Metabolomics Approaches for the Comprehensive Evaluation of Fermented Foods: A Review
Abstract
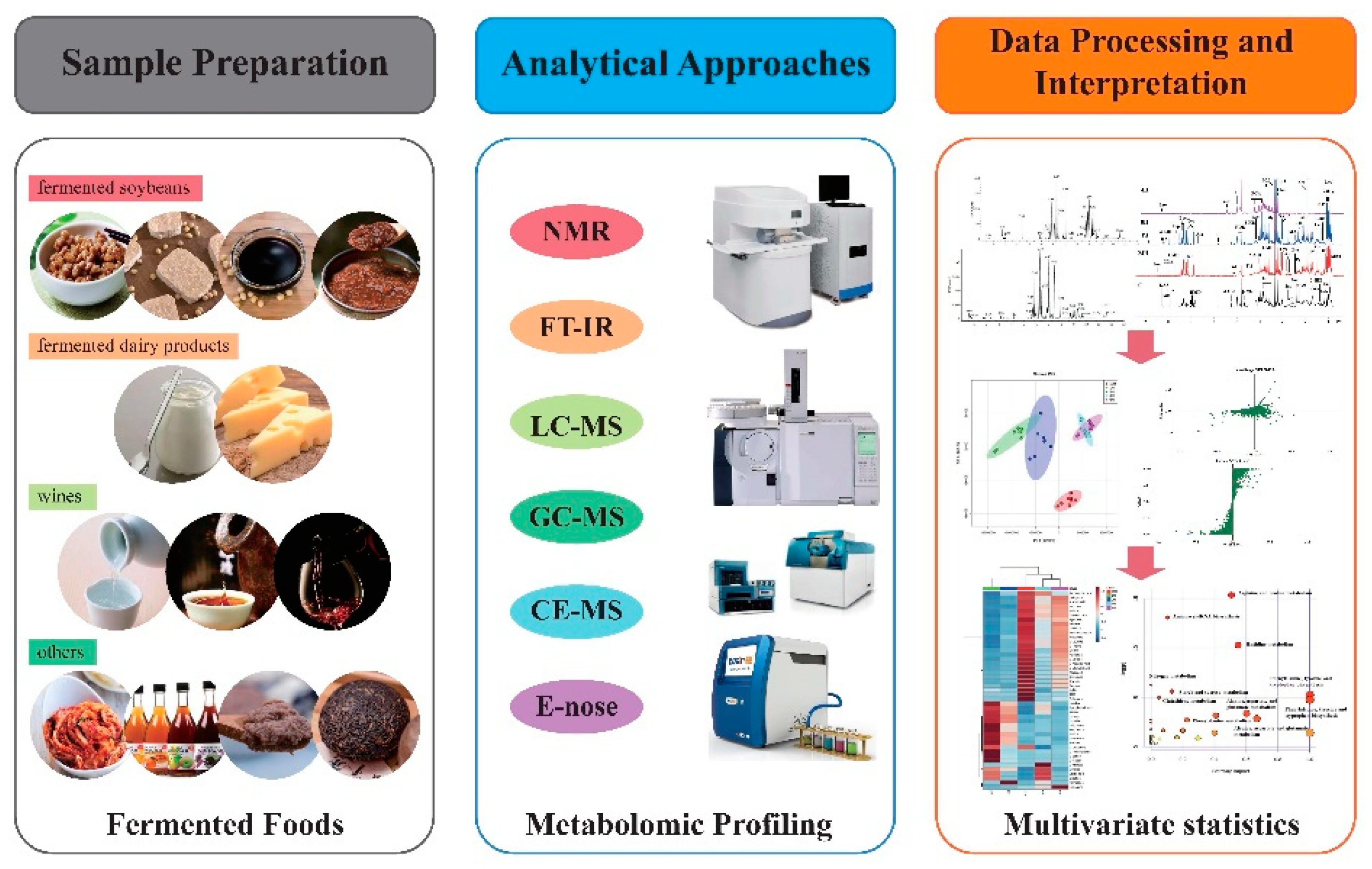
:1. Introduction
2. Methodology
3. Metabolomics Workflow
3.1. Metabolomic Approaches Based on Different Separations and Instrumentation
3.1.1. NMR-Based Metabolomics
3.1.2. FT-IR-Based Metabolomics
3.1.3. GC-MS/LC-MS-Based Metabolomics
3.1.4. CE-MS-Based Metabolomics
3.1.5. Electronic Nose-Based Metabolomics
3.2. Metabolomic Analyses Based on Data Interpretation and Multivariate Statistics
| Biobanks | Role | References |
|---|---|---|
| MestreNova | Data processing prediction, publication, verification; data storage and retrieval | [97] |
| Progenesis QI | Data processing and normalization; qualitative, quantitative and identification of small molecules with significant changes | [98] |
| SIMCA | Multivariate statistical analysis; pattern recognition of PCA, PLS-DA, OPLS-DA | [97,99,100,101] |
| RStudio | Multivariate tool; heatmap of metabolites and their concentration changes | [97] |
| HMDB | Physicochemical and biological properties; biomarker discovery; metabolic pathway information | [97,98,99,100,101] |
| METLIN | Metabolite identification; metabolite structure; links to other databases | [98,100,101] |
| PubChem | Biological properties of organic small molecule; metabolite structure; links to other databases | [100] |
| MetaboAnalyst | Data analysis, visualization, and functional annotation; multivariate analysis; metabolite significance; pathway identification | [98,99,101] |
| KEGG | Metabolic pathway; metabolite interactions; delivery of gene to metabolite function | [98,99,101] |
| Mev | Hierarchical cluster analysis; heatmap of metabolites and their concentration changes | [100] |
| Cytoscape | Interaction network visualization: correlation analysis linked to gene, protein, and metabolite expression | [99] |
4. Applications of Fermented Foods
4.1. Flavor
| Fermented Foods | Microorganisms | Metabolomic Analysis | References | |
|---|---|---|---|---|
| Techniques | Compounds/Properties Analyzed | |||
| Dajiang | Yeast, Aspergillus, Mucor, Rhizopus, Lactobacillus, Tetragenococcus | HS-SPME/GC-MS | Alcohols, esters, phenolic acids, aldehydes, ketones | [78] |
| Red sufu | Monascus purpureus, Aspergilus oryzae, Actinomucor elegans | GC-MS, GC-MS-O, E-nose | Amino acids, organic acids | [119] |
| Natto | Bacillus subtilis | GC-MS; NMR | Amino acids, organic acids, pyrazines; ammonia | [120,121,122] |
| Cheese | Lactic acid bacteria | HS-SPME/GC-MS, FT-IR, E-nose | Lactose, lactate and citrate, amino acids, fatty acids | [113] |
| Huangjiu | Yeast | GC/GC-MS, GC-O | Esters, linalool, neroidol, geranyl acetone, 2-pentyl-furan, methanethiol | [112,115] |
| Baijiu | Yeast, Lactobacillus, Acetobacter | GC×GC-TOF/MS | Aromatic compounds, pyrazines | [116] |
| Red wine | Yeast | HS-SPME-Arrow-GC-MS/MS | Piperitone, mintlactone, menthyl acetate, neomenthyl acetate | [79] |
| Vinegar | Acetobacter, Lactobacillus | HS-SPME/GC-MS | Ethyl acetate, phenylethyl alcohol, acetoin, acetic acid | [124] |
| Pu-erh tea | Monascus purpureus, Bacillus, Rasamsonia, Lichtheimia, Debaryomyces | HS-SPME/GC-MS | β-damascenone, methoxybenzene, 2,4-nonadienal, terpinene, linalool | [114,117] |
| Siniperca chuatsi | Psychrilyobacter, Fusobacterium, Vibrio | HS-SPME/GC-MS | Alcohols, hydrocarbons, nitrogen compounds | [123] |
| Shrimp paste | Salimicrobium, Lentibacillus, Lactobacillus, Tetragenococcus | HS-SPME/GC-MS | Alcohols, aldehydes, nitrogen compounds | [125] |
4.2. Nutrition and Function
| Fermented Foods | Microorganisms | Metabolomic Analysis | References | |
|---|---|---|---|---|
| Techniques | Compounds | |||
| Meju | Bacillus sp., Mucor sp., Aspergillus sp. | UPLC-Q/TOF-MS | Small peptides, amino acids, GABA | [132] |
| Doenjang | Penicillium glabrum, Aspergillus oryzae | GC-TOF-MS, UPLC-Q/TOF-MS | Amino acids, organic acids, sugars and sugar alcohols, isoflavones | [133,134,135] |
| Cheonggukujang (or miso, natto) | Bacillus subtilis, Mucor sp., Bacillus sp., Aspergillus sp.; E. faecium | UPLC-Q/TOF-MS | Phenolic compounds, peptides, GABA | [131,136] |
| Cheese | Lactic acid bacteria | NMR | Lactose, uridine diphosphate-hexose, amino acids, organic acids, | [137] |
| Yogurt | Lactic acid bacteria | UPLC-Triple/TOF-MS | Lipids, lipid-like molecules, small peptides, amino acids, GABA | [98,139] |
| Wines | Yeasts, Lactobacillus, Acetobacter | NMR, FT-IR; GC/GC-TOF/MS; HPLC-MS; | Polyphenols, amino acids, ethanol, resveratrol, stilbenes | [49,116,146,147] |
| Cabbage vinegar | Lactobacillus, Acetobacter | NMR, GC-MS | Organic acids, alcohols, sulfides (dimethyl sulfide, dimethyl disulfide, and dimethyl trisulfide) | [148] |
| Pu-erh tea | Aspergillus pallidofulvus, Aspergillus sesamicola, Penicillium manginii | UHPLC-Q/TOF-MS | Phenolic compounds, amino acids | [105] |
| Fermented fish sauce | Firmicutes, Proteobacteria, Fusobacteria | UHPLC-Q/TOF-MS | Small peptides, amino acids | [23] |
| Pickle nozawana | Latilactobacillus curvatus, Levilactobacillus brevis | NMR, GC-MS | Organic acids, GABA, choline, 2,3-butanedione, acetoin, ethyl acetate | [149] |
4.3. Safety
| Fermented Foods | Microorganisms | Metabolomic Analysis | References | |
|---|---|---|---|---|
| Techniques | Compounds/Components Analyzed | |||
