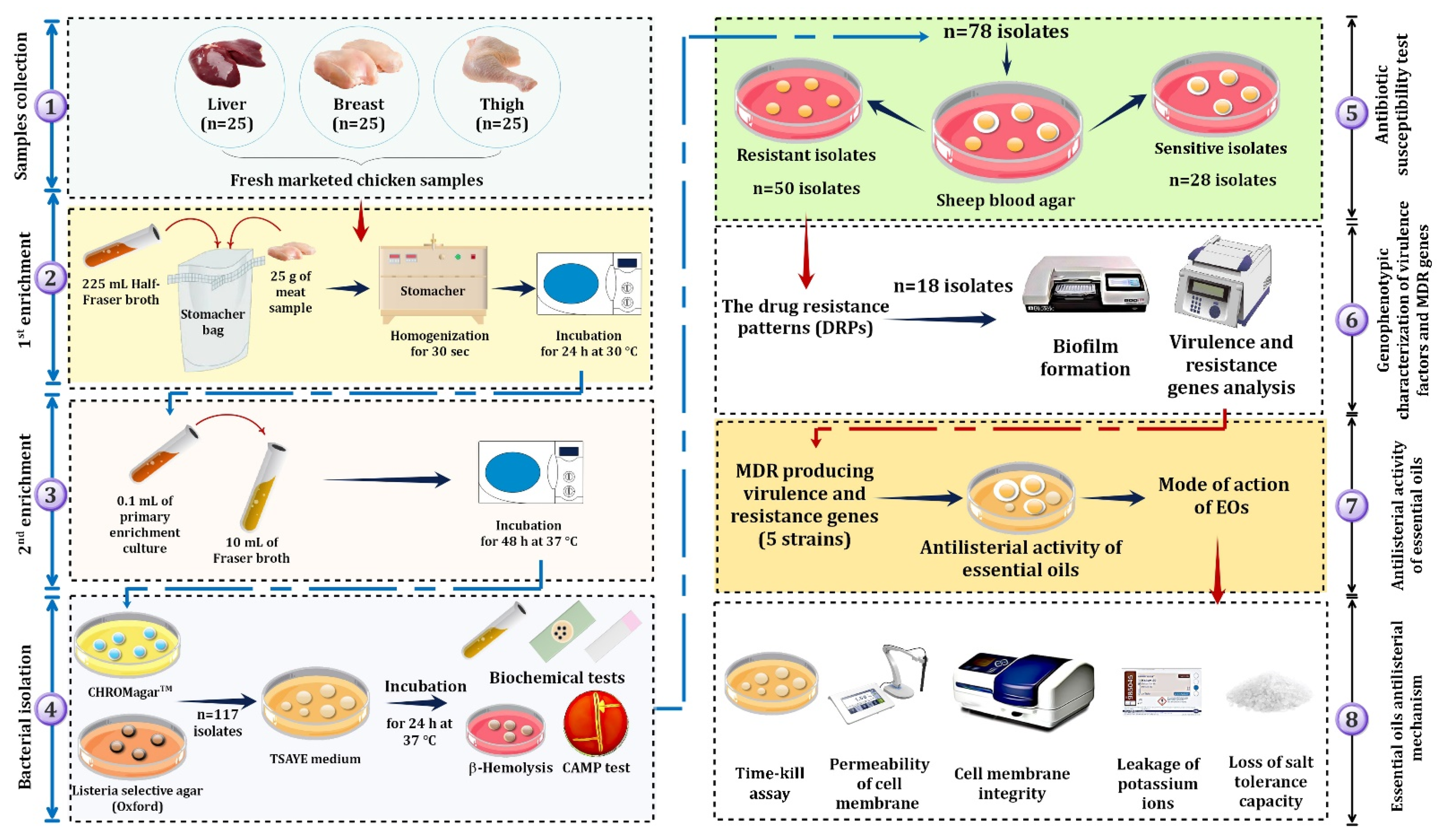
2.1. Sampling
2.2. Isolation of L. monocytogenes
2.3. Antibiotics Susceptibility Test
2.4. Molecular Identification, Resistance, and Virulence Genes Detection
2.5. Biofilm Quantification
2.6. Gas Chromatography/Mass Spectrometry (GC/MS) Analysis of EOs
2.7. Antilisterial Activity of EOs
2.8. Detection of Essential Oil Bioactivity
2.8.1. Time–Kill Assay
2.8.2. Cytoplasmic Membrane Permeability
2.8.3. Potassium Ion Leakage Assay
2.8.4. Cell Membrane Integrity Assay
2.8.5. Loss of Salt Tolerance Capacity Assay
2.9. Statistical Analysis
3. Results and Discussion
3.1. Prevalence and Phenotypic Characterization of L. monocytogenes
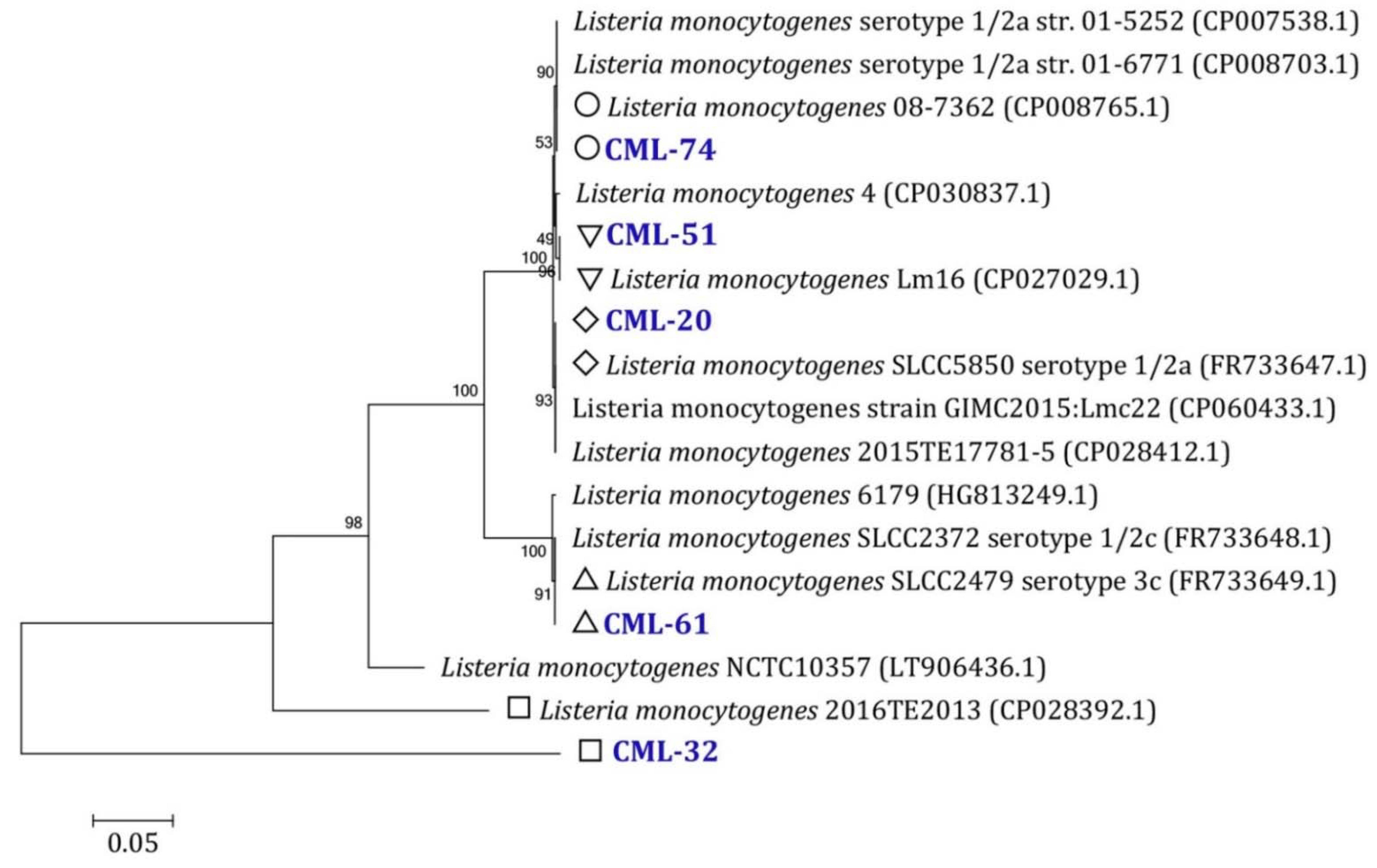
3.2. L. monocytogenes Drug Resistance and Their Drug Resistance Patterns
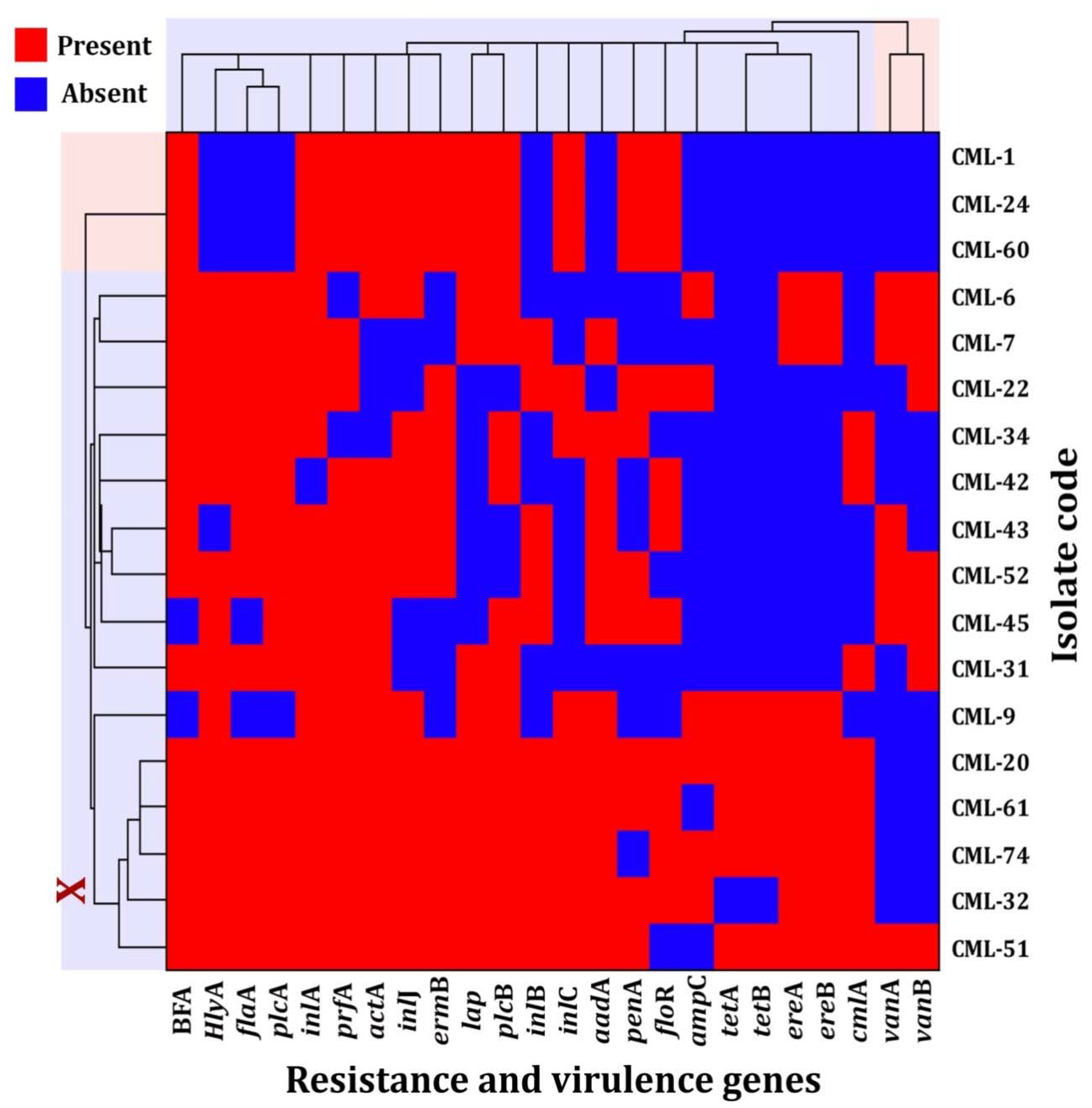
3.3. Drug Resistance Genes and Virulence Factors
3.4. L. monocytogenes Biofilm-Forming Ability
3.5. Essential Oil Antilisterial Activity
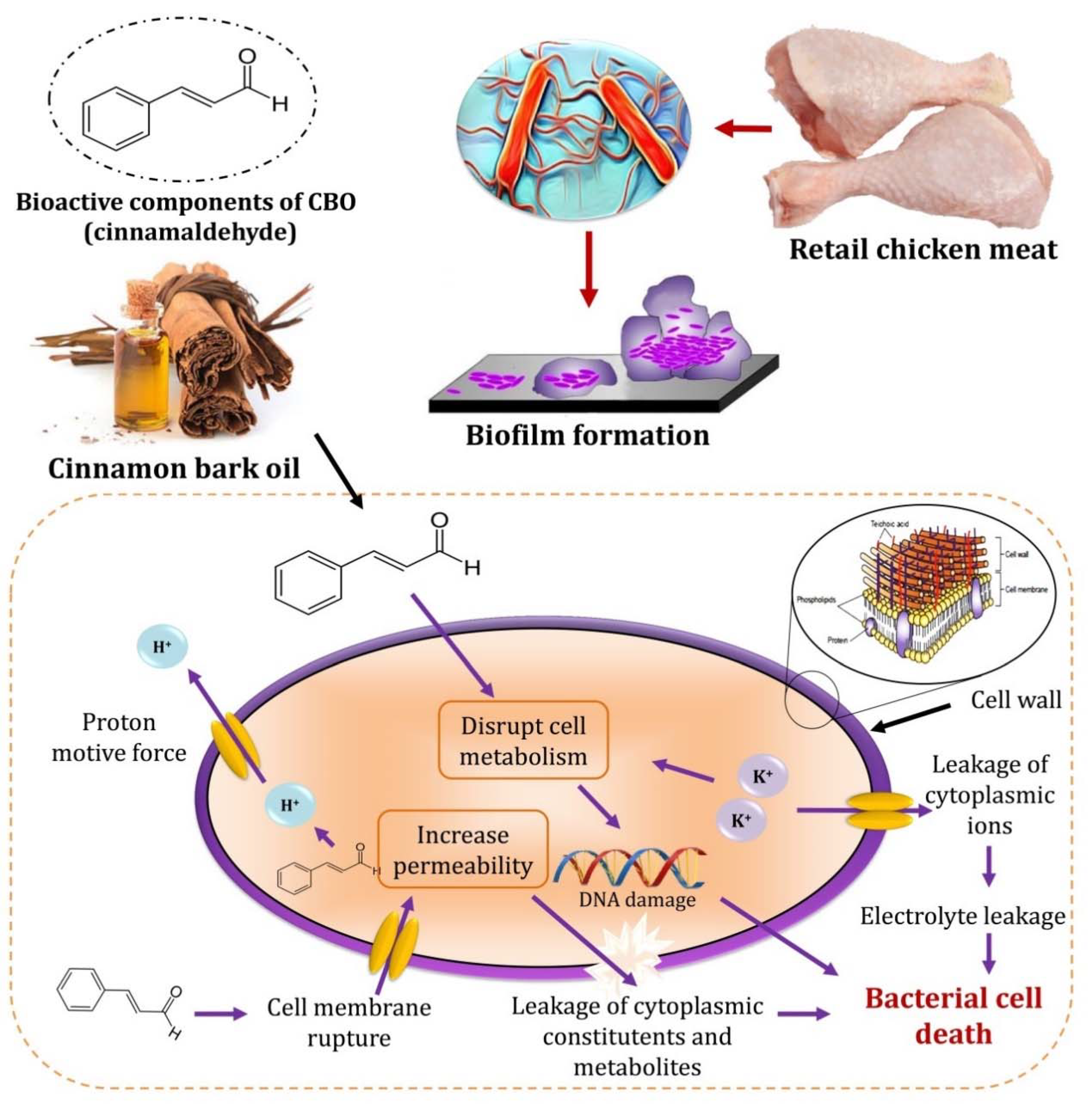
3.6. CBO Bioactivity as an Antilisterial Agent
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farber, J.M.; Zwietering, M.; Wiedmann, M.; Schaffner, D.; Hedberg, C.W.; Harrison, M.A.; Hartnett, E.; Chapman, B.; Donnelly, C.W.; Goodburn, K.E.; et al. Alternative approaches to the risk management of Listeria monocytogenes in low risk foods. Food Control 2021, 123, 107601. [Google Scholar] [CrossRef]
- Yu, H.; Guo, W.; Lu, X.; Xu, H.; Yang, Q.; Tan, J.; Zhang, W. Reduced graphene oxide nanocomposite based electrochemical biosensors for monitoring foodborne pathogenic bacteria: A review. Food Control 2021, 127, 108117. [Google Scholar] [CrossRef]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of Its physiology and pathogenesis. Nat. Rev. Microbiol. 2017, 16, 32–46. [Google Scholar] [CrossRef]
- Huang, Y.-T.; Kuo, Y.-W.; Lee, M.-R.; Tsai, Y.-H.; Teng, L.-J.; Tsai, M.-S.; Liao, C.-H.; Hsueh, P.-R. Clinical and molecular epidemiology of human listeriosis in Taiwan. Int. J. Infect. Dis. 2021, 104, 718–724. [Google Scholar] [CrossRef]
- Ghosh, P.; Halvorsen, E.M.; Ammendolia, D.A.; Mor-Vaknin, N.; O’Riordan, M.X.D.; Brumell, J.H.; Markovitz, D.M.; Higgins, D.E. Invasion of the brain by Listeria monocytogenes is mediated by InlF and host cell Vimentin. MBio 2018, 9, e00160-18. [Google Scholar] [CrossRef]
- Bouymajane, A.; Rhazi Filali, F.; Oulghazi, S.; Lafkih, N.; Ed-Dra, A.; Aboulkacem, A.; El Allaoui, A.; Ouhmidou, B.; Moumni, M. Occurrence, antimicrobial resistance, serotyping and virulence genes of Listeria monocytogenes Isolated from foods. Heliyon 2021, 7, e06169. [Google Scholar] [CrossRef] [PubMed]
- Barreca, S.; La Bella, S.; Maggio, A.; Licata, M.; Buscemi, S.; Leto, C.; Pace, A.; Tuttolomondo, T. Flavouring extra-virgin olive oil with aromatic and medicinal plants essential oils stabilizes oleic acid composition during photo-oxidative stress. Agriculture 2021, 11, 266. [Google Scholar] [CrossRef]
