The Bacterial Diversity of Spontaneously Fermented Dairy Products Collected in Northeast Asia
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Sample Collection
2.2. DNA Extraction, PCR Amplification, and PacBio SMRT Sequencing
2.3. Bioinformatics Analysis
2.4. Statistical Analyses
3. Results
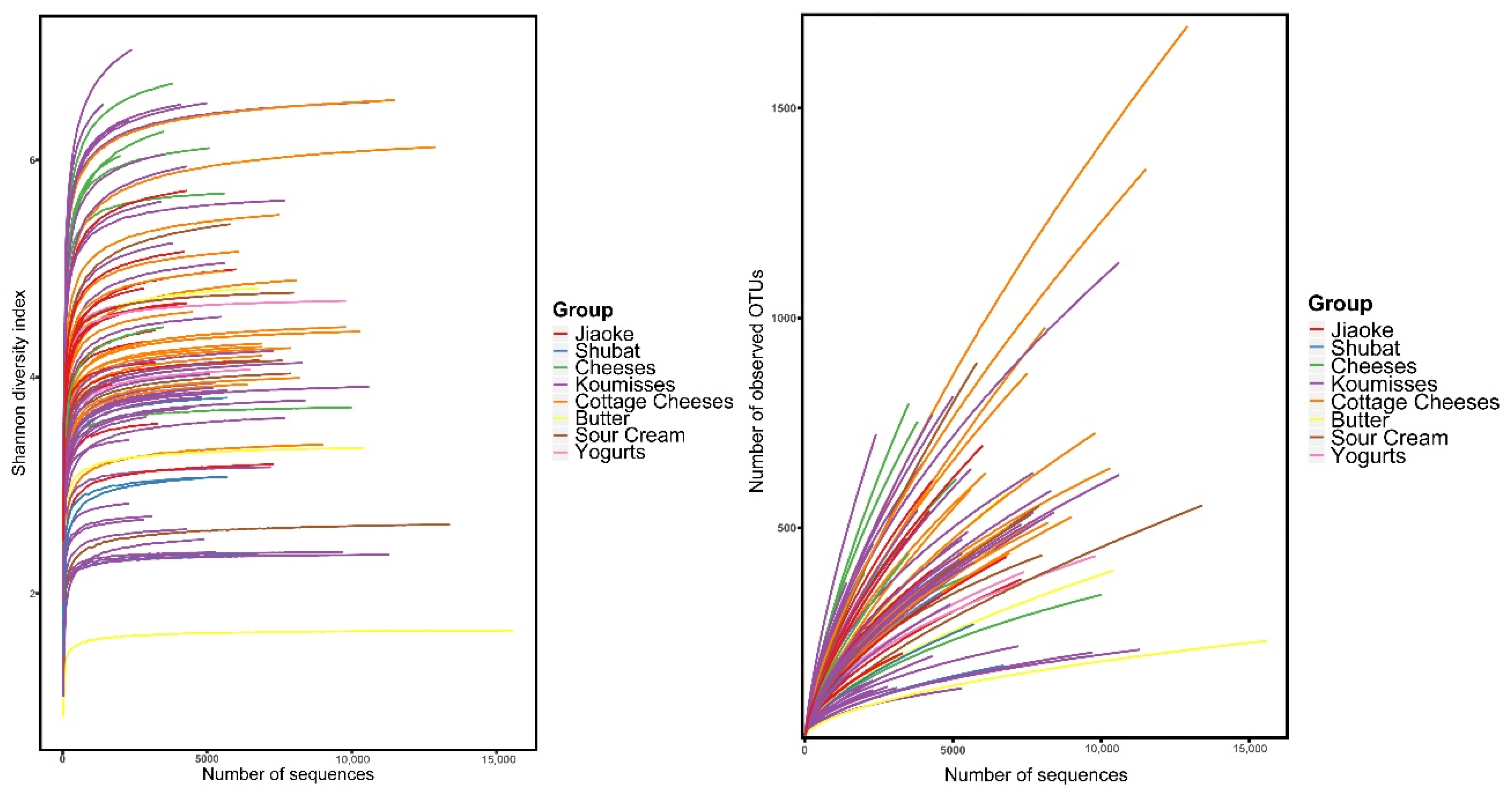