| Doenjang | Bacillus subtilis, Rhizopus, Mucor, Aspergillus sp. | GC-MS | Soyasaponins | [134] |
| Cheonggukjang | Bacillus sp. | GC-TOF-MS, CE-TOF-MS | Soyasaponins | [21] |
| Koji | Aspergillus oryzae, Bacillus amyloliquefaciens | GC-TOF-MS, UHPLC-MS/MS | Soyasaponins | [151] |
| Vinegar | Bacillus subtilis, Rizhopus, Mucor, Aspergillus sp. | GC-MS | Benzoic acid, sorbic acid, dehydroacetic acid, ethyl paraben | [153] |
| Fermented fish sauce | Firmicutes, Proteobacteria, Fusobacteria | UHPLC-Q/TOF-MS | Trimethylamine N-oxide, putrescine, cadaverine | [23] |
| Shrimp paste, Pickles | Lentibacillus, Lactobacillus, Latilactobacillus, Tetragenococcus | GC-FID | Benzoic acid, sorbic acid, propionic acid | [154] |
| Cheese | Lactic acid bacteria | GC/LC-MS, NMR, FI-TR | Pathogenic bacteria and its metabolites | [155] |
5. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Powell, I.B. Food fermentations: Microorgan-isms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Anyogu, A.; Olukorede, A.; Anumudu, C.; Onyeaka, H.; Areo, E.; Adewale, O.; Odimba, J.N.; Nwaiwu, O. Microorganisms and food safety risks associated with indigenous fermented foods from Africa. Food Control. 2021, 129, 108227. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.; Foligne, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Terefe, N.S.; Augustin, M.A. Fermentation for tailoring the technological and health related functionality of food products. Crit. Rev. Food Sci. Nutr. 2019, 60, 2887–2913. [Google Scholar] [CrossRef] [PubMed]
- Gille, D.; Schmid, A.; Walther, B.; Vergères, G. Fermented Food and Non-Communicable Chronic Diseases: A Review. Nutrients 2018, 10, 448. [Google Scholar] [CrossRef] [Green Version]
- Jayachandran, M.; Xu, B. An insight into the health benefits of fermented soy products. Food Chem. 2018, 271, 362–371. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, J.; Liu, E.; Yang, M.; Chen, S.; Hu, F.; Ma, H.; Liu, Z.; Yu, X. Enhancing the taste of raw soy sauce using low intensity ultrasound treatment during moromi fermentation. Food Chem. 2019, 298, 124928. [Google Scholar] [CrossRef]
- Dong, Z.Y.; Liu, Y.; Xu, M.; Zhang, T.H.; Ren, H.; Liu, W.; Li, M.Y. Accelerated aging of grape pomace vinegar by using additives combined with physical methods. J. Food Process. Eng. 2020, 43, e13398. [Google Scholar] [CrossRef]
- Nwaiwu, O.; Itumoh, M. Chemical Contaminants Associated with Palm Wine from Nigeria Are Potential Food Safety Hazards. Beverages 2017, 3, 16. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Adebiyi, J.A.; Gbashi, F.; Phoku, J.Z.; Kayitesi, E. Fermented pulse-based food products in developing nations as functional foods and ingredients. In Functional Food-Improve Health through Adequate Food; Hueda, M.C., Ed.; IntechOpen: London, UK, 2017; Chapter 5; pp. 77–109. [Google Scholar] [CrossRef] [Green Version]
- Campbell-Platt, G. Fermented foods-a world perspective. Food Res Int. 1994, 27, 253–257. [Google Scholar] [CrossRef]
- Gupta, S.; Abu-Ghannam, N. Probiotic Fermentation of Plant Based Products: Possibilities and Opportunities. Crit. Rev. Food Sci. Nutr. 2012, 52, 183–199. [Google Scholar] [CrossRef]
- Pereira, G.; Neto, D.P.D.C.; Junqueira, A.C.D.O.; Karp, S.G.; Letti, L.A.J.; Júnior, A.I.M.; Soccol, C.R. A Review of Selection Criteria for Starter Culture Development in the Food Fermentation Industry. Food Rev. Int. 2019, 36, 135–167. [Google Scholar] [CrossRef]
- Dorđevi’c, T.M.; Šiler-Marinković, S.S.; Dimitrijević-Branković, S.I. Effect of fermentation on antioxidant properties of some cereals and pseudo cereals. Food Chem. 2010, 119, 957–963. [Google Scholar] [CrossRef]
- Saharan, P.; Sadh, P.K.; Duhan, J.S. Comparative assessment of effect of fermentation on phenolics, flavanoids and free radical scavenging activity of commonly used cereals. Biocatal. Agric. Biotechnol. 2017, 12, 236–240. [Google Scholar] [CrossRef]
- Thirunathan, P.; Manickavasagan, A. Processing methods for reducing alpha-galactosides in pulses. Crit. Rev. Food Sci. Nutr. 2018, 59, 3334–3348. [Google Scholar] [CrossRef] [PubMed]
- Gänzle, M. Food fermentations for improved digestibility of plant foods-an essential ex-situ digestion step in agricultural societies? Curr. Opin. Food Sci. 2020, 32, 124–132. [Google Scholar] [CrossRef]
- Xiang, H.; Sun-Waterhouse, D.; Waterhouse, G.; Cui, C.; Ruan, Z. Fermentation-enabled wellness foods: A fresh perspective. Food Sci. Hum. Wellness 2019, 8, 203–243. [Google Scholar] [CrossRef]
- González-Peña, D.; Brennan, L. Recent Advances in the Application of Metabolomics for Nutrition and Health. Annu. Rev. Food Sci. Technol. 2019, 10, 479–519. [Google Scholar] [CrossRef]
- Cook, P.W.; Nightingale, K.K. Use of omics methods for the advancement of food quality and food safety. Anim. Front. 2018, 8, 33–41. [Google Scholar] [CrossRef]
- Kim, J.Y.; Choi, J.N.; John, K.M.; Kusano, M.; Oikawa, A.; Saito, K.; Lee, C.H. GC-TOF-MS- and CE-TOF-MS-based metabolic pro-filing of cheonggukjang (fastfermented bean paste) during fermentation and its correlation with metabolic pathways. J. Agr Food Chem. 2012, 60, 9746–9753. [Google Scholar] [CrossRef]
