Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains Cultivation
2.2. Probiotic Properties and Safety Assessment
2.2.1. Survival in Simulated Gastric Juice and Small Intestinal Condition
2.2.2. Bile Salt Hydrolase (BSH) Activity
2.2.3. Hemolytic Activity
2.2.4. Determination of Antibiotic Susceptibility
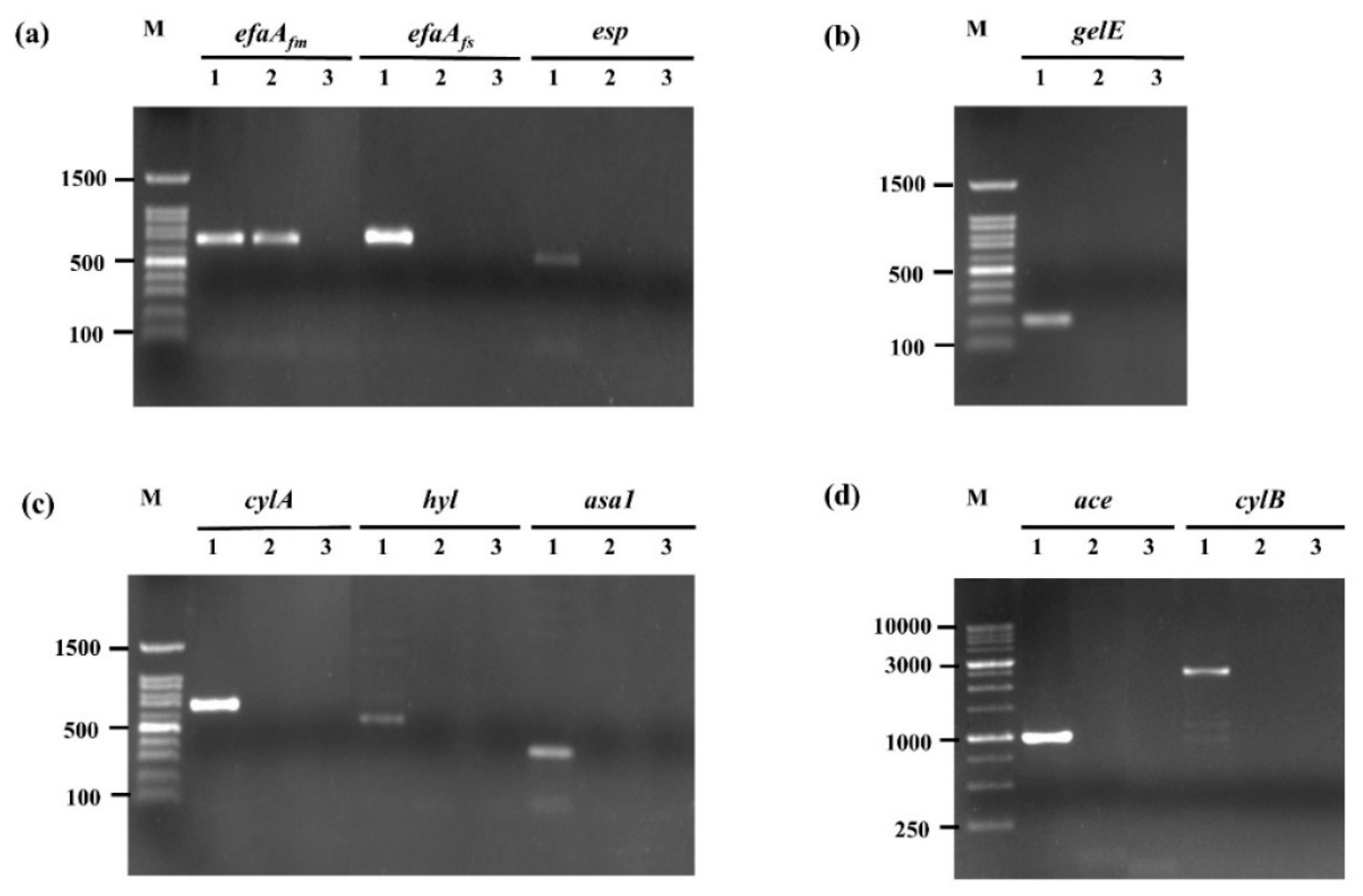
2.2.5. Evaluation of Virulence-Associated Genes