- Puvača, N.; Milenković, J.; Galonja Coghill, T.; Bursić, V.; Petrović, A.; Tanasković, S.; Pelić, M.; Ljubojević Pelić, D.; Miljković, T. Antimicrobial activity of selected essential oils against selected pathogenic bacteria: In vitro study. Antibiotics 2021, 10, 546. [Google Scholar] [CrossRef]
- Andrade-Ochoa, S.; Chacón-Vargas, K.F.; Sánchez-Torres, L.E.; Rivera-Chavira, B.E.; Nogueda-Torres, B.; Nevárez-Moorillón, G.V. Differential antimicrobial effect of essential oils and their main components: Insights based on the cell membrane and external structure. Membranes 2021, 11, 405. [Google Scholar] [CrossRef]
- Gyawali, R.; Ibrahim, S.A. Natural products as antimicrobial agents. Food Control 2014, 46, 412–429. [Google Scholar] [CrossRef]
- Negi, P.S. Plant extracts for the control of bacterial growth: Efficacy, stability and safety issues for food application. Int. J. Food Microbiol. 2012, 156, 7–17. [Google Scholar] [CrossRef]
- Ju, J.; Xu, X.; Xie, Y.; Guo, Y.; Cheng, Y.; Qian, H.; Yao, W. Inhibitory effects of cinnamon and clove essential oils on mold growth on baked foods. Food Chem. 2018, 240, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.J.; Sowbhagya, H.B. Nanoencapsulation of Spice Oils. In Handbook of Food Bioengineering, Soft Chemistry and Food Fermentation; Academic Press: Cambridge, MA, USA, 2017; pp. 179–207. [Google Scholar] [CrossRef]
- Cherrat, L.; Dumas, E.; Bakkali, M.; Degraeve, P.; Laglaoui, A.; Oulahal, N. Effect of essential oils on cell viability, membrane integrity and membrane fluidity of listeria innocua and Escherichia coli. J. Essent. Oil Bear. Plants 2016, 19, 155–166. [Google Scholar] [CrossRef]
- Somrani, M.; Inglés, M.-C.; Debbabi, H.; Abidi, F.; Palop, A. Garlic, onion, and cinnamon essential oil anti-biofilms’ effect against Listeria monocytogenes. Foods 2020, 9, 567. [Google Scholar] [CrossRef]
- Hussain, Z.; Li, X.; Zhang, D.; Hou, C.; Ijaz, M.; Bai, Y.; Xiao, X.; Zheng, X. Influence of adding cinnamon bark oil on meat quality of ground lamb during storage at 4 °C. Meat Sci. 2021, 171, 108269. [Google Scholar] [CrossRef]
- Mellado-García, P.; Puerto, M.; Pichardo, S.; Llana-Ruiz-Cabello, M.; Moyano, R.; Blanco, A.; Jos, A.; Cameán, A.M. Toxicological evaluation of an allium-based commercial product in a 90-day feeding study in sprague–dawley rats. Food Chem. Toxicol. 2016, 90, 18–29. [Google Scholar] [CrossRef]
- Ariza, J.J.; García-López, D.; Sánchez-Nieto, E.; Guillamón, E.; Baños, A.; Martínez-Bueno, M. Antilisterial effect of a natural formulation based on citrus extract in ready-to-eat foods. Foods 2021, 10, 1475. [Google Scholar] [CrossRef]
- Cock, I.E.; Van Vuuren, S.F. The traditional use of Southern African medicinal plants in the treatment of viral respiratory diseases: A review of the ethnobotany and scientific evaluations. J. Ethnopharmacol. 2020, 262, 113194. [Google Scholar] [CrossRef]
- British Standards Institution (BSI). BS EN ISO 11290-2 Microbiology of Food and Animal Feeding Stuffs ± Horizontal Method for the Detection and Enumeration of Listeria monocytogenes-Part 2: Enumeration Method; BSI: London, UK, 1998. [Google Scholar] [CrossRef]