- Lee, D.E.; Shin, G.R.; Lee, S.; Jang, E.S.; Shin, H.W.; Moon, B.S.; Lee, C.H. Metabolomics reveal that amino acids are the main con-tributors to antioxidant activity in wheat and rice gochujangs (korean fermented red pepper paste). Food Res. Int. 2016, 87, 10–17. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Li, L.; Yang, X.; Chen, S.; Wu, Y.; Zhao, Y.; Wang, J.; Wei, Y.; Yang, D. Application of UHPLC-Q/TOF-MS-based metabolomics in the evaluation of metabolites and taste quality of Chinese fish sauce (Yu-lu) during fermentation. Food Chem. 2019, 296, 132–141. [Google Scholar] [CrossRef]
- He, S.; Wang, Y.; Xie, J.; Gao, H.; Li, X.; Huang, Z. 1H NMR-based metabolomic study of the effects of flavonoids on citrinin production by Monascus. Food Res. Int. 2020, 137, 109532. [Google Scholar] [CrossRef]
- Zhao, G.; Zhao, W.; Han, L.; Ding, J.; Chang, Y. Metabolomics analysis of sea cucumber (Apostichopus japonicus) in different geo-graphical origins using UPLC-Q-TOF/MS. Food Chem. 2020, 333, 127453. [Google Scholar] [CrossRef]
- Fuhrer, T.; Zamboni, N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015, 31, 73–78. [Google Scholar] [CrossRef]
- González-Peña, D.; Dudzik, D.; Colina-Coca, C.; Ancos, B.D.; García, A.; Barbas, C.; Sánchez-Moreno, C. Evaluation of onion as a functional ingredient in the prevention of metabolic impairments associated to diet-induced hypercholesterolaemia using a multiplat-form approach based on LC-MS, CE-MS and GC-MS. J. Funct Foods 2015, 19, 363–375. [Google Scholar] [CrossRef]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Rudaz, S. Implementation of liquid chromatography-high resolution mass spec-trometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim Acta. 2020, 1105, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Mapelli, V.; Olsson, L.; Nielsen, J. Metabolic footprinting in microbiology: Methods and applications in functional genomics and biotechnology. Trends Biotechnol. 2008, 26, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Milburn, M.V.; Ryals, J.A.; Lonergan, S.C.; Mitchell, M.W.; Wulff, J.E.; Alexander, D.C.; Evans, A.M.; Bridgewater, B.; Miller, L.; et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc. Natl. Acad. Sci. USA 2015, 112, E4901–E4910. [Google Scholar] [CrossRef] [Green Version]
- Medina, S.; Dominguez-Perles, R.; Gil, J.; Ferreres, F.; Gil-Izquierdo, A. Metabolomics and the Diagnosis of Human Diseases -A Guide to the Markers and Pathophysiological Pathways Affected. Curr. Med. Chem. 2014, 21, 823–848. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.; O’Gorman, A.; Brennan, L. Metabolomics as a tool in nutritional research. Curr. Opin. Lipidol. 2015, 26, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef]
- Uawisetwathana, U.; Karoonuthaisiri, N. Metabolomics for rice quality and traceability: Feasibility and future aspects. Curr. Opin. Food Sci. 2019, 28, 58–66. [Google Scholar] [CrossRef]
- Hu, C.; Xu, G. Mass-spectrometry-based metabolomics analysis for foodomics. TrAC Trends Anal. Chem. 2013, 52, 36–46. [Google Scholar] [CrossRef]
- Mozzi, F.; Ortiz, M.E.; Bleckwedel, J.; De Vuyst, L.; Pescuma, M. Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Res. Int. 2013, 54, 1152–1161. [Google Scholar] [CrossRef]
- Wishart, D.S. NMR metabolomics: A look ahead. J. Magn. Reson. 2019, 306, 155–161. [Google Scholar] [CrossRef]
- Larive, C.; Barding, G.A.; Dinges, M. NMR Spectroscopy for Metabolomics and Metabolic Profiling. Anal. Chem. 2014, 87, 133–146. [Google Scholar] [CrossRef]
- Ha, D.; Paulsen, J.; Sun, N.; Song, Y.Q.; Ham, D. Scalable NMR spectroscopy with semico nductor chips. Proc. Natl. Acad. Sci. USA 2014, 111, 11955–11960. [Google Scholar] [CrossRef] [Green Version]
- Sundekilde, U.K.; Larsen, L.B.; Bertram, H.C. NMR-Based Milk Metabolomics. Metabolites 2013, 3, 204–222. [Google Scholar] [CrossRef]
- Peterson, A.L.; Waterhouse, A.L. 1H NMR: A novel approach to determining the thermodynamic properties of acetaldehyde con-densation reactions with glycerol, (+)-catechin, and glutathione in model wine. J. Agric. Food Chem. 2016, 64, 6869–6878. [Google Scholar] [CrossRef] [PubMed]
- Noh, M.F.M.; Gunasegavan, R.D.-N.; Khalid, N.M.; Balasubramaniam, V.; Mustar, S.; Rashed, A.A. Molecules Recent techniques in nutrient analysis for food composition database. Molecules 2020, 25, 4567. [Google Scholar] [CrossRef]
- Singh, D.; Lee, S.; Lee, C.H. Metabolomics for empirical delineation of the traditional Korean fermented foods and beverages. Trends Food Sci. Technol. 2017, 61, 103–115. [Google Scholar] [CrossRef]
- Rocchetti, G.; O’Callaghan, T.F. Application of metabolomics to assess milk quality and traceability. Curr. Opin. Food Sci. 2021, 40, 168–178. [Google Scholar] [CrossRef]
- Trimigno, A.; Marincola, F.C.; Dellarosa, N.; Picone, G.; Laghi, L. Definition of food quality by NMR-based foodomics. Curr. Opin. Food Sci. 2015, 4, 99–104. [Google Scholar] [CrossRef]
- Rochfort, S. Metabolomics Reviewed: A New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef]
- Yang, Z. Online hyphenated liquid chromatography–nuclear magnetic resonance spectroscopy–mass spectrometry for drug metabolite and nature product analysis. J. Pharm. Biomed. Anal. 2006, 40, 516–527. [Google Scholar] [CrossRef]
- Tabago, M.K.A.G.; Calingacion, M.N.; Garcia, J. Recent advances in NMR-based metabolomics of alcoholic beverages. Food Chem. Mol. Sci. 2020, 2, 100009. [Google Scholar] [CrossRef]
- Li, Y.; Teng, Z.; Parkin, K.L.; Wang, Q.; Zhang, Q.; Luo, W.; Ma, D.; Zhao, M. Identification of bioactive metabolites dihydrocana-densolide, kojic acid, and vanillic acid in soy sauce using GC-MS, NMR spectroscopy, and single-crystal x-ray diffraction. J. Agric. Food Chem. 2014, 62, 8392–8401. [Google Scholar] [CrossRef]
