Diversity of Listeria monocytogenes Strains Isolated from Food Products in the Central European Part of Russia in 2000–2005 and 2019–2020
Abstract
:1. Introduction
2. Materials and Methods
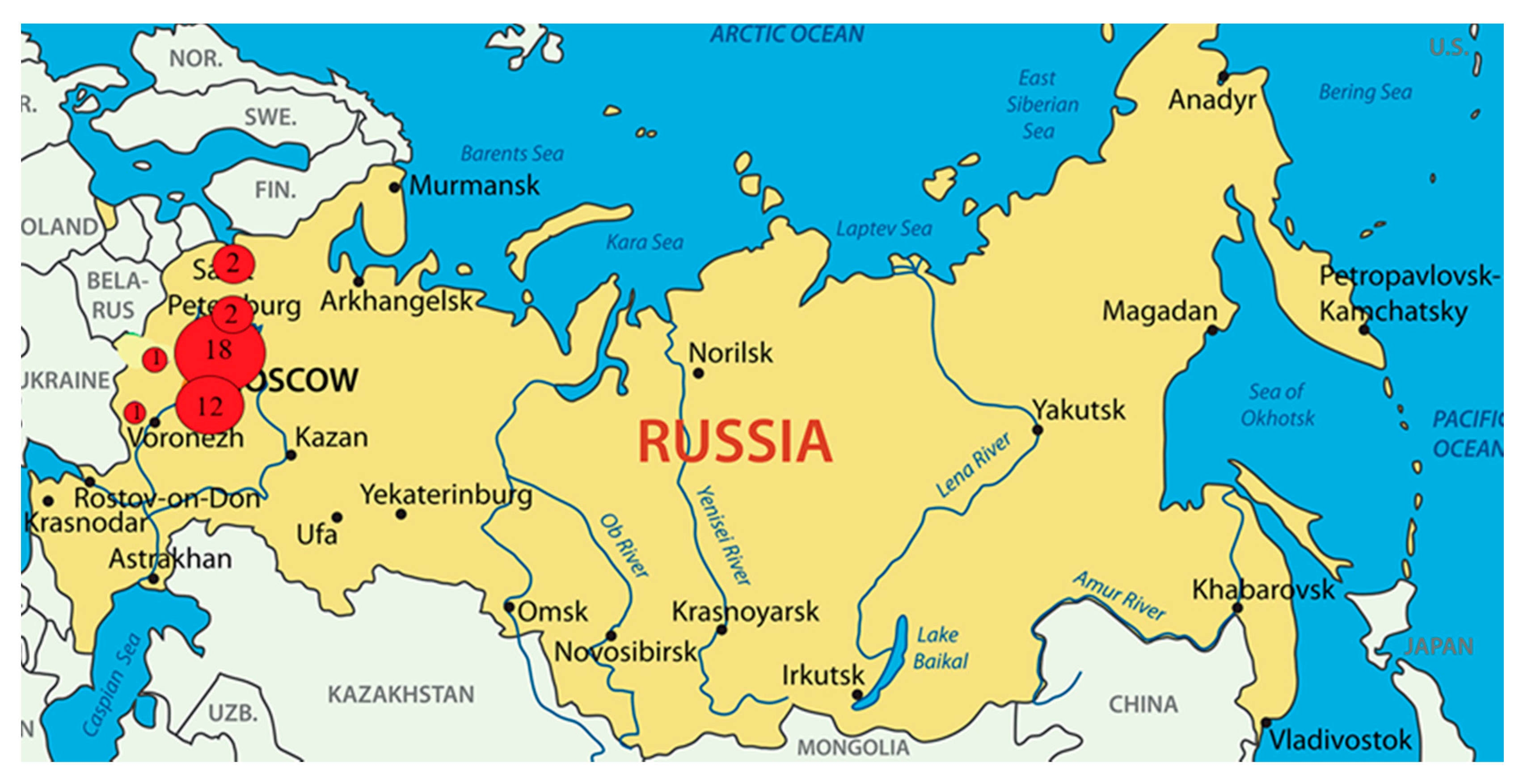
2.1. Researched Sources from Meat Products in 2019
2.2. Culturing Conditions of Bacterial Strains
2.3. Antibiotic Resistance Assays
2.4. L. monocytogenes Virulence in HEp-2 Cells
2.5. Multiplex PCR Assay to Serotype L. monocytogenes
2.6. PCR
2.7. Sequencing of PCR Products
2.8. Sequence Analysis
2.9. Calculation of Diversity Indexes
2.10. Statistics
3. Results
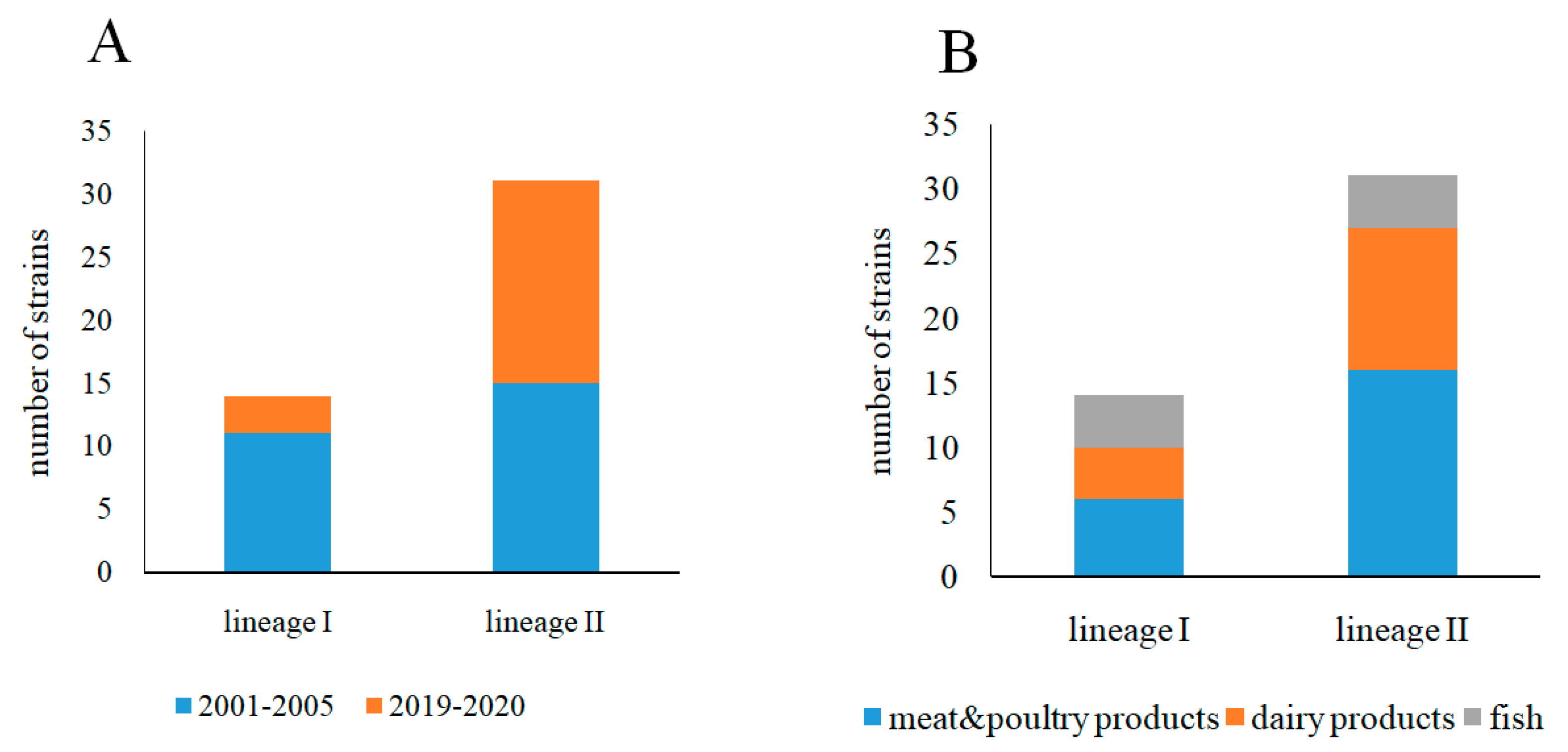
3.1. Phylogenetic Characterization of L. monocytogenes Strains Isolated from Raw Food Products in 2001–2005 and 2019–2020
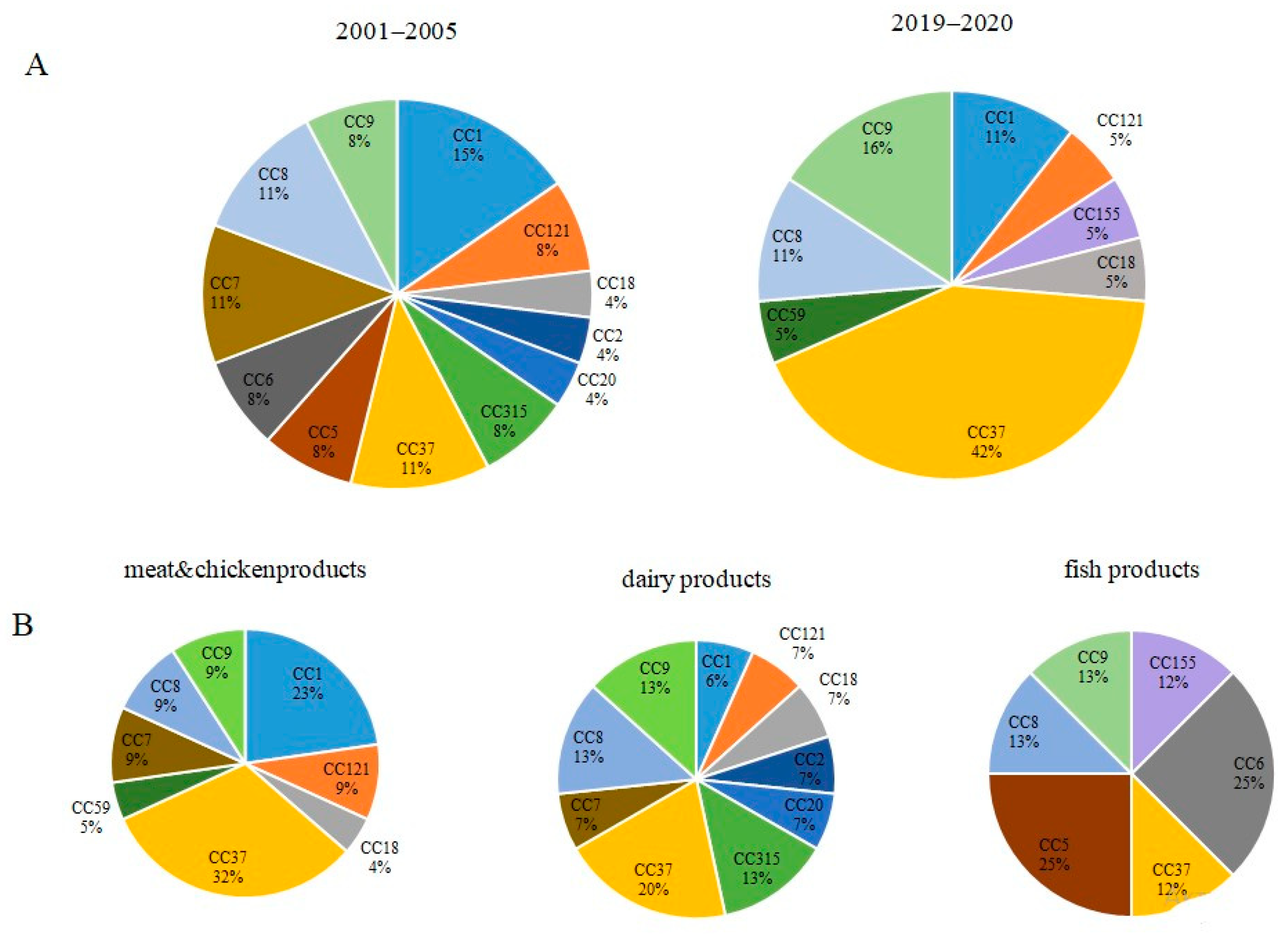
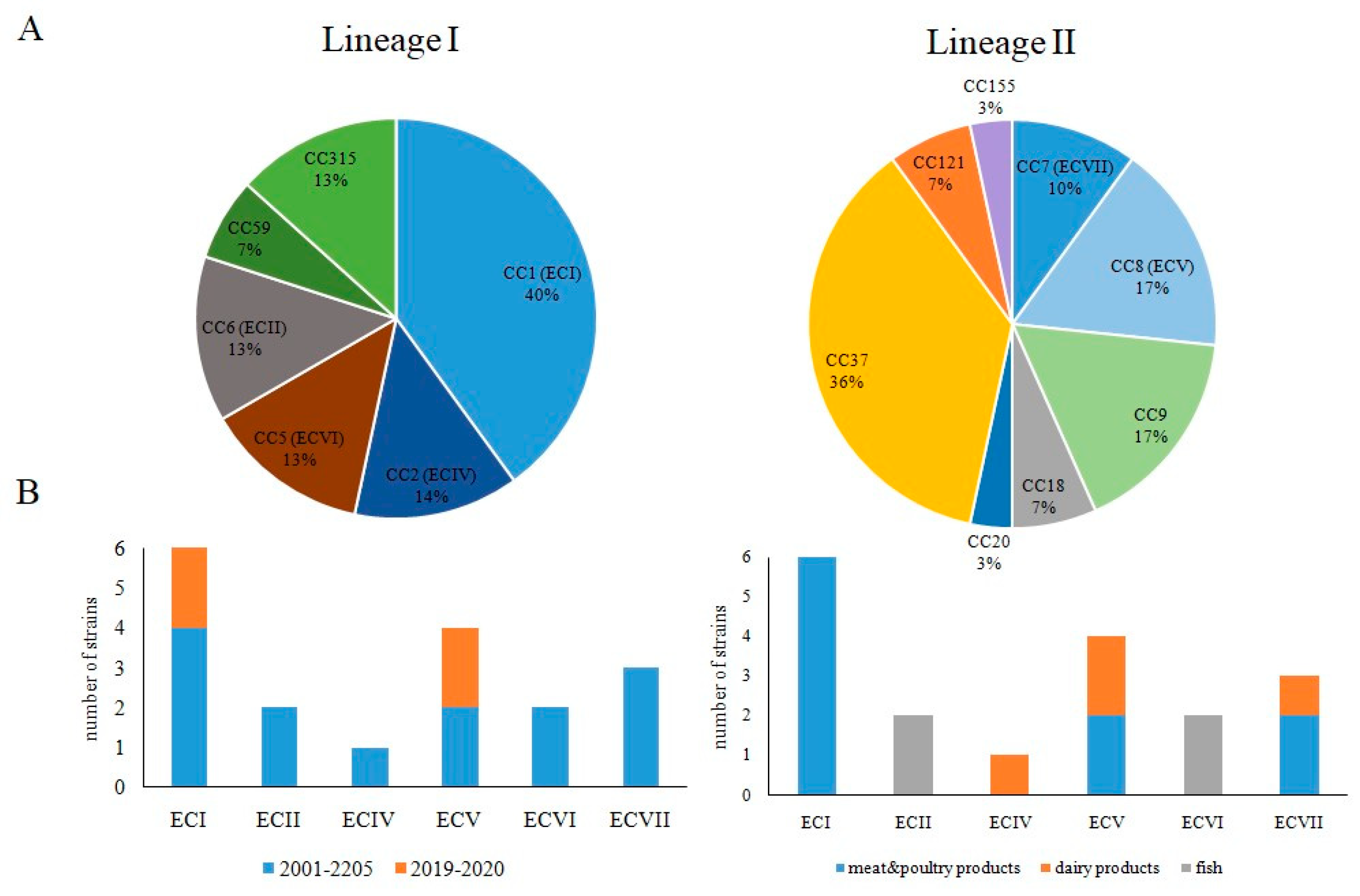
3.2. Occurrence of Worldwide Distributed Epidemic Clones among Studied Strains
3.3. MvLST Typing Did Not Increase Resolution of the MLST Scheme but Showed Similarity of Invasion Factors InlA and InlB in Strains Belonging to Distinct CCs
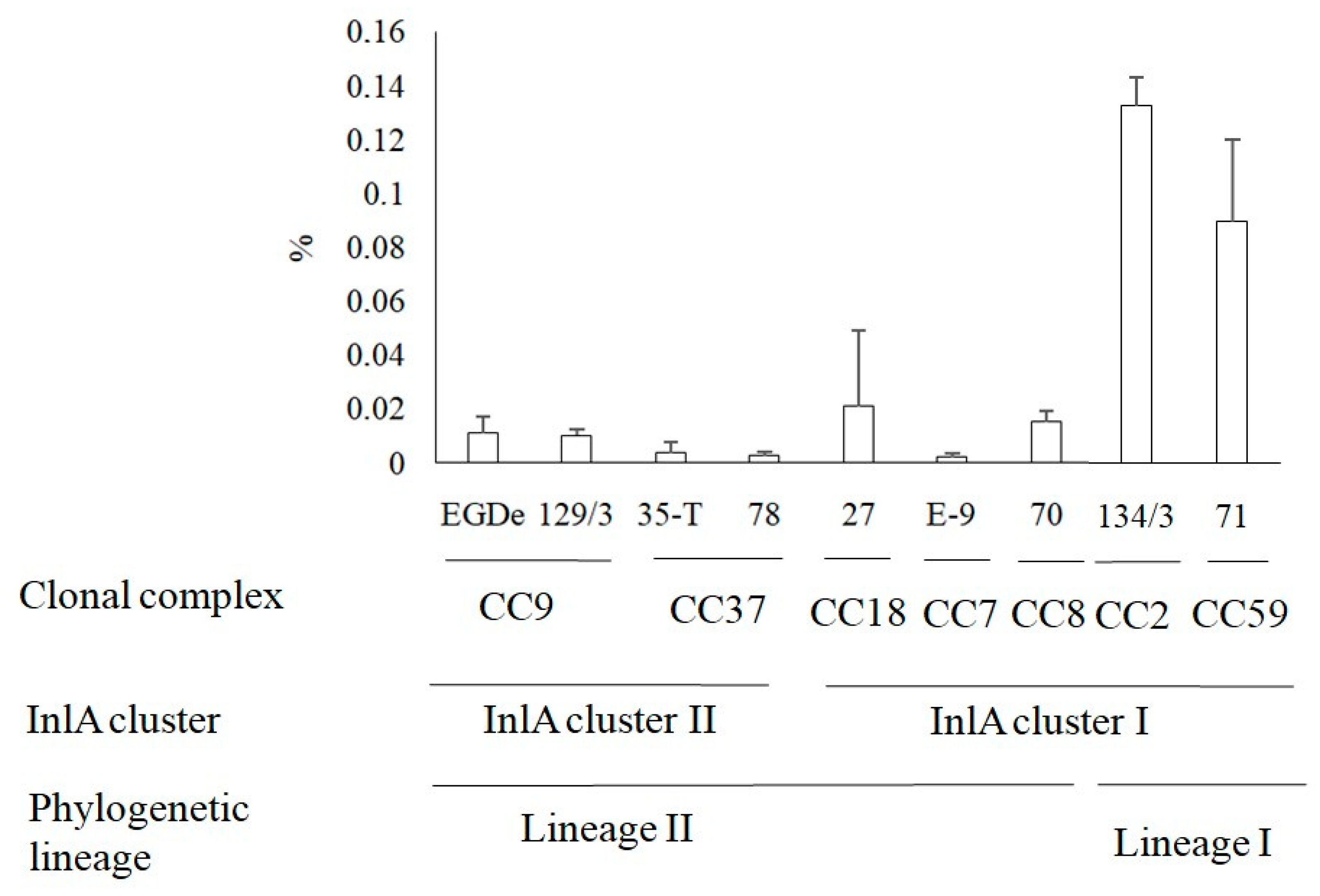
3.4. Invasion of L. monocytogenes into Human HEp-2 Cells
3.5. Antibiotics Resistance of L. monocytogenes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Allerberger, F.; Wagner, M. Listeriosis: A resurgent foodborne infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- Schlech, W.F. Epidemiology and clinical manifestations of listeria monocytogenes infection. Microbiol. Spectr. 