- Seeliger, H.P.; Jones, D. Listeria. In Bergey’s Manual of Systematic Bacteriology; Sneath, P.H.A., Nair, N.S., Sharpe, N.E., Holt, J.G., Eds.; Williams and Wilkins: Baltimore, MD, USA, 1986; pp. 1235–1245. [Google Scholar]
- Honjoh, K.; Fujihara, K.; Haraguchi, T.; Ono, Y.; Kobayashi, H.; Hiwaki, H.; Kamikado, H.; Jang, S.S.; Ryu, S.; Miyamoto, T. Subtyping of Listeria monocytogenes based on nucleotide polymorphism in the ClpC, InlA, HlyA, and PlcA genes and rapid identification of L. monocytogenes genetically similar to clinical isolates. Food Sci. Technol. Res. 2008, 14, 557–564. [Google Scholar] [CrossRef]
- Morvan, A.; Moubareck, C.; Leclercq, A.; Hervé-Bazin, M.; Bremont, S.; Lecuit, M.; Courvalin, P.; Le Monnier, A. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 2010, 54, 2728–2731. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, CLSI Supplement M100, 27th ed.; Clinical Laboratory Standards Institute: Wayne, NJ, USA, 2017; pp. 15–20. [Google Scholar] [CrossRef]
- Odjadjare, E.E.O.; Obi, L.C.; Okoh, A.I. Municipal wastewater effluents as a source of Listerial pathogens in the aquatic milieu of the Eastern Cape province of South Africa: A concern of public health importance. Int. J. Environ. Res. Public Health 2010, 7, 2376–2394. [Google Scholar] [CrossRef]
- Osundiya, O.O.; Oladele, R.O.; Oduyebo, O.O. Multiple antibiotic resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. Afr. J. Clin. Exp. Microbiol. 2013, 14, 164–168. [Google Scholar] [CrossRef]
- Osman, K.M.; Samir, A.; Abo-Shama, U.H.; Mohamed, E.H.; Orabi, A.; Zolnikov, T. Determination of virulence and antibiotic resistance pattern of biofilm producing Listeria species isolated from retail raw milk. BMC Microbiol. 2016, 16, 263. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef]
- Di Ciccio, P.; Meloni, D.; Festino, A.R.; Conter, M.; Zanardi, E.; Ghidini, S.; Vergara, A.; Mazzette, R.; Ianieri, A. Longitudinal study on the sources of Listeria monocytogenes contamination in cold-smoked salmon and its processing environment in Italy. Int. J. Food Microbiol. 2012, 158, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.I.; Kroll, R.G. Polymerase chain reaction amplification of the FlaA gene for the rapid identification of Listeria spp. Lett. Appl. Microbiol. 1995, 20, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Furrer, B.; Candrian, U.; Hoefelein, C.; Luethy, J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J. Appl. Bacteriol. 1991, 70, 372–379. [Google Scholar] [CrossRef]
- Notermans, S.H.; Dufrenne, J.; Leimeister-Wächter, M.; Domann, E.; Chakraborty, T. Phosphatidylinositol-specific phospholipase C activity as a marker to distinguish between pathogenic and nonpathogenic Listeria species. Appl. Environ. Microbiol. 1991, 57, 2666–2670. [Google Scholar] [CrossRef]