- Lee, S.H.; Jung, J.Y.; Jeon, C.O. Bacterial community dynamics and metabolite changes in myeolchi-aekjeot, a Korean traditional fermented fish sauce, during fermentation. Int. J. Food Microbiol. 2015, 203, 15–22. [Google Scholar] [CrossRef]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Kasprzyk, I.; Depciuch, J.; Grabek-Lejko, D.; Parlinska-Wojtan, M. FTIR-ATR spectroscopy of pollen and honey as a tool for unifloral honey authentication. The case study of rape honey. Food Control. 2018, 84, 33–40. [Google Scholar] [CrossRef]
- Wang, F.; Shao, C.; Chen, Q.; Meng, T.; Li, C. Application on sensory prediction of Chinese moutai-flavor liquor based on ATR-FTIR. E3S Web Conf. 2019, 79, 03001. [Google Scholar] [CrossRef]
- Puxeu, M.; Andorra, I.; De Lamo-Castellví, S.; Ferrer-Gallego, R. Determination of Nutrient Supplementation by Means of ATR-FTIR Spectroscopy during Wine Fermentation. Fermentation 2019, 5, 58. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Hernandez, C.; Salvo-Comino, C.; Martin-Pedrosa, F.; Garcia-Cabezon, C.; Rodriguez-Mendez, M. Analysis of red wines using an electronic tongue and infrared spectroscopy. Correlations with phenolic content and color parameters. LWT Food Sci. Technol. 2019, 118, 108785. [Google Scholar] [CrossRef]
- Krähmer, A.; Böttcher, C.; Gudi, G.; Stürtz, M.; Schulz, H. Application of ATR-FTIR spectroscopy for profiling of non-structural carbohydrates in onion (Allium cepa L.) bulbs. Food Chem. 2021, 360, 129978. [Google Scholar] [CrossRef] [PubMed]
- Anjos, O.; Santos, A.J.A.; Estevinho, L.M.; Caldeira, I. FTIR-ATR spectroscopy applied to quality control of grape-derived spirits. Food Chem. 2016, 205, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozulku, G.; Yildirim, R.M.; Toker, O.S.; Karasu, S.; Durak, M.Z. Rapid detection of adulteration of cold pressed sesame oil adultered with hazelnut, canola, and sunflower oils using ATR-FTIR spectroscopy combined with chemometric. Food Control. 2017, 82, 212–216. [Google Scholar] [CrossRef]
- Galvin-King, P.; Haughey, S.A.; Elliott, C.T. Garlic adulteration detection using NIR and FTIR spectroscopy and chemometrics. J. Food Compos. Anal. 2020, 96, 103757. [Google Scholar] [CrossRef]
- Labaky, P.; Dahdouh, L.; Ricci, J.; Wisniewski, C.; Pallet, D.; Louka, N.; Grosmaire, L. Impact of ripening on the physical properties of mango purees and application of simultaneous rheometry and in situ FTIR spectroscopy for rapid identification of biochemical and rheological changes. J. Food Eng. 2021, 300, 110507. [Google Scholar] [CrossRef]
- Kumar, K.; Giehl, A.; Patz, C.-D. Chemometric assisted Fourier Transform Infrared (FTIR) Spectroscopic analysis of fruit wine samples: Optimizing the initialization and convergence criteria in the non-negative factor analysis algorithm for developing a robust classification model. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 209, 22–31. [Google Scholar] [CrossRef]
- Li, Z.; Wang, P.-P.; Huang, C.-C.; Shang, H.; Pan, S.-Y.; Li, X.-J. Application of Vis/NIR Spectroscopy for Chinese Liquor Discrimination. Food Anal. Methods 2013, 7, 1337–1344. [Google Scholar] [CrossRef]
- Wang, L.; Sun, D.W.; Pu, H.; Cheng, J.H. Quality analysis, classification, and authentication of liquid foods by near-infrared spectroscopy: A review of recent research developments. Crit. Rev. Food Sci. 2017, 57, 1524–1538. [Google Scholar] [CrossRef]
- Brunius, C.; Shi, L.; Landberg, R. Large-scale untargeted LC-MS metabolomics data correction using between-batch feature alignment and cluster-based within-batch signal intensity drift correction. Metabolomics 2016, 12, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2015, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Goodacre, R. An introduction to liquid chromatography-mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem. Anal. 2010, 21, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Mihailova, A.; Kelly, S.D.; Chevallier, O.P.; Elliott, C.T.; Maestroni, B.M.; Cannavan, A. High-resolution mass spectrometry-based metabolomics for the discrimination between organic and conventional crops: A review. Trends Food Sci. Technol. 2021, 110, 142–154. [Google Scholar] [CrossRef]
- Song, H.; Liu, J. GC-O-MS technique and its applications in food flavor analysis. Food Res. Int. 2018, 114, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Dervishi, E.; Zhang, G.; Dunn, S.M.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. GC–MS Metabolomics Identifies Metabolite Alterations That Precede Subclinical Mastitis in the Blood of Transition Dairy Cows. J. Proteome Res. 2016, 16, 433–446. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography–Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Moros, G.; Chatziioannou, A.C.; Gika, H.G.; Raikos, N.; Theodoridis, G. Investigation of the derivatization conditions for GC–MS metabolomics of biological samples. Bioanalysis 2017, 9, 53–65. [Google Scholar] [CrossRef]
- Vinaixa, M.; Schymanski, E.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Constantinou, M.; Louca-Christodoulou, D.; Agapiou, A. Method validation for the determination of 314 pesticide residues using tandem MS systems (GC–MS/MS and LC-MS/MS) in raisins: Focus on risk exposure assessment and respective processing factors in real samples (a pilot survey). Food Chem. 2021, 360, 129964. [Google Scholar] [CrossRef]