2019, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Russini, V.; Spaziante, M.; Zottola, T.; Fermani, A.G.; Di Giampietro, G.; Blanco, G.; Fabietti, P.; Marrone, R.; Parisella, R.; Parrocchia, S.; et al. A Nosocomial outbreak of invasive listeriosis in an italian hospital: Epidemiological and genomic features. Pathogens 2021, 10, 591. [Google Scholar] [CrossRef]
- Desai, A.N.; Anyoha, A.; Madoff, L.C.; Lassmann, B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: A review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef] [Green Version]
- Lecuit, M. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin. Microbiol. Infect. 2005, 11, 430–436. [Google Scholar] [CrossRef] [Green Version]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bai, L.; Fu, P.; Han, H.; Liu, J.; Guo, Y. The epidemiology of listeria monocytogenes in China. Foodborne Pathog. Dis. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Voronina, O.; Ryzhova, N.; Kunda, M.; Kurnaeva, M.; Semenov, A.; Aksenova, E.; Egorova, I.Y.; Kolbasov, D.; Ermolaeva, S.; Alexander, G.L. Diversity and pathogenic potential of listeria monocytogenes isolated from environmental sources in the russian federation. Int. J. Mod. Emg. Res. 2015, 5, 5–13. [Google Scholar]
- Wacheck, S.; Fredriksson-Ahomaa, M.; König, M.; Stolle, A.; Stephan, R. Wild boars as an important reservoir for foodborne pathogens. Foodborne Pathog. Dis. 2010, 7, 307–312. [Google Scholar] [CrossRef]
- Adgamov, R.; Zaytseva, E.; Thiberge, J.-M.; Brisse, S.; Ermolaev, S. Genetically Related Listeria Monocytogenes Strains Isolated from Lethal Human Cases and Wild Animals. Genet. Divers. Microorg. 2012, 235–250. [Google Scholar]
- Maury, M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlier, C.; Perrodeau, E.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.; Moura, A.; Goffinet, F.; et al. Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef]
- Lomonaco, S.; Nucera, D.; Filipello, V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 2015, 35, 172–183. [Google Scholar] [CrossRef]
- Haase, J.K.; Didelot, X.; Lecuit, M.; Korkeala, H.; Achtman, M.; Leclercq, A.; Grant, K.; Wiedmann, M.; Apfalter, P. The ubiquitous nature of Listeria monocytogenes clones: A large-scale Multilocus Sequence Typing study. Environ. Microbiol. 2014, 16, 405–416. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [Green Version]
- Cantinelli, T.; Chenal-Francisque, V.; Diancourt, L.; Frezal, L.; Leclercq, A.; Wirth, T.; Lecuit, M.; Brisse, S. “Epidemic clones” of listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 2013, 51, 3770–3779. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Jayarao, B.M.; Knabel, S.J. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 2004, 70, 913–920. [Google Scholar] [CrossRef] [Green Version]