- Cooray, K.J.; Nishibori, T.; Xiong, H.; Matsuyama, T.; Fujita, M.; Mitsuyama, M. Detection of multiple virulence-associated genes of Listeria monocytogenes by PCR in artificially contaminated milk samples. Appl. Environ. Microbiol. 1994, 60, 3023–3026. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.; Gonzalez-Zorn, B.; Vega, Y.; Chico-Calero, I.; Vazquez-Boland, J.-A. A Role for ActA in epithelial cell invasion by Listeria monocytogenes. Cell. Microbiol. 2001, 3, 853–864. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Austin, F.W.; Ainsworth, A.J. A multiplex PCR for species-and virulence-specific determination of Listeria monocytogenes. J. Microbiol. Methods 2007, 71, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.E.I.; Knabel, S.J. Multiplex PCR assay simplifies serotyping and sequence typing of Listeria monocytogenes associated with human outbreaks. J. Food Prot. 2005, 68, 1907–1910. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.; Kuhnert, P.; Boerlin, P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 2003, 91, 73–84. [Google Scholar] [CrossRef]
- Okamoto, K.; Gotoh, N.; Nishino, T. Extrusion of penem antibiotics by multicomponent efflux systems MexAB-OprM, MexCD-OprJ, and MexXY-OprM of Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2002, 46, 2696–2699. [Google Scholar] [CrossRef]
- Antignac, A. Polymorphism of Neisseria meningitidis PenA gene associated with reduced susceptibility to Penicillin. J. Antimicrob. Chemother. 2001, 47, 285–296. [Google Scholar] [CrossRef]
- Keyes, K.; Hudson, C.; Maurer, J.J.; Thayer, S.; White, D.G.; Lee, M.D. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob. Agents Chemother. 2000, 44, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Gebreyes, W.A.; Altier, C. Molecular characterization of multidrug-resistant Salmonella enterica subsp. Enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 2002, 40, 2813–2822. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, G.; Verbrugge, D.; Chasseur-Libotte, M.-L.; Moens, W.; Collard, J.-M. PCR typing of tetracycline resistance determinants (Tet A–E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 2000, 32, 77–85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sutcliffe, J.; Tait-Kamradt, A.; Wondrack, L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to Macrolides but sensitive to Clindamycin: A common resistance pattern mediated by an Efflux System. Antimicrob. Agents Chemother. 1996, 40, 1817–1824. [Google Scholar] [CrossRef]
- Lee, B.-H.; Hébraud, M.; Bernardi, T. Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front. Microbiol. 2017, 8, 2221. [Google Scholar] [CrossRef]
- Dudek-Wicher, R.; Paleczny, J.; Kowalska-Krochmal, B.; Szymczyk-Ziółkowska, P.; Pachura, N.; Szumny, A.; Brożyna, M. Activity of liquid and volatile fractions of essential oils against biofilm formed by selected reference strains on polystyrene and hydroxyapatite surfaces. Pathogens 2021, 10, 515. [Google Scholar] [CrossRef]
- Diao, W.-R.; Hu, Q.-P.; Feng, S.-S.; Li, W.-Q.; Xu, J.-G. Chemical composition and antibacterial activity of the essential oil from green Huajiao (Zanthoxylum schinifolium) against selected foodborne pathogens. J. Agric. Food Chem. 2013, 61, 6044–6049. [Google Scholar] [CrossRef]