- Li, W.; Chen, Y.P.; Blank, I.; Li, F.; Liu, Y. GC×GC-TOF-MS and GC-IMS based volatile profile characterization of the chinese dry-cured hams from different regions. Food Res. Int. 2021, 142, 110222. [Google Scholar] [CrossRef] [PubMed]
- Magagna, F.; Cordero, C.; Cagliero, C.; Liberto, E.; Rubiolo, P.; Sgorbini, B.; Bicchi, C. Black tea volatiles fingerprinting by compre-hensive two-dimensional gas chromatography-Mass spectrometry combined with high concentration capacity sample preparation techniques: Toward a fully automated sensomic assessment. Food Chem. 2017, 225, 276–287. [Google Scholar] [CrossRef]
- Kuś, P.M.; Rola, R. LC-QQQ-MS/MS methodology for determination of purine and pyrimidine derivatives in unifloral honeys and application of chemometrics for their classification. Food Chem. 2021, 348, 129076. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Li, M.; Zhao, Y.; Zhang, Y.; Mu, D.; Hu, X.; You, S.; Wu, J.; Wu, R. Metatranscriptome-based investigation of flavor-producing core microbiota in different fermentation stages of dajiang, a traditional fermented soybean paste of Northeast China. Food Chem. 2020, 343, 128509. [Google Scholar] [CrossRef]
- Lisanti, M.T.; Laboyrie, J.; Marchand-Marion, S.; de Revel, G.; Moio, L.; Riquier, L.; Franc, C. Minty aroma compounds in red wine: Development of a novel automated HS-SPME-arrow and gas chromatography-tandem mass spectrometry quantification method. Food Chem. 2021, 361, 130029. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; Somsen, G.W.; De Jong, G.J. CE-MS for metabolomics: Developments and applications in the period 2012–2014. Electrophoresis 2014, 36, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Drouin, N.; Pezzatti, J.; Gagnebin, Y.; González-Ruiz, V.; Schappler, J.; Rudaz, S. Effective mobility as a robust criterion for compound annotation and identification in metabolomics: Toward a mobility-based library. Anal. Chim. Acta 2018, 1032, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Hatano, N.; Nishiumi, S.; Irino, Y.; Izumi, Y.; Takenawa, T.; Azuma, T. Diagnosis of gastroenterological diseases by metabolome analysis using gas chromatography–mass spectrometry. J. Gastroenterol. 2011, 47, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Miggiels, P.; Wouters, B.; van Westen, G.J.; Dubbelman, A.-C.; Hankemeier, T. Novel technologies for metabolomics: More for less. TrAC Trends Anal. Chem. 2018, 120, 115323. [Google Scholar] [CrossRef]
- Montona, M.R.N.; Soga, T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J. Chromatogr. A. 2008, 1168, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ramautar, R. CE-MS for metabolomics: Developments and applications in the period 2018–2020. Electrophoresis 2020, 42, 381–401. [Google Scholar] [CrossRef]
- Wilson, A.D. Advances in Electronic-Nose Technologies for the Detection of Volatile Biomarker Metabolites in the Human Breath. Metabolites 2015, 5, 140–163. [Google Scholar] [CrossRef]
- Rocchi, R.; Mascini, M.; Faberi, A.; Sergi, M.; Compagnone, D.; Di Martino, V.; Carradori, S.; Pittia, P. Comparison of IRMS, GC-MS and E-Nose data for the discrimination of saffron samples with different origin, process and age. Food Control. 2019, 106, 106736. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhastia, M.; Apetreib, C.; Lozanoc, J.; Anyogu, A. Potential use of electronic noses, electronic tongues and biosensors as multisensor systems for spoilage examination in foods. Trends. Food Sci. Technol. 2018, 80, 71–92. [Google Scholar] [CrossRef]
- Tan, J.; Xu, J. Applications of electronic nose (e-nose) and electronic tongue (e-tongue) in food quality-related properties determination: A review. Artif. Intell. Agric. 2020, 4, 104–115. [Google Scholar] [CrossRef]
- Xu, S.; Zhou, Z.; Tian, L.; Lu, H.; Luo, X.; Lan, Y. Study of the similarity and recognition between volatiles of brown rice plant hoppers and rice stem based on the electronic nose. Comput. Electron. Agric. 2018, 152, 19–25. [Google Scholar] [CrossRef]
- Yang, C.; Ding, W.; Ma, L.; Jia, R. Discrimination and characterization of different intensities of goaty flavor in goat milk by means of an electronic nose. J. Dairy Sci. 2015, 98, 55–67. [Google Scholar] [CrossRef]
- Wasilewski, T.; Migon, D.; Gebicki, J.; Kamysz, W. Critical review of electronic nose and tongue instruments prospects in pharma-ceutical analysis. Anal. Chim Acta. 2019, 1077, 14–29. [Google Scholar] [CrossRef]
- Mohd Ali, M.; Hashim, N.; Aziz, S.A.; Lasekan, O. Principles and recent advances in electronic nose for quality inspection of agricultural and food products. Trends. Food Sci. Tech. 2020, 99, 1–10. [Google Scholar] [CrossRef]
- Xu, J.; Liu, K.; Zhang, C. Electronic nose for volatile organic compounds analysis in rice aging. Trends Food Sci. Technol. 2021, 109, 83–93. [Google Scholar] [CrossRef]
- Jo, D.; Kim, G.-R.; Yeo, S.-H.; Jeong, Y.-J.; Noh, B.S.; Kwon, J.-H. Analysis of aroma compounds of commercial cider vinegars with different acidities using SPME/GC-MS, electronic nose, and sensory evaluation. Food Sci. Biotechnol. 2013, 22, 1559–1565. [Google Scholar] [CrossRef]