- Voronina, O.; Kunda, M.; RyzhYushchukova, N.; Kutuzova, A.; Aksenova, E.; Karpova, T.; Tartakovskij, I.; Yushchuk, N.; Klimova, E.; Karetkina, G.; et al. Listeriosis: Genotyping as a key for identification a possible source of infection. Clin. Microbiol. Antimicrob. Chemother. 2019, 21, 261–273. [Google Scholar] [CrossRef]
- Psareva, E.; Egorova, I.; Liskova, E.; Razheva, I.; Gladkova, N.; Sokolova, E.; Potemkin, E.; Zhurilov, P.; Mikhaleva, T.; Blokhin, A.; et al. Retrospective study of listeria monocytogenes isolated in the territory of inner eurasia from 1947 to 1999. Pathogens 2019, 8, 184. [Google Scholar] [CrossRef] [Green Version]
- Orsi, R.H.; denBakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, T.; Osborne, J.; Kathariou, S. Listeria monocytogenes source distribution analysis indicates regional heterogeneity and ecological niche preference among serotype 4b clones. MBio 2018, 9, e00396-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, A.; Burall, L. Serotype to genotype: The changing landscape of listeriosis outbreak investigations. Food Microbiol. 2018, 75, 18–27. [Google Scholar] [CrossRef]