3.1. Bacterial Alpha Diversity Analysis
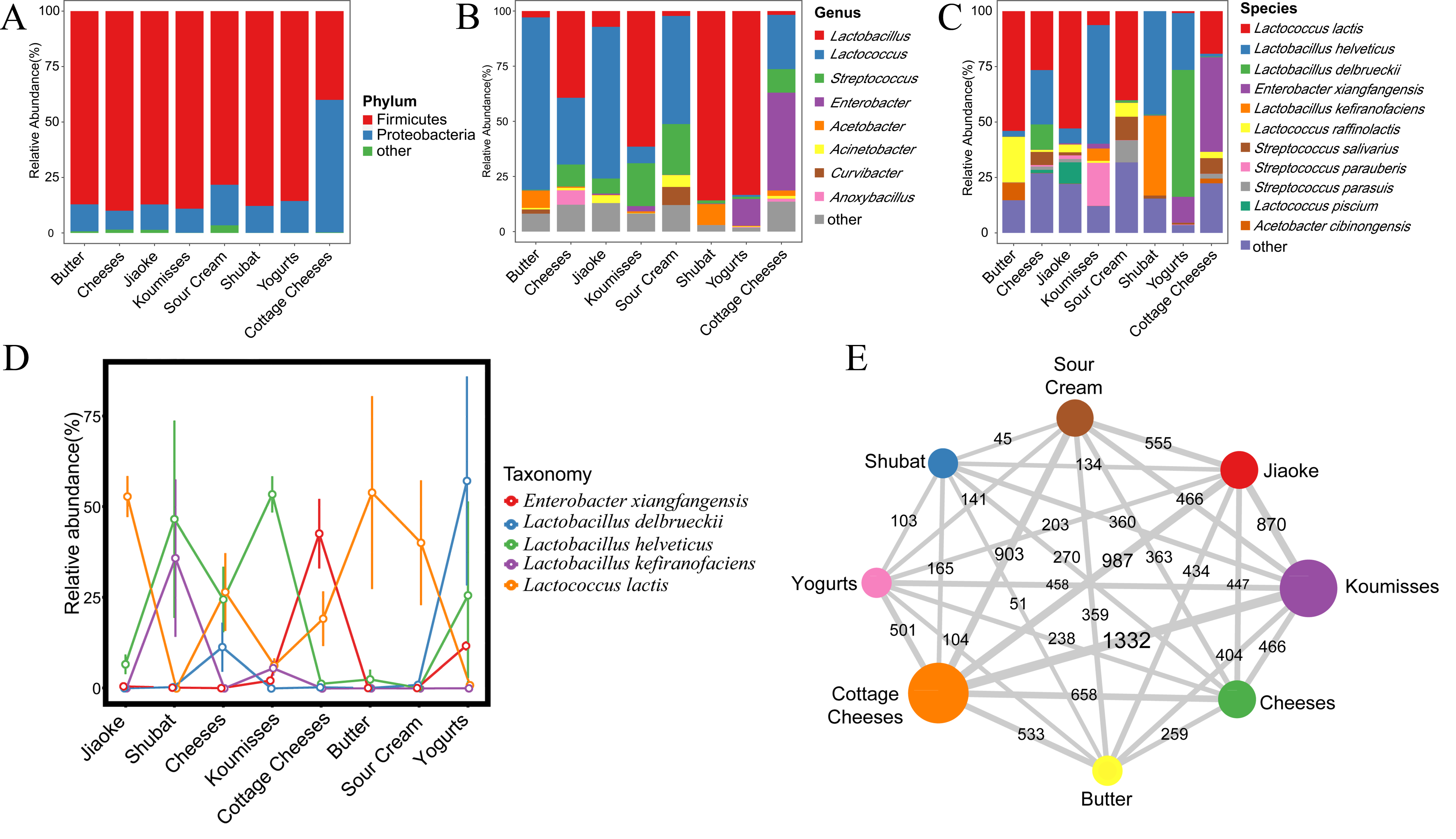
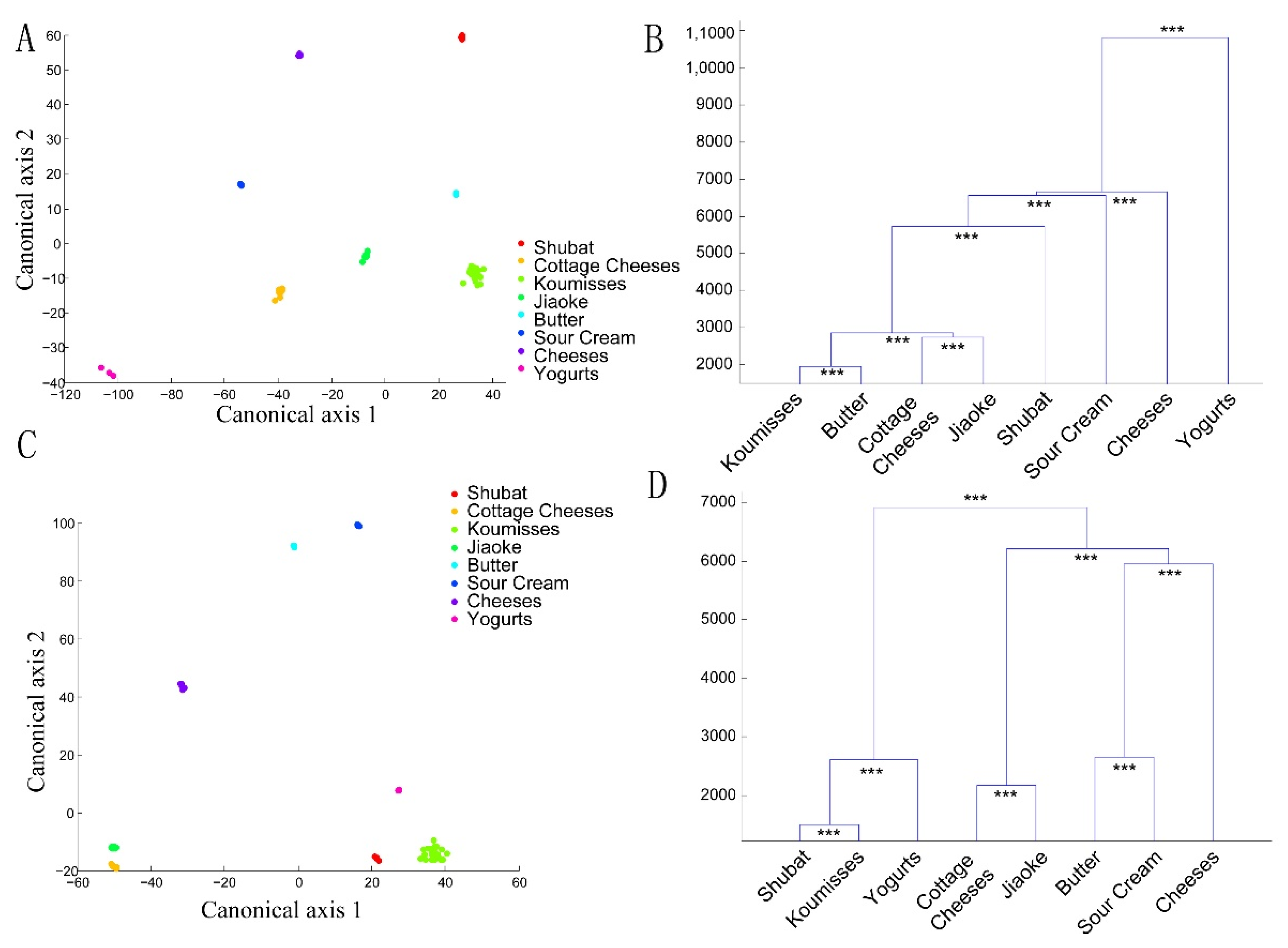
3.2. Composition of the Bacterial Structure in Spontaneously Fermented Dairy Products
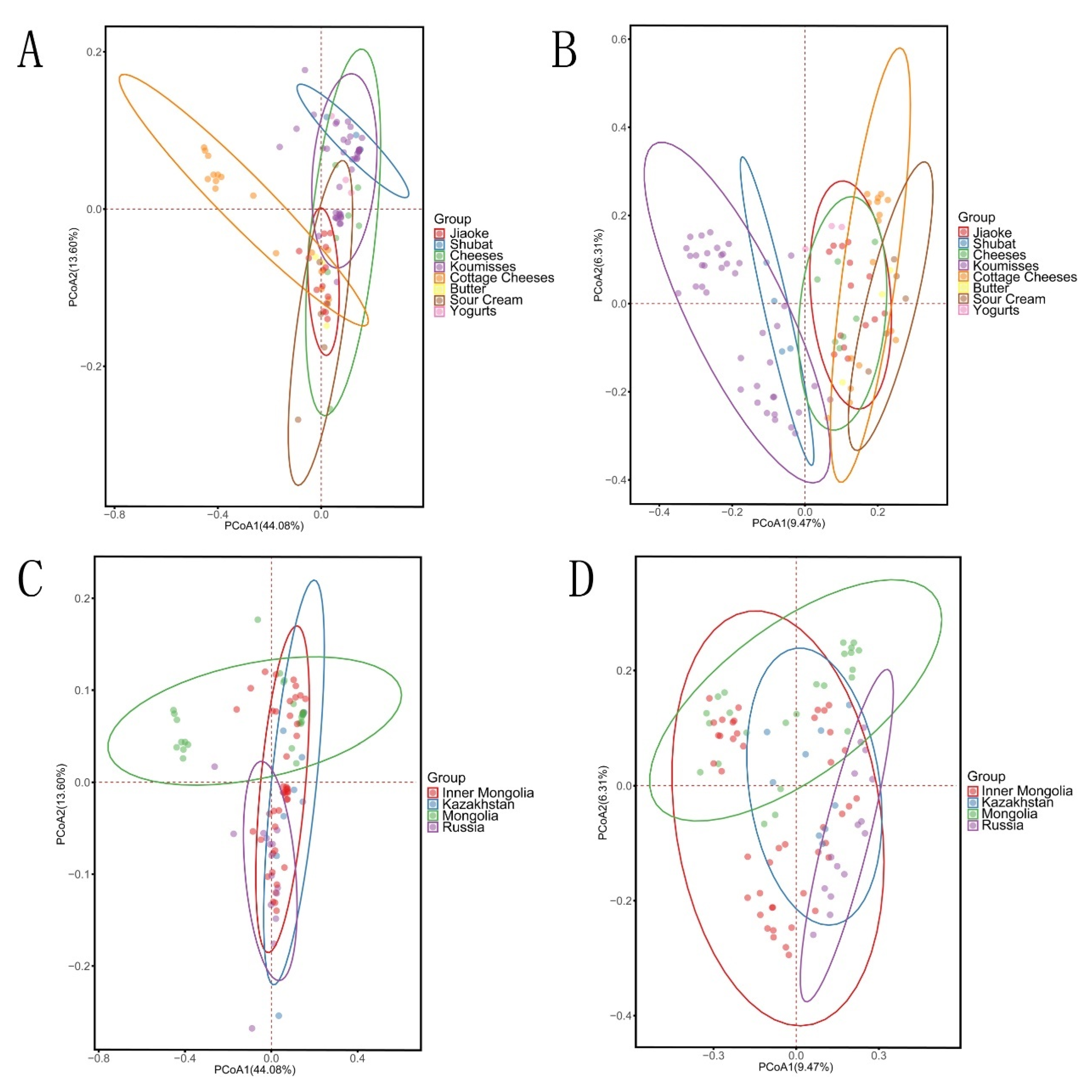
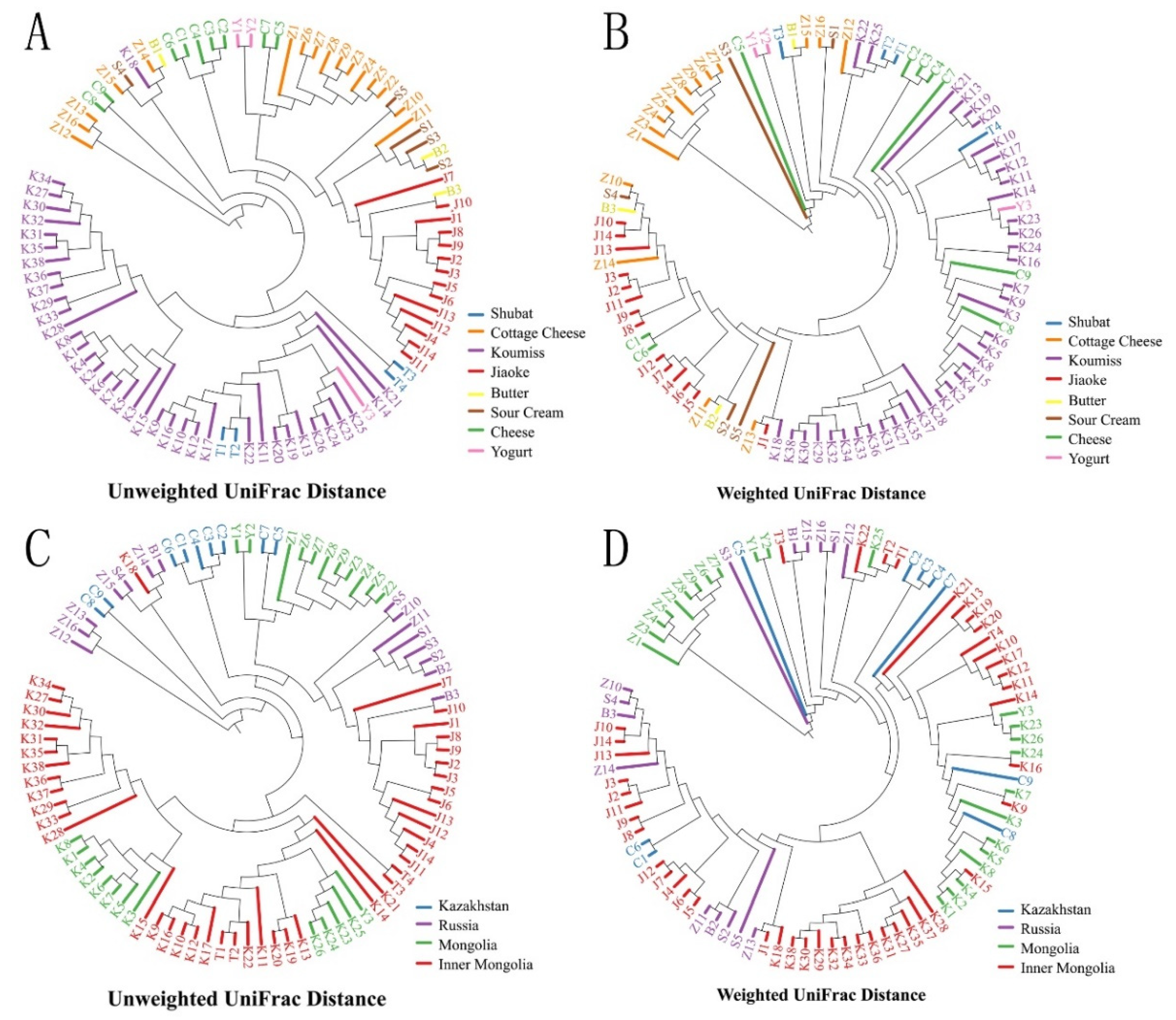
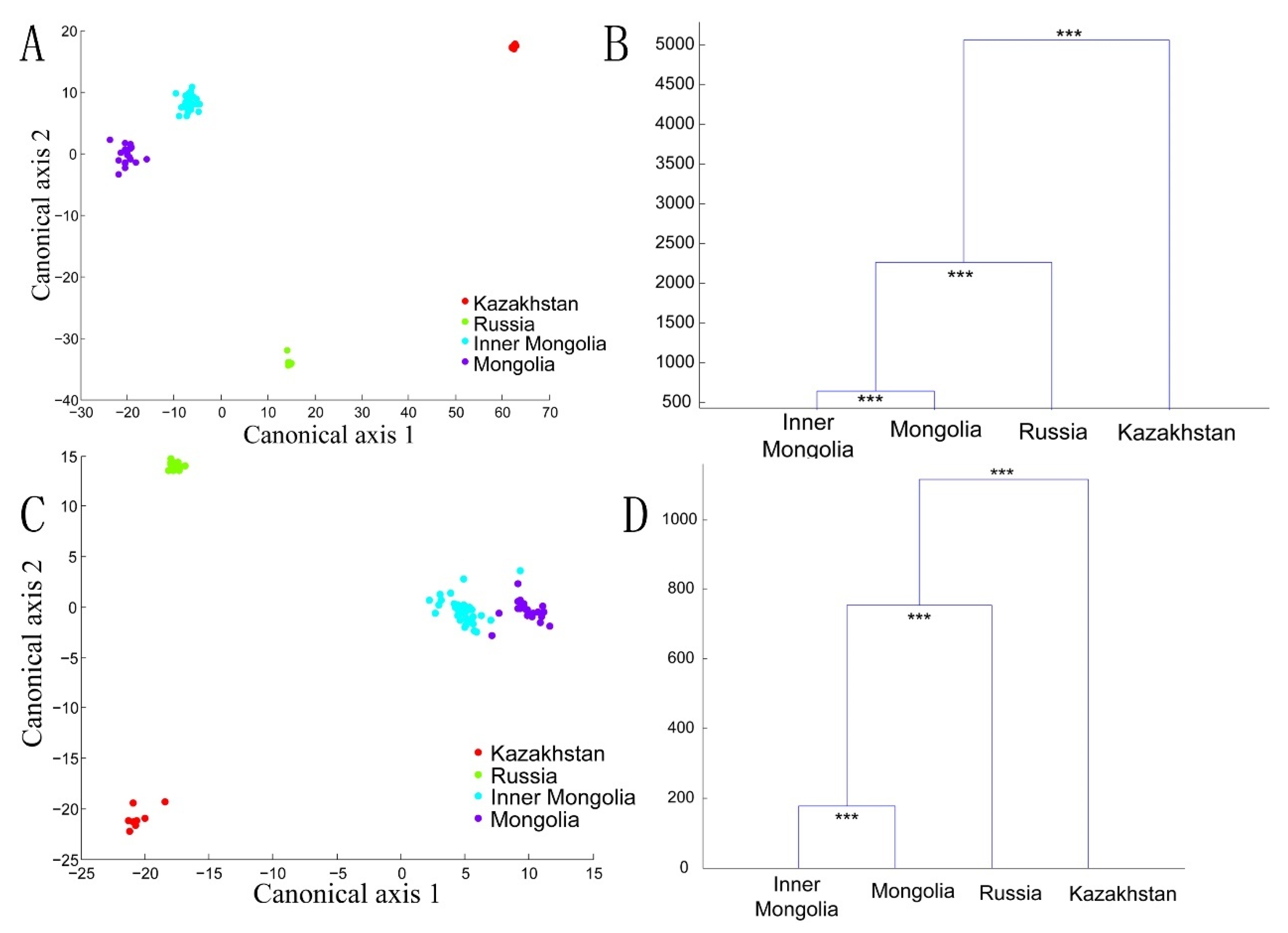
3.3. Bacterial Structures of the Spontaneously Fermented Dairy Products
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
The list of Abbreviations
| Abbreviation | Full name |
| PCR | Polymerase Chain Reaction |
| OTU | Operational Taxonomic Unit |
| RDP | Remote Desktop Protocol |
| L. delbrueckii | Lactobacillus delbrueckii |
| E. xiangfangensis | Enterobacter xiangfangensis |
| L. kefiranofaciens | Lactobacillus kefiranofaciens |
| L. lactis | Lactobacillus lactis |
| L. helveticus | Lactobacillus helveticus |
| Lac. raffinolactis | Lactococcus raffinolactis |
| Lac. lactis | Lactococcus lactis |
| S. salivarius | Streptococcus salivarius |
| S. parauberis | Streptococcus parauberis |
| S. vestibularis | Streptococcus vestibularis |
| A. baumannii | Acinetobacter baumannii |
References
- Akuzawa, R.; Miura, T.; Surono, I.S. Asian fermented milks. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 507–511. [Google Scholar]
- Zhao, B. Nanfang Daily. Available online: http://epaper.southcn.com/nfdaily/html/2017-10/11/content_7673391.htm (accessed on 11 October 2017).