2.3. Inhibition of Adhesion on Caco-2 Cell Model
2.3.1. Cell Culture
2.3.2. Inhibition of Enteropathogenic Bacterial Adhesion
2.4. Immunomodulation Associated with Inflammatory Cytokines
2.5. Antimicrobial Activity against Pathogenic Bacteria
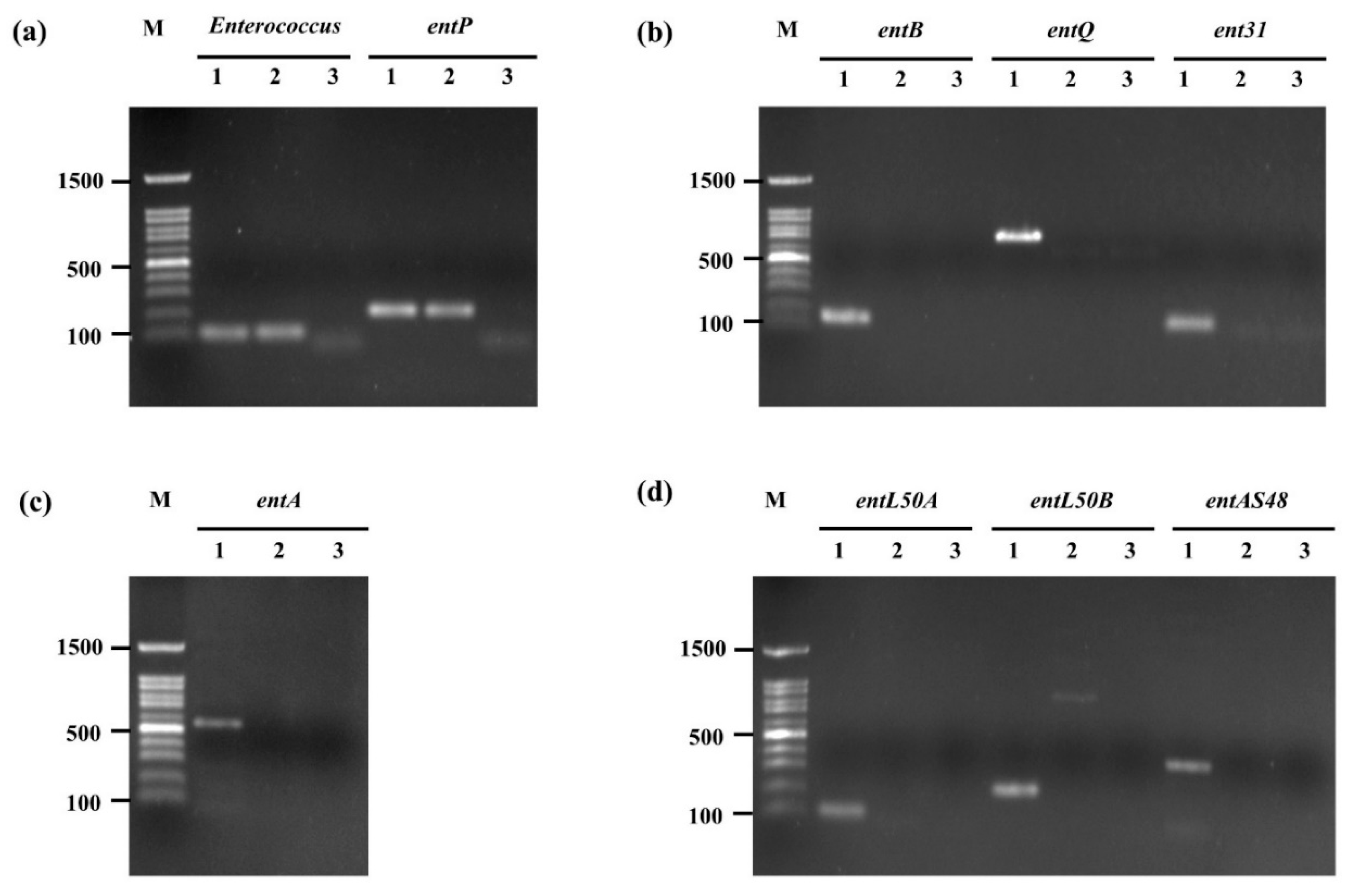
2.6. Confirmation of Bacteriocin-Encoding Gene
2.7. Partial Purification of AMPs
2.8. Molecular Mass and Amino Acid Sequence Determination of AMPs