- Hong, Y.; Noh, B.-S.; Kim, H.-Y. Discrimination of doenjang samples using a mass spectrometry-based electronic nose and human sensory preference testing. Food Sci. Biotechnol. 2015, 24, 31–36. [Google Scholar] [CrossRef]
- Lalaleo, L.; Hidalgo, D.; Valle, M.; Calero-Cáceres, W.; Lamuela-Raventós, R.M.; Becerra-Martínez, E. Differentiating, evaluating, and classifying three quinoa ecotypes by washing, cooking and germination treatments, using 1H NMR-based metabolomic approach. Food Chem. 2020, 331, 127351. [Google Scholar] [CrossRef]
- Peng, C.; Yao, G.; Sun, Y.; Guo, S.; Wang, J.; Mu, X.; Sun, Z.; Zhang, H. Comparative effects of the single and binary probiotics of Lacticaseibacillus casei Zhang and Bifidobacterium lactis V9 on the growth and metabolomic profiles in yogurts. Food Res. Int. 2021, 110603. [Google Scholar] [CrossRef]
- Raja, G.; Jung, Y.; Jung, S.H.; Kim, T.-J. 1H-NMR-based metabolomics for cancer targeting and metabolic engineering—A review. Process. Biochem. 2020, 99, 112–122. [Google Scholar] [CrossRef]
- Shi, B.; Ding, H.; Wang, L.; Wang, C.; Tian, X.; Fu, Z.; Zhang, L.; Han, L. Investigation on the stability in plant metabolomics with a special focus on freeze-thaw cycles: LC–MS and NMR analysis to Cassiae Semen (Cassia obtusifolia L.) seeds as a case study. J. Pharm. Biomed. Anal. 2021, 204, 114243. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Gao, F.; Chang, R.; Zhang, X.; Zhong, J.; Wen, J.; Wu, J.; Zhou, T. Metabolomics based comprehensive investigation of Gardeniae Fructus induced hepatotoxicity. Food Chem. Toxicol. 2021, 153, 112250. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.; Afonso, T.; Maraschin, M.; Rocha, M. WebSpecmine: A Website for Metabolomics Data Analysis and Mining. Metabolites 2019, 9, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.; Xu, Y.; Correa, E.; Turner, M.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.H.; Lee, E.H.; Yoon, H.S.; Lee, J.H.; Kim, J.Y.; Kang, K.; Park, J.-S.; Jin, J.B.; Ko, G.; Pan, C.-H. Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system. Food Chem. 2018, 263, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, X.; Zheng, C.; Zhou, B.; Xu, C.; Xia, T. Comparison of characteristic components in tea-leaves fermented by Aspergillus pallidofulvus PT-3, Aspergillus sesamicola PT-4 and Penicillium manginii PT-5 using LC-MS metabolomics and HPLC analysis. Food Chem. 2021, 350, 129228. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Sun, Y.; Dai, Y. ACE-Inhibitory Peptide Isolated from Fermented Soybean Meal as Functional Food. Int. J. Food Eng. 2013, 9, 1–8. [Google Scholar] [CrossRef]
- Tasdemir, S.S.; Sanlier, N. An insight into the anticancer effects of fermented foods: A review. J. Funct. Foods 2020, 75, 104281. [Google Scholar] [CrossRef]
- Dey, T.B.; Chakraborty, S.; Jain, K.K.; Sharma, A.; Kuhad, R.C. Antioxidant phenolics and their microbial production by submerged and solid state fermentation process: A review. Trends Food Sci. Technol. 2016, 53, 60–74. [Google Scholar] [CrossRef]
- Ferri, M.; Serrazanetti, D.I.; Tassoni, A.; Baldissarri, M.; Gianotti, A. Improving the functional and sensorial profile of cereal-based fermented foods by selecting Lactobacillus plantarum strains via a metabolomics approach. Food Res. Int. 2016, 89, 1095–1105. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, C.; Zhu, Y.; Zhou, C.; Xiong, Z.; Eweys, A.S.; Zhou, H.; Dong, Y.; Xiao, X. Metabolomics strategy for revealing the components in fermented barley extracts with lactobacillus plantarum dy-1—Sciencedirect. Food Res. Int. 2021, 139, 109808. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Tan, Y.; Wang, H.; Yang, F.; Chen, L.; Hao, F.; Lv, X.; Du, H.; Xu, Y. Effect of Pichia on shaping the fermentation microbial community of sauce-flavor Baijiu. Int. J. Food Microbiol. 2020, 336, 108898. [Google Scholar] [CrossRef]
- Chen, G.-M.; Huang, Z.-R.; Wu, L.; Wu, Q.; Guo, W.-L.; Zhao, W.-H.; Liu, B.; Zhang, W.; Rao, P.-F.; Lv, X.-C.; et al. Microbial diversity and flavor of Chinese rice wine (Huangjiu): An overview of current research and future prospects. Curr. Opin. Food Sci. 2021, 42, 37–50. [Google Scholar] [CrossRef]
- Khattab, A.R.; Guirguis, H.A.; Tawfik, S.M.; Farag, M.A. Cheese ripening: A review on modern technologies towards flavor en-hancement, process acceleration and improved quality assessment. Trends Food Sci. Tech. 2019, 88, 343–360. [Google Scholar] [CrossRef]
- Zhou, Z.; Jian, D.; Gong, M.; Zhu, S.; Li, G.; Zhang, S.; Zhong, F.; Mao, J. Characterization of the key aroma compounds in aged Zhenjiang aromatic vinegar by gas chromatography-olfactometry-mass spectrometry, quantitative measurements, aroma recombination and omission experiments. Food Res. Int. 2020, 136, 109434. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yuan, C.; Gao, X.; Kang, Y.; Huang, M.; Wu, J.; Liu, Y.; Zhang, J.; Li, H.; Zhang, Y. Characterization of key aroma compounds in Huangjiu from northern China by sensory-directed flavor analysis. Food Res. Int. 2020, 134, 109238. [Google Scholar] [CrossRef]
- Yang, L.; Fan, W.; Xu, Y. GC × GC-TOF/MS and UPLC-Q-TOF/MS based untargeted metabolomics coupled with physicochemical properties to reveal the characteristics of different type daqus for making soy sauce aroma and flavor type baijiu. LWT 2021, 146, 111416. [Google Scholar] [CrossRef]