- Fuller, R. Probiotics: An Overview Human Healh: The Contribution of Microorganisms; Gibson, S.A.W., Ed.; Springer: New York, NY, USA, 1992; pp. 63–73. [Google Scholar]
- Hou, Q.; Li, C.; Liu, Y.; Li, W.; Chen, Y.; Siqinbateer; Bao, Y.; Saqila, W.; Zhang, H.; Menghe, B. Koumiss consumption modulates gut microbiota, increases plasma high density cholesterol, decreases immunoglobulin G and albumin. J. Funct. Foods 2019, 52, 469–478. [Google Scholar] [CrossRef]
- Marshall, V.M.E.; Tamime, A.Y. Physiology and biochemistry of fermented milks. In Microbiology and Biochemistry of Cheese and Fermented Milk, 2nd ed.; Law, B.A., Ed.; Blackie Academic & Professional: London, UK, 1997; pp. 153–192. [Google Scholar]
- Oberman, H.; Libudzisz, Z. Fermented milks. In Microbiology of Fermented Foods; Blackie Academic & Professional: London, UK, 1998; pp. 308–350. [Google Scholar]
- Motato, K.E.; Milani, C.; Ventura, M.; Valencia, F.E.; Rúas-Madiedo, P.; Delgado, S. Bacterial diversity of the colombian fermented milk “suero costeño” assessed by culturing and high-throughput sequencing and dgge analysis of 16s rrna gene amplicons. Food Microbiol. 2017, 68, 129. [Google Scholar] [CrossRef]
- Lappa, I.K.; Gantzias, C.; Manolopoulou, E.; Brandt, E.D.; Georgalaki, M. Maldi-tof ms insight into the biodiversity of staka, the artisanal cretan soured cream. Int. Dairy J. 2021, 116, 104969. [Google Scholar] [CrossRef]
- Zhong, Z.; Hou, Q.; Kwok, L.; Zheng, Y.; Sun, Z.; Menghe, B.; Zhang, H. Bacterial microbiota compositions of naturally fermented milk are shaped by both geographic origin and sample type. J. Dairy Sci. 2016, 99, 7832–7841. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Hou, Q.; Hui, W.; Kwok, L.; Zhang, H.; Zhang, W. Assessment of the microbial diversity of Chinese Tianshan tibicos by single molecule, real-time sequencing technology. Food Sci. Biotechnol. 2018, 28, 139–145. [Google Scholar] [CrossRef]
- Cao, J.; Yang, J.; Hou, Q.; Xu, H.; Zheng, Y.; Zhang, H.; Zhang, L. Assessment of bacterial profiles in aged, home-made Sichuan paocai brine with varying titratable acidity by PacBio SMRT sequencing technology. Food Control 2017, 78, 14–23. [Google Scholar] [CrossRef]
- Mosher, J.J.; Bernberg, E.L.; Shevchenko, O.; Kan, J.; Kaplan, L.A. Efficacy of a 3rd generation high-throughput sequencing platform for analyses of 16S rRNA genes from environmental samples. J. Microbiol. 2013, 95, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Erica Sodergren, B.M.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Bandela, A.M.; Cardenas, E.; Garrity, G.M.; Tiedje, J.M. The ribosomal database project (RDP-II): Introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007, 35, D169–D172. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Xu, H.; Zheng, Y.; Xi, X.; Kwok, L.; Sun, Z.; Zhang, H.; Zhang, W. Evaluation of bacterial contamination in raw milk, ultra-high temperature milk and infant formula using single molecule, real-time sequencing technology. J. Dairy Sci. 2015, 98, 8464–8472. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2006, 71, 8228–8235. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Hujer, A.M.; Hujer, K.M.; Decker, B.K.; Rather, P.N.; Bonomo, R.A. Global Challenge of Multidrug-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 3471–3484. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gu, F.; He, J.; Xiao, J.; Chen, Q.; Ruan, H.; He, G. Metagenome analysis of bacterial diversity in Tibetan kefir grains. Eur. Food Res. Technol. 2013, 236, 549–556. [Google Scholar] [CrossRef]