- Li, Z.-H.; Cai, M.; Liu, Y.-S.; Sun, P.-L.; Luo, S.-L. Antibacterial activity and mechanisms of essential oil from Citrus medica L. Var. Sarcodactylis. Molecules 2019, 24, 1577. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Li, X.; Yuan, L.; He, H. Effect of the surface activity on the antibacterial activity of octadecanoyl acetal sodium sulfite series. Colloids Surf. A Physicochem. Eng. Asp. 2005, 268, 85–89. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Sharma, A.; Baek, K.-H. Antibacterial mode of action of Cudrania tricuspidata fruit essential oil, affecting membrane permeability and surface characteristics of food-borne pathogens. Food Control. 2013, 32, 582–590. [Google Scholar] [CrossRef]
- Carson, C.F.; Mee, B.J.; Riley, T.V. Mechanism of action of Melaleuca alternifolia (Tea Tree) oil on staphylococcus aureus determined by time-kill, lysis, leakage, and salt tolerance assays and electron microscopy. Antimicrob. Agents Chemother. 2002, 46, 1914–1920. [Google Scholar] [CrossRef]
- Miksusanti; Jenie, B. S.L.; Priosoeryanto, B.P.; Syarief, R.; Rekso, G.T. Mode of action Temu Kunci (Kaempferia pandurata) essential oil on E. Coli K1.1 cell determined by leakage of material cell and salt Tolerance assays. HAYATI J. Biosci. 2008, 15, 56–60. [Google Scholar] [CrossRef]
- Elmali, M.; Can, H.Y.; Yaman, H. Prevalence of Listeria monocytogenes in poultry meat. Food Sci. Technol. 2015, 35, 672–675. [Google Scholar] [CrossRef][Green Version]
- Ceylan, Z.G.; Demirkaya, A.K.; Adigüzel, G. Incidence of Listeria monocytogenes in retail chicken meat and establishing relationship with some bacteria by logistic regression. J. Food Qual. 2008, 31, 121–130. [Google Scholar] [CrossRef]
- Escudero-Gilete, M.L.; González-Miret, M.L.; Temprano, R.M.; Heredia, F.J. Application of a multivariate concentric method system for the location of Listeria monocytogenes in a poultry slaughterhouse. Food Control 2007, 18, 69–75. [Google Scholar] [CrossRef]
- Goh, S.G.; Kuan, C.H.; Loo, Y.Y.; Chang, W.S.; Lye, Y.L.; Soopna, P.; Tang, J.Y.H.; Nakaguchi, Y.; Nishibuchi, M.; Afsah-Hejri, L.; et al. Listeria monocytogenes in retailed raw chicken meat in Malaysia. Poult. Sci. 2012, 91, 2686–2690. [Google Scholar] [CrossRef]
- Gufe, C.; Canaan Hodobo, T.; Mbonjani, B.; Majonga, O.; Marumure, J.; Musari, S.; Jongi, G.; Makaya, P.V.; Machakwa, J. Antimicrobial profiling of bacteria isolated from fish sold at informal market in Mufakose, Zimbabwe. Int. J. Microbiol. 2019, 2019, 8759636. [Google Scholar] [CrossRef] [PubMed]
- Mpondo, L.; Ebomah, K.E.; Okoh, A.I. Multidrug-resistant Listeria species shows abundance in environmental waters of a Key District municipality in South Africa. Int. J. Environ. Res. Public Health 2021, 18, 481. [Google Scholar] [CrossRef] [PubMed]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Teuber, M. Spread of antibiotic resistance with food-borne pathogens. Cell. Mol. Life Sci. 1999, 56, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Actualités et Perspectives 2014 Du Comité Antibiogramme de la Société Française de Microbiologie (CA-SFM). Option/Bio 2014, 25, 13. Available online: https://www.sciencedirect.com/science/article/abs/pii/S0992594514716411?via%3Dihub (accessed on 1 September 2021). [CrossRef]