- Pushkareva, V.; Ermolaeva, S.A.; Gintsburg, A.L. The role of technogenic and biological factors in spread of foodborne bacterial infections. Zhurnal Mikrobiol. Epidemiol. I Immunobiol. 2014, 5, 111–118. [Google Scholar]
- Lopez-Valladares, G.; Danielsson-Tham, M.L.; Tham, W. Implicated food products for listeriosis and changes in serovars of listeria monocytogenes affecting humans in recent decades. Foodborne Pathog. Dis. 2018, 15, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Hernando, A.; Prieto, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control 2012, 3, 37–41. [Google Scholar] [CrossRef]
- Charpentier, E.; Courvalin, P. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 1999, 43, 2103–2108. [Google Scholar] [CrossRef] [Green Version]
- Stessl, B.; Szakmary-Brändle, K.; Vorberg, U.; Schoder, D.; Wagner, M. Temporal analysis of the Listeria monocytogenes population structure in floor drains during reconstruction and expansion of a meat processing plant. Int. J. Food Microbiol. 2020, 314, 108360. [Google Scholar] [CrossRef]
- Lomonaco, S.; Verghese, B.; Gerner-Smidt, P.; Tarr, C.; Gladney, L.; Joseph, L.; Katz, L.; Turnsek, M.; Frace, M.; Chen, Y.; et al. Novel epidemic clones of Listeria monocytogenes, US, 2011. Emerg. Infect. Dis. 2013, 19, 147–150. [Google Scholar] [CrossRef]