- Parker, M.; Zobrist, S.; Donahue, C.; Edick, C.; Mansen, K.; Nadjari, M.H.Z.; Heerikhuisen, M.; Sybesma, W.; Molenaar, D.; Diallo, A.M.; et al. Naturally Fermented Milk From Northern Senegal: Bacterial Community Composition and Probiotic Enrichment With Lactobacillus rhamnosus. Front. Microbiol. 2018, 9, 2218. [Google Scholar] [CrossRef] [PubMed]
- Araújo, C.; Moraes, M.S.; Costa, L.E.O.; Nascimento, J.S. Short communication: Multidrug-resistant Acinetobacter baumannii-calcoaceticus complex isolated from infant milk formula and utensils in a nursery in Rio de Janeiro, Brazil. J. Dairy Sci. 2015, 98, 2303–2306. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Bai, X.; Li, W.; Gao, X.; Zhang, F.; Sun, Z.; Zhang, H. Design of Primers for Evaluation of Lactic Acid Bacteria Populations in Complex Biological Samples. Front. Microbiol. 2018, 9, 2045. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Hou, X.G.; Sultana, F.; Miura, H.; Ezaki, T. Determination of 16S rRNA Sequences of Streptococcus mitis and Streptococcus gordonii and Phylogenetic Relationships among Members of the Genus Streptococcus. Int. J. Syst. Bacteriol. 1995, 45, 406–408. [Google Scholar] [CrossRef] [PubMed]
- Delorme, C.; Abraham, A.; Renault, P.; Guédon, E. Genomics of Streptococcus salivarius, a major human commensal. Infect. Genet. Evol. 2015, 33, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Sun, Z.; Liu, W.; Xi, X.; Song, Y.; Xu, H.; Lv, Q.; Bao, Q.; Menghe, B.; Sun, T. Multilocus sequence typing of Streptococcus thermophilusfrom naturally fermented dairy foods in China and Mongolia. BMC Microbiol. 2015, 15, 236. [Google Scholar] [CrossRef] [PubMed]
- Delorme, C.; Poyart, C.; Ehrlich, S.D.; Renault, P. Extent of horizontal gene transfer in evolution of Streptococci of the salivarius group. J. Bacteriol. 2007, 189, 1330–1341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of Microorganisms in Global Fermented Foods and Beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef]
- Bao, Q.; Yu, J.; Liu, W.; Qing, M.; Wang, W.; Chen, X.; Wang, F.; Li, M.; Wang, H.; Lv, Q.; et al. Predominant lactic acid bacteria in traditional fermented yak milk products in the Sichuan Province of China. Dairy Sci. Technol. 2012, 92, 309–319. [Google Scholar] [CrossRef]
- Ghosh, T.; Beniwal, A.; Semwal, A.; Navani, N.K. Mechanistic insights into probiotic properties of lactic acid bacteria associated with ethnic fermented dairy products. Front. Microbiol. 2019, 10, 502. [Google Scholar] [CrossRef] [PubMed]
- Gesudu, Q.; Zheng, Y.; Xi, X.; Hou, Q.C.; Xu, H.; Huang, W.; Zhang, H.; Menghe, B.; Liu, W. Investigating bacterial population structure and dynamics in traditional koumiss from Inner Mongolia using single molecule real-time sequencing. J. Dairy Sci. 2016, 99, 7852–7863. [Google Scholar] [CrossRef]
- Yahya, M.A.; Alhaj, O.A.; Al-Khalifah, A.S. Antihypertensive Effect of Fermented Skim Camel (Camelus dromedarius) Milk on Spontaneously Hypertensive Rats. Nutr. Hosp. 2017, 34, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yu, J.; Sun, Z.; Song, Y.; Wang, X.; Wang, H.; Wuren, T.; Zha, M.; Bilige, M.; Zhang, H. Relationships between functional genes in Lactobacillus delbrueckii ssp. bulgaricus isolates and phenotypic characteristics associated with fermentation time and flavor production in yogurt elucidated using multilocus sequence typing. J. Dairy Sci. 2015, 99, 89–103. [Google Scholar] [CrossRef]