- Morosi, S.; Francisci, D.; Baldelli, F. A case of Rhombencephalitis caused by Listeria monocytogenes successfully treated with linezolid. J. Infect. 2006, 52, e73–e75. [Google Scholar] [CrossRef] [PubMed]
- Conter, M.; Paludi, D.; Zanardi, E.; Ghidini, S.; Vergara, A.; Ianieri, A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 2009, 128, 497–500. [Google Scholar] [CrossRef]
- Chen, M.; Cheng, J.; Wu, Q.; Zhang, J.; Chen, Y.; Xue, L.; Lei, T.; Zeng, H.; Wu, S.; Ye, Q.; et al. Occurrence, antibiotic resistance, and population diversity of Listeria monocytogenes isolated from fresh aquatic products in China. Front. Microbiol. 2018, 9, 2215. [Google Scholar] [CrossRef]
- Srinivasan, V.; Nam, H.M.; Nguyen, L.T.; Tamilselvam, B.; Murinda, S.E.; Oliver, S.P. Prevalence of antimicrobial resistance genes In Listeria monocytogenes isolated from dairy farms. Foodborne Pathog. Dis. 2005, 2, 201–211. [Google Scholar] [CrossRef]
- Van Stelten, A.; Roberts, A.R.; Manuel, C.S.; Nightingale, K.K. Listeria monocytogenes isolates carrying virulence-attenuating mutations in internalin A are commonly isolated from ready-to-eat food processing plant and retail environments. J. Food Prot. 2016, 79, 1733–1740. [Google Scholar] [CrossRef]
- Rugna, G.; Carra, E.; Bergamini, F.; Franzini, G.; Faccini, S.; Gattuso, A.; Morganti, M.; Baldi, D.; Naldi, S.; Serraino, A.; et al. Distribution, virulence, genotypic characteristics and antibiotic resistance of Listeria monocytogenes isolated over one-year monitoring from two pig slaughterhouses and processing plants and their fresh hams. Int. J. Food Microbiol. 2021, 336, 108912. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Rump, L.; Zhang, Y.; Chen, Y.; Wang, X.; Meng, J. Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 2013, 35, 58–64. [Google Scholar] [CrossRef]
- Pang, X.; Wong, C.; Chung, H.-J.; Yuk, H.-G. Biofilm formation of Listeria monocytogenes and its resistance to Quaternary ammonium compounds in a simulated salmon processing environment. Food Control 2019, 98, 200–208. [Google Scholar] [CrossRef]
- Kumar, S.; Parvathi, A.; George, J.; Krohne, G.; Karunasagar, I.; Karunasagar, I. A study on the effects of some laboratory-derived genetic mutations on biofilm formation by Listeria monocytogenes. World J. Microbiol. Biotechnol. 2008, 25, 527–531. [Google Scholar] [CrossRef]
- Price, R.; Jayeola, V.; Niedermeyer, J.; Parsons, C.; Kathariou, S. The Listeria monocytogenes key virulence determinants Hly and PrfA are involved in biofilm formation and Aggregation but not colonization of fresh produce. Pathogens 2018, 7, 18. [Google Scholar] [CrossRef]
- Piercey, M.J.; Hingston, P.A.; Truelstrup Hansen, L. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef]
- Mahfuzul Hoque, M.D.; Bari, M.L.; Inatsu, Y.; Juneja, V.K.; Kawamoto, S. Antibacterial activity of guava (Psidium guajava L.) and neem (Azadirachta indica A. Juss.) extracts against foodborne pathogens and spoilage bacteria. Foodborne Pathog. Dis. 2007, 4, 481–488. [Google Scholar] [CrossRef]