- Muhterem-Uyar, M.; Ciolacu, L.; Wagner, K.; Wagner, M.; Schmitz-Esser, S.; Stessl, B. New aspects on listeria monocytogenes st5-ecvi predominance in a heavily contaminated cheese processing environment. Front. Microbiol. 2018, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, J.; Chang, Z.; Liu, X.; Chen, W.; Yu, Y.; Wang, X.; Dong, Q.; Ye, Y.; Zhang, X. Listeria monocytogenes contamination characteristics in two ready-to-eat meat plants from 2019 to 2020 in Shanghai. Front. Microbiol. 2021, 12, 729114. [Google Scholar] [CrossRef]
- Harrand, A.S.; Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H. Evolution of listeria monocytogenes in a food processing plant involves limited single-nucleotide substitutions but considerable diversification by gain and loss of prophages. Appl. Environ. Microbiol. 2020, 86, e02493-19. [Google Scholar] [CrossRef] [PubMed]
- Voronina, O.; Tartakovsky, I.; Yuyshchuk, N.; Ryzhova, N.; Kunda, M.; Aksenova, E.; Kutuzova, A.; Melkumyan, A.; Karpova, T.; Gruzdeva, O.; et al. Analysis of sporadic cases of invasivelisteriosis in a metropolis. J. Microbiol. Epidemiol. Immunobiol. 2020, 97, 547–555. [Google Scholar]
- Shamtsyan, M. Food legislation and its harmonization in Russia. J. Sci. Food Agric. 2014, 94, 1966–1969. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psareva, E.K.; Liskova, E.A.; Razheva, I.V.; Yushina, Y.K.; Grudistova, M.A.; Gladkova, N.A.; Potemkin, E.A.; Zhurilov, P.A.; Sokolova, E.V.; Andriyanov, P.A.; et al. Diversity of Listeria monocytogenes Strains Isolated from Food Products in the Central European Part of Russia in 2000–2005 and 2019–2020. Foods 2021, 10, 2790. https://doi.org/10.3390/foods10112790