- Deng, X.; Huang, G.; Tu, Q.; Zhou, H.; Li, Y.; Shi, H.; Wu, X.; Ren, H.; Huang, K.; He, X.; et al. Evolution analysis of flavor-active compounds during artificial fermentation of Pu-erh tea. Food Chem. 2021, 357, 129783. [Google Scholar] [CrossRef]
- Park, M.K.; Kim, Y.S. Comparative metabolic expressions of fermented soybeans according to different microbial starters. Food Chem. 2019, 305, 125461. [Google Scholar] [CrossRef]
- Wang, P.; Ma, X.; Wang, W.; Xu, D.; Sun, Y. Characterization of flavor fingerprinting of red sufu during fermentation and the com-parison of volatiles of typical products. Food Sci. Hum. Well. 2019, 8, 375–384. [Google Scholar] [CrossRef]
- Leejeerajumnean, A.; Duckham, S.C.; Owens, J.D.; Ames, J.M. Volatile compounds in Bacillus-fermented soybeans. J. Sci. Food Agric. 2001, 81, 525–529. [Google Scholar] [CrossRef]
- Kimura, K.; Yokoyama, S. Trends in the application of Bacillus in fermented foods. Curr. Opin. Biotech. 2018, 56, 36–42. [Google Scholar] [CrossRef]
- Gao, Y.X.; Xu, B.; Fan, H.R.; Zhang, M.R.; Zhang, L.J.; Lu, C.; Na Zhang, N.; Fan, B.; Wang, F.Z.; Li, S. 1H NMR-based chemometric metabolomics characterization of soymilk fermented by Bacillus subtilis BSNK-5. Food Res. Int. 2020, 138, 109686. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, Y.; Wu, Y.; Li, C.; Li, L.; Zhao, Y.; Hu, X.; Wei, Y.; Huang, H. Comparison of the microbial community and flavor compounds in fermented mandarin fish (Siniperca chuatsi): Three typical types of Chinese fermented mandarin fish products. Food Res. Int. 2021, 144, 110365. [Google Scholar] [CrossRef]
- Fang, G.-Y.; Chai, L.-J.; Zhong, X.-Z.; Jiang, Y.-J. Deciphering the succession patterns of bacterial community and their correlations with environmental factors and flavor compounds during the fermentation of Zhejiang rosy vinegar. Int. J. Food Microbiol. 2021, 341, 109070. [Google Scholar] [CrossRef]
- Che, H.; Yu, J.; Sun, J.; Lu, K.; Xie, W. Bacterial composition changes and volatile compounds during the fermentation of shrimp paste: Dynamic changes of microbial communities and flavor composition. Food Biosci. 2021, 101169. [Google Scholar] [CrossRef]
- Thakur, K.; Tomar, S.K.; Wei, Z.-J. Comparative mRNA Expression Profiles of Riboflavin Biosynthesis Genes in Lactobacilli Isolated from Human Feces and Fermented Bamboo Shoots. Front. Microbiol. 2017, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kandasamy, S.; Kavitake, D.; Shetty, P.H. Probiotic characterization and antioxidant properties of Weissella confusa KR780676, isolated from an Indian fermented food. LWT 2018, 97, 53–60. [Google Scholar] [CrossRef]
- Thakur, K.; Tomar, S.K.; De, S. Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 2015, 9, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Zyalçin, B.; Sanlier, N. The effect of diet components on cancer with epigenetic mechanisms. Trends Food Sci. Tech. 2020, 102, 138–145. [Google Scholar] [CrossRef]
- Crespo, L.; Gaglio, R.; Martínez, F.G.; Martin, G.M.; Franciosi, E.; Madrid-Albarrán, Y.; Settanni, L.; Mozzi, F.; Pescum, M. Bioaccumulation of selenium-by fruit origin lactic acid bacteria in tropical fermented fruit juices. LWT Food Sci. Technol. 2021, 151, 112103. [Google Scholar] [CrossRef]
- Daliri, B.M.; Tyagi, A.; Ofosu, F.K.; Chelliah, R.; Kim, J.H.; Kim, J.R.; Yoo, D.; Oh, D.H. A discovery-based metabolomic approach using UHPLC Q-TOF MS/MS unveils a plethora of prospective antihypertensive compounds in Korean fermented soybeans. LWT Food Sci. Technol. 2021, 137, 110399. [Google Scholar] [CrossRef]
- Kang, H.J.; Yang, H.J.; Kim, M.J.; Han, E.S.; Kim, H.J.; Kwon, D.Y. Metabolomic analysis of meju during fermentation by ultraper-formance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS). Food Chem. 2011, 127, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Lyu, G.; Luan, Y.; Yang, H.; Zhao, Z. Metabolomic study of the soybean pastes fermented by the single species Penicillium glabrum GQ1-3 and Aspergillus oryzae HGPA20. Food Chem. 2019, 295, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Namgung, H.-J.; Park, H.-J.; Cho, I.H.; Choi, H.-K.; Kwon, D.-Y.; Shim, S.-M.; Kim, Y.-S. Metabolite profiling of doenjang, fermented soybean paste, during fermentation. J. Sci. Food Agric. 2010, 90, 1926–1935. [Google Scholar] [CrossRef]
- Kim, S.S.; Kwak, H.S.; Kim, M.J. The effect of various salinity levels on metabolomic profiles, antioxidant capacities and sensory attributes of doenjang, a fermented soybean paste. Food Chem. 2020, 328, 127176. [Google Scholar] [CrossRef]
- Jang, H.-H.; Noh, H.; Kim, H.-W.; Cho, S.-Y.; Kim, H.-J.; Lee, S.-H.; Lee, S.-H.; Gunter, M.J.; Ferrari, P.; Scalbert, A.; et al. Metabolic tracking of isoflavones in soybean products and biosamples from healthy adults after fermented soybean consumption. Food Chem. 2020, 330, 127317. [Google Scholar] [CrossRef]
- Piras, C.; Marincola, F.C.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Viale, S.; Pisano, M.B. A NMR metabolomics study of the ripening process of the Fiore Sardo cheese produced with autochthonous adjunct cultures. Food Chem. 2013, 141, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Huang, T.; Guo, S.; Wang, Y.; Wang, J.; Kwok, L.-Y.; Dan, T.; Zhang, H.; Bilige, M. Probiotic Lactobacillus casei Zhang improved the properties of stirred yogurt. Food Biosci. 2020, 37, 100718. [Google Scholar] [CrossRef]