- Patra, J.K.; Baek, K.-H. Anti-Listerial activity of four seaweed essential oils against Listeria monocytogenes. Jundishapur J. Microbiol. 2016, 9, e31784. [Google Scholar] [CrossRef]
- Huang, S.; Liu, B.; Ge, D.; Dai, J. Effect of combined treatment with supercritical CO2 and rosemary on microbiological and physicochemical properties of ground pork stored at 4 °C. Meat Sci. 2017, 125, 114–120. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Sharma, A.; Baek, K.-H. Antibacterial mechanism of action of taxus cuspidata stem essential oil against selected foodborne pathogens. J. Food Saf. 2013, 33, 348–359. [Google Scholar] [CrossRef]
- Zhou, K.; Zhou, W.; Li, P.; Liu, G.; Zhang, J.; Dai, Y. Mode of action of Pentocin 31-1: An antilisteria bacteriocin produced by Lactobacillus Pentosus from Chinese traditional ham. Food Control 2008, 19, 817–822. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Wang, Y.; Jiang, P.; Quek, S. Antibacterial activity and mechanism of cinnamon essential oil against Escherichia Coli and Staphylococcus aureus. Food Control 2016, 59, 282–289. [Google Scholar] [CrossRef]
- Vasconcelos, N.G.; Croda, J.; Simionatto, S. Antibacterial mechanisms of cinnamon and its constituents: A review. Microb. Pathog. 2018, 120, 198–203. [Google Scholar] [CrossRef]
- Chen, E.; Wu, S.; McClements, D.J.; Li, B.; Li, Y. Influence of PH and Cinnamaldehyde on the physical stability and lipolysis of whey protein isolate-stabilized emulsions. Food Hydrocoll. 2017, 69, 103–110. [Google Scholar] [CrossRef]
- Boughendjioua, H.; Djeddi, S. Study of the organoleptic and physicochemical properties of cinnamon essential oil (Cinnamomum zeylanicum). Am. J. Life Sci. Res. 2018, 6, 123–130. [Google Scholar]
- Goñi, P.; López, P.; Sánchez, C.; Gómez-Lus, R.; Becerril, R.; Nerín, C. Antimicrobial activity in the vapour phase of a combination of cinnamon and clove essential oils. Food Chem. 2009, 116, 982–989. [Google Scholar] [CrossRef]
- Ojagh, S.M.; Rezaei, M.; Razavi, S.H.; Hosseini, S.M.H. Effect of chitosan coatings enriched with cinnamon oil on the quality of refrigerated rainbow trout. Food Chem. 2010, 120, 193–198. [Google Scholar] [CrossRef]


| Antibiotics | Concentration (μg/Disc) | Resistant | Intermediate | Susceptible | |||
|---|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | No. | (%) | ||
| Ampicillin (AMP) | 10 | 22 | 28.2 | 4 | 5.13 | 52 | 66.67 |
| Chloramphenicol (CHL) | 30 | 29 | 37.2 | 2 | 2.56 | 47 | 60.26 |
| Ciprofloxacin (CIP) | 5 | 29 | 37.2 | 5 | 6.41 | 44 | 56.41 |
| Clindamycin (CLI) | 2 | 36 | 46.2 | 1 | 1.28 | 41 | 52.56 |
| Erythromycin (ERY) | 15 | 48 | 61.5 | 5 | 6.41 | 25 | 32.05 |
| Gentamicin (GEN) | 120 | 57 | 73.1 | 7 | 8.97 | 14 | 17.95 |
| Imipenem (IPM) | 10 | 29 | 37.2 | 2 | 2.56 | 47 | 60.26 |
| Linezolid (LNZ) | 30 | 29 | 37.2 | 5 | 6.41 | 44 | 56.41 |
| Nalidixic acid (NAL) | 30 | 29 | 37.2 | 5 | 6.41 | 44 | 56.41 |
| Oxacillin (OXA) | 1 | 29 | 37.2 | 4 | 5.13 | 45 | 57.69 |