- Shiby, V.K.; Mishra, H.N. Fermented Milks and Milk Products as Functional Foods—A Review. Crit. Rev. Food Sci. Nutr. 2013, 53, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Li, R.; Liu, F.; Xu, H.; Li, B.; Yuan, Y.; Saris, P.E.J.; Qiao, M. Mu insertion in feuD triggers the increase in nisin immunity in Lactococcus lactis subsp. lactis N8. J. Appl. Microbiol. 2016, 120, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Sayeed, M.A.; Hossen, A.; Saha, R.; Jakaria, M. Antimicrobial activities of isolated probiotics and their metabolites against some pathogenic microorganisms. IIUC Stud. 2017, 14, 21–28. [Google Scholar] [CrossRef]
- Tamime, Y. Fermented milks: A historical food with modern applications—A review. Eur. J. Clin. Nutr. 2002, 56, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Stanton, C.; Ross, R.P.; Fitzgerald, G.F.; Van Sinderen, D. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 2005, 16, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.M.; Crispie, F.; Daari, K.; O’Sullivan, O.; Martin, J.C.; Arthur, C.T.; Claesson, M.J.; Scott, K.P.; Cotter, P.D. Strain-Level Metagenomic Analysis of the Fermented Dairy Beverage Nunu Highlights Potential Food Safety Risks. Appl. Environ. Microbiol. 2017, 83, e01144-17. [Google Scholar] [CrossRef] [PubMed]
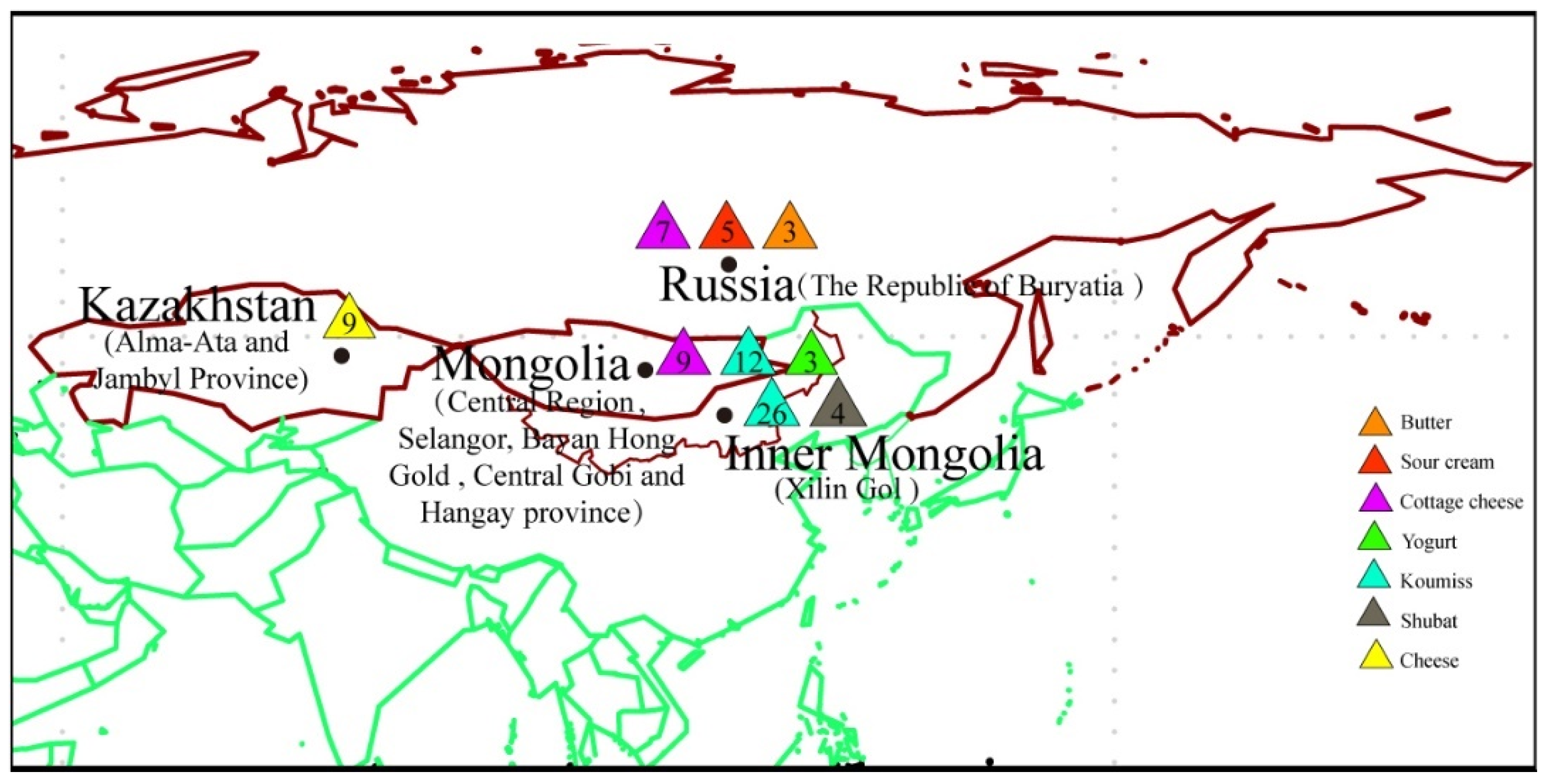
| Types of Sample | Sample ID | Number of Samples | Sampling Location |
|---|---|---|---|
| Jiaoke | J1-J14 | 14 | Inner Mongolia |
| Cheese | C1-C9 | 9 | Kazakhstan |
| Cottage cheese | Z10-Z16 | 7 | Russia |
| Butter | B1-B3 | 3 | Russia |
| Sour cream | S1-S5 | 5 | Russia |
| Koumiss | K9-K22 | 14 | Inner Mongolia |
| Koumiss | K27-K38 | 12 | Inner Mongolia |
| Koumiss | K23-K26 | 4 | Mongolia |
| Koumiss | K1-K8 | 8 | Mongolia |
| Shubat | T1-T4 | 4 | Inner Mongolia |
| Yogurt | Y1-Y3 | 3 | Mongolia |
| Cottage cheese | Z1-Z9 | 9 | Mongolia |
| Item | Jiaoke | Cheeses | Koumisses | Cottage Cheeses | Yogurts | Butter | Sour Cream | Shubat | p-Value 1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Index | ||||||||||
| Chao1 | 507.55 ± 200.59 | 758.50 ± 389.64 | 542.44 ± 318.33 | 661.16 ± 274.07 | 392.04 ± 99.49 | 351.97 ± 244.08 | 608.39 ± 375.97 | 333.53 ± 138.22 | 0.18 | |
| Observed species | 188.91 ± 52.51 | 276.13 ± 104.95 | 204.98 ± 97.15 | 217.75 ± 55.18 | 160.26 ± 12.01 | 124.96 ± 69.84 | 189.86 ± 68.03 | 127.83 ± 45.33 | 0.03 | |
| Shannon | 4.33 ± 0.61 | 5.38 ± 1.03 | 4.11 ± 1.30 | 4.43 ± 0.73 | 4.35 ± 0.39 | 3.13 ± 1.50 | 3.99 ± 0.89 | 2.96 ± 0.55 | 0.01 | |
| Simpson | 0.80 ± 0.07 | 0.90 ± 0.07 | 0.77 ± 0.12 | 0.82 ± 0.07 | 0.87 ± 0.05 | 0.64 ± 0.26 | 0.75 ± 0.16 | 0.67 ± 0.09 | 0.01 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Peng, C.; Kwok, L.-y.; Zhang, H. The Bacterial Diversity of Spontaneously Fermented Dairy Products Collected in Northeast Asia. Foods 2021, 10, 2321. https://doi.org/10.3390/foods10102321
Yu Z, Peng C, Kwok L-y, Zhang H. The Bacterial Diversity of Spontaneously Fermented Dairy Products Collected in Northeast Asia. Foods. 2021; 10(10):2321. https://doi.org/10.3390/foods10102321
Chicago/Turabian StyleYu, Zhongjie, Chuantao Peng, Lai-yu Kwok, and Heping Zhang. 2021. "The Bacterial Diversity of Spontaneously Fermented Dairy Products Collected in Northeast Asia" Foods 10, no. 10: 2321. https://doi.org/10.3390/foods10102321
APA StyleYu, Z., Peng, C., Kwok, L.-y., & Zhang, H. (2021). The Bacterial Diversity of Spontaneously Fermented Dairy Products Collected in Northeast Asia. Foods, 10(10), 2321. https://doi.org/10.3390/foods10102321