- Yang, S.; Yan, D.; Zou, Y.; Mu, D.; Li, X.; Shi, H.; Luo, X.; Yang, M.; Yue, X.; Wu, R.; et al. Fermentation temperature affects yogurt quality: A metabolomics study. Food Biosci. 2021, 42, 101104. [Google Scholar] [CrossRef]
- Wu, S.; Li, N.; Li, S.-J.; Bhandari, B.; Yang, B.-L.; Chen, X.D.; Mao, Z.-H. Effects of Incubation Temperature, Starter Culture Level and Total Solids Content on the Rheological Properties of Yogurt. Int. J. Food Eng. 2009, 5, 3. [Google Scholar] [CrossRef]
- Alaa, A.E.; Sally, S.; Samia, E.; Hany, E. Developing functional yogurt rich in bioactive peptides and gamma-aminobutyric acid related to cardiovascular health. LWT Food Sci. Technol. 2018, 98, 390–397. [Google Scholar] [CrossRef]
- Shi, M.; Mathai, M.L.; Xu, G.; McAinch, A.J.; Su, X.Q. The effects of supplementation with blueberry, cyanidin-3-O-β-glucoside, yoghurt and its peptides on obesity and related comorbidities in a diet-induced obese mouse model. J. Funct. Foods. 2019, 56, 92–101. [Google Scholar] [CrossRef]
- Tufariello, M.; Rizzuti, A.; Palombi, L.; Ragone, R.; Capozzi, V.; Gallo, V.; Mastrorilli, P.; Grieco, F. Non-targeted metabolomic approach as a tool to evaluate the chemical profile of sparkling wines fermented with autochthonous yeast strains. Food Control. 2021, 126, 108099. [Google Scholar] [CrossRef]
- Weiskirchen, S.; Weiskirchen, R. Resveratrol: How much wine do you have to drink to stay healthy? Adv. Nutr. 2016, 7, 706–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benbouguerra, N.; Hornedo-Ortega, R.; Garcia, F.; El Khawand, T.; Saucier, C.; Richard, T. Stilbenes in grape berries and wine and their potential role as anti-obesity agents: A review. Trends Food Sci. Technol. 2021, 112, 362–381. [Google Scholar] [CrossRef]
- Pugajeva, I.; Pērkons, I.; Górnaś, P. Identification and determination of stilbenes by Q-TOF in grape skins, seeds, juice and stems. J. Food Compos. Anal. 2018, 74, 44–52. [Google Scholar] [CrossRef]
- Li, R.-Y.; Zheng, X.-W.; Zhang, X.; Yan, Z.; Wang, X.-Y.; Han, B.-Z. Characterization of bacteria and yeasts isolated from traditional fermentation starter (Fen-Daqu) through a 1H NMR-based metabolomics approach. Food Microbiol. 2018, 76, 11–20. [Google Scholar] [CrossRef]
- Ishihara, S.; Inaoka, T.; Nakamura, T.; Kimura, K.; Tomita, S. Nuclear magnetic resonance- and gas chromatography/mass spectrometry-based metabolomic characterization of water-soluble and volatile compound profiles in cabbage vinegar. J. Biosci Bioeng. 2018, 126, 53–62. [Google Scholar] [CrossRef]
- Tomita, S.; Watanabe, J.; Kuribayashi, T.; Tanaka, S.; Kawahara, T. Metabolomic evaluation of different starter culture effects on water-soluble and volatile compound profiles in nozawana pickle fermentation. Food Chem. Mol. Sci. 2021, 2, 100019. [Google Scholar] [CrossRef]
- Drake, M.A.; Delahunty, C.M. Chapter 20—Sensory character of cheese and its evaluation. In Cheese, 4th ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 517–545. [Google Scholar] [CrossRef]
- Seo, H.S.; Lee, S.; Singh, D.; Shin, H.W.; A Cho, S.; Lee, C.H. Untargeted metabolite profiling for koji-fermentative bioprocess unravels the effects of varying substrate types and microbial inocula. Food Chem. 2018, 266, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Kasprowicz-Potocka, M.; Zaworska, A.; Gulewicz, P.; Nowak, P.; Frankiewicz, A. The effect of fermentation of high alkaloid seeds of Lupinus angustifolius var. Karo by Saccharomyces cerevisieae, Kluyveromyces lactis, and Candida utilis on the chemical and microbial composition of products. J. Food Process. Preserv. 2017, 42, e13487. [Google Scholar] [CrossRef]
- Ding, M.; Liu, W.; Peng, J.; Liu, X.; Tang, Y. Simultaneous determination of seven preservatives in food by dispersive liquid-liquid microextraction coupled with gas chromatography-mass spectrometry. Food Chem. 2018, 269, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Tungkijanansin, N.; Alahmad, W.; Nhujak, T.; Varanusupakul, P. Simultaneous determination of benzoic acid, sorbic acid, and pro-pionic acid in fermented food by headspace solid-phase microextraction followed by GC-FID. Food Chem. 2020, 329, 127161. [Google Scholar] [CrossRef] [PubMed]
- Oyedeji, A.B.; Green, E.; Adebiyi, J.A.; Ogundele, O.M.; Gbashi, S.; Adefisoye, M.A.; Oyeyinka, S.A.; Adebo, O.A. Metabolomic approaches for the determination of metabolites from pathogenic microorganisms: A review. Food Res. Int. 2020, 140, 110042. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Hou, L.; Gao, J.; Li, D.; Tian, Z.; Fan, B.; Wang, F.; Li, S. Metabolomics Approaches for the Comprehensive Evaluation of Fermented Foods: A Review. Foods 2021, 10, 2294. https://doi.org/10.3390/foods10102294
Gao Y, Hou L, Gao J, Li D, Tian Z, Fan B, Wang F, Li S. Metabolomics Approaches for the Comprehensive Evaluation of Fermented Foods: A Review. Foods. 2021; 10(10):2294. https://doi.org/10.3390/foods10102294
Chicago/Turabian StyleGao, Yaxin, Lizhen Hou, Jie Gao, Danfeng Li, Zhiliang Tian, Bei Fan, Fengzhong Wang, and Shuying Li. 2021. "Metabolomics Approaches for the Comprehensive Evaluation of Fermented Foods: A Review" Foods 10, no. 10: 2294. https://doi.org/10.3390/foods10102294
APA StyleGao, Y., Hou, L., Gao, J., Li, D., Tian, Z., Fan, B., Wang, F., & Li, S. (2021). Metabolomics Approaches for the Comprehensive Evaluation of Fermented Foods: A Review. Foods, 10(10), 2294. https://doi.org/10.3390/foods10102294