| Rifampicin (RIF) | 5 | 22 | 28.2 | 6 | 7.69 | 50 | 64.10 |
| Tetracycline (TET) | 30 | 48 | 61.5 | 6 | 7.69 | 24 | 30.77 |
| Trimethoprim (TMP) | 5 | 36 | 46.2 | 5 | 6.41 | 37 | 47.44 |
| Vancomycin (VAN) | 30 | 22 | 28.2 | 4 | 5.13 | 52 | 66.67 |
| Pattern Code. | Antimicrobial Resistance Pattern | MAR Index | Strains No. |
|---|---|---|---|
| P1 | W, X, Y, AMP, OXA, TET, TMP | 0.93 | 5 |
| P1a | X, Y, Z, AMP, LNZ, NAL, OXA | 0.93 | 3 |
| P1b | W, X, Y, Z, AMP | 0.93 | 3 |
| P1c | W, X, Y, Z, OXA | 0.93 | 3 |
| P2 | X, Y, LNZ, NAL, TET | 0.64 | 2 |
| P3 | AMP, CHL, CLI, IPM, NAL, RIF, TET, VAN | 0.57 | 2 |
| P3a | Y, Z, AMP, CLI | 0.57 | 1 |
| P3b | X, GEN, OXA, RIF, TET, TMP | 0.57 | 2 |
| P3c | W, Y, CHL, CIP | 0.57 | 3 |
| P4 | AMP, CIP, IPM, OXA, RIF, TMP, VAN | 0.50 | 2 |
| P4a | Z, AMP, CLI, Y, NAL | 0.50 | 2 |
| P4b | AMP, CLI, IPM, NAL, RIF, TET, VAN | 0.50 | 2 |
| P4c | CIP, CLI, ERY, GEN, LNZ, TET, VAN | 0.50 | 2 |
| P5 | CHL, ERY, GEN, NAL, OXA, TET | 0.43 | 4 |
| P5a | ERY, GEN, LNZ, OXA, TET, TMP | 0.43 | 4 |
| P6 | ERY, GEN, LNZ, TET, TMP | 0.36 | 3 |
| P7 | GEN, OXA, TET, TMP | 0.29 | 5 |
| P8 | CHL, ERY, GEN | 0.2 | 2 |
| No. | Essential Oil | Code | Main Constituent * | RT | Content (%) |
|---|---|---|---|---|---|
| 1 | Cinnamon bark oil | CBO | Cinnamaldehyde | 29.79 | 63.4 |
| Cinnamyl acetate | 21.73 | 15.2 | |||
| 2 | Thyme (wild) oil | TWO | Carvacrol | 18.09 | 67.2 |
| 3 | Thyme (red) oil | TRO | Thymol | 21.02 | 17.4 |
| γ-terpinene | 10.11 | 22.8 | |||
| 4 | Thyme (geraniol) oil | TGO | Geraniol | 12.45 | 28.6 |
| Geranyl acetate | 16.33 | 51.7 | |||
| 5 | Coriander oil | CRO | Linalool | 25.25 | 62.2 |
| 6 | Lavender (true) oil | LTO | Linalool | 14.24 | 31.4 |
| Linalyl acetate | 17.41 | 35.2 | |||
| 7 | Rosemary oil | RO | 1,8-cineole | 12.43 | 28.5 |
| α-pinene | 7.07 | 19.3 | |||
| Camphor | 18.01 | 16.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morshdy, A.E.M.A.; Al-Mogbel, M.S.; Mohamed, M.E.M.; Elabbasy, M.T.; Elshafee, A.K.; Hussein, M.A. Bioactivity of Essential Oils for Mitigation of Listeria monocytogenes Isolated from Fresh Retail Chicken Meat. Foods 2021, 10, 3006. https://doi.org/10.3390/foods10123006
Morshdy AEMA, Al-Mogbel MS, Mohamed MEM, Elabbasy MT, Elshafee AK, Hussein MA. Bioactivity of Essential Oils for Mitigation of Listeria monocytogenes Isolated from Fresh Retail Chicken Meat. Foods. 2021; 10(12):3006. https://doi.org/10.3390/foods10123006
Chicago/Turabian StyleMorshdy, Alaa Eldin M. A., Mohammed S. Al-Mogbel, Mohamed E. M. Mohamed, Mohamed Tharwat Elabbasy, Azza K. Elshafee, and Mohamed A. Hussein. 2021. "Bioactivity of Essential Oils for Mitigation of Listeria monocytogenes Isolated from Fresh Retail Chicken Meat" Foods 10, no. 12: 3006. https://doi.org/10.3390/foods10123006
APA StyleMorshdy, A. E. M. A., Al-Mogbel, M. S., Mohamed, M. E. M., Elabbasy, M. T., Elshafee, A. K., & Hussein, M. A. (2021). Bioactivity of Essential Oils for Mitigation of Listeria monocytogenes Isolated from Fresh Retail Chicken Meat. Foods, 10(12), 3006. https://doi.org/10.3390/foods10123006