Psareva EK, Liskova EA, Razheva IV, Yushina YK, Grudistova MA, Gladkova NA, Potemkin EA, Zhurilov PA, Sokolova EV, Andriyanov PA, et al. Diversity of Listeria monocytogenes Strains Isolated from Food Products in the Central European Part of Russia in 2000–2005 and 2019–2020. Foods. 2021; 10(11):2790. https://doi.org/10.3390/foods10112790
Chicago/Turabian StylePsareva, Ekaterina K., Elena A. Liskova, Irina V. Razheva, Yulia K. Yushina, Maria A. Grudistova, Nadezda A. Gladkova, Eugene A. Potemkin, Pavel A. Zhurilov, Elena V. Sokolova, Pavel A. Andriyanov, and et al. 2021. "Diversity of Listeria monocytogenes Strains Isolated from Food Products in the Central European Part of Russia in 2000–2005 and 2019–2020" Foods 10, no. 11: 2790. https://doi.org/10.3390/foods10112790
APA StylePsareva, E. K., Liskova, E. A., Razheva, I. V., Yushina, Y. K., Grudistova, M. A., Gladkova, N. A., Potemkin, E. A., Zhurilov, P. A., Sokolova, E. V., Andriyanov, P. A., Voronina, O. L., Kolbasov, D. V., & Ermolaeva, S. A. (2021). Diversity of Listeria monocytogenes Strains Isolated from Food Products in the Central European Part of Russia in 2000–2005 and 2019–2020. Foods, 10(11), 2790. https://doi.org/10.3390/foods10112790