2.9. Enzyme Sensitivity, Physicochemical Treatments and Long-Term Storage of AMPs
2.10. Statistical Analysis
3. Results
3.1. Probiotic Properties and Safety Assessment
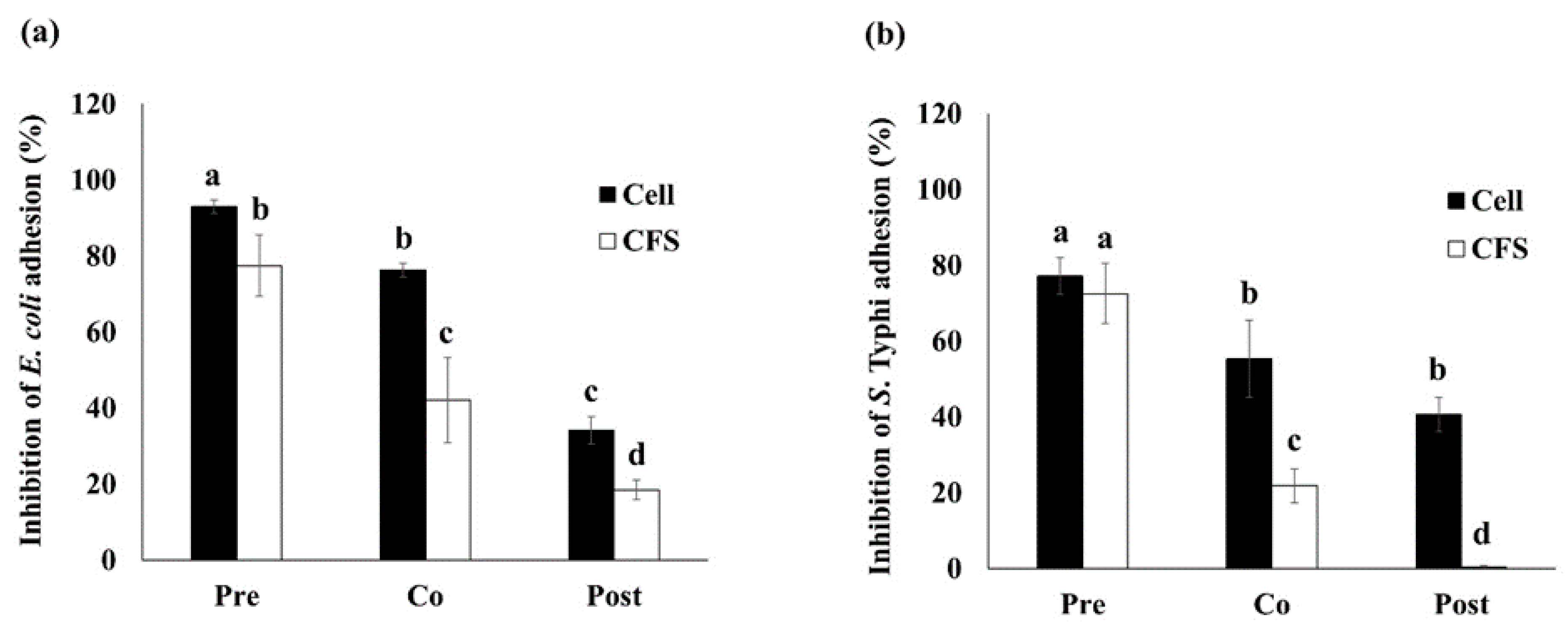
3.2. Inhibition of Adhesion on Caco-2 Cell Model
3.3. Immunomodulation Associated with Inflammatory Cytokines
3.4. Antimicrobial Activity against Pathogenic Bacteria
3.5. Confirmation of Bacteriocin-Encoding Gene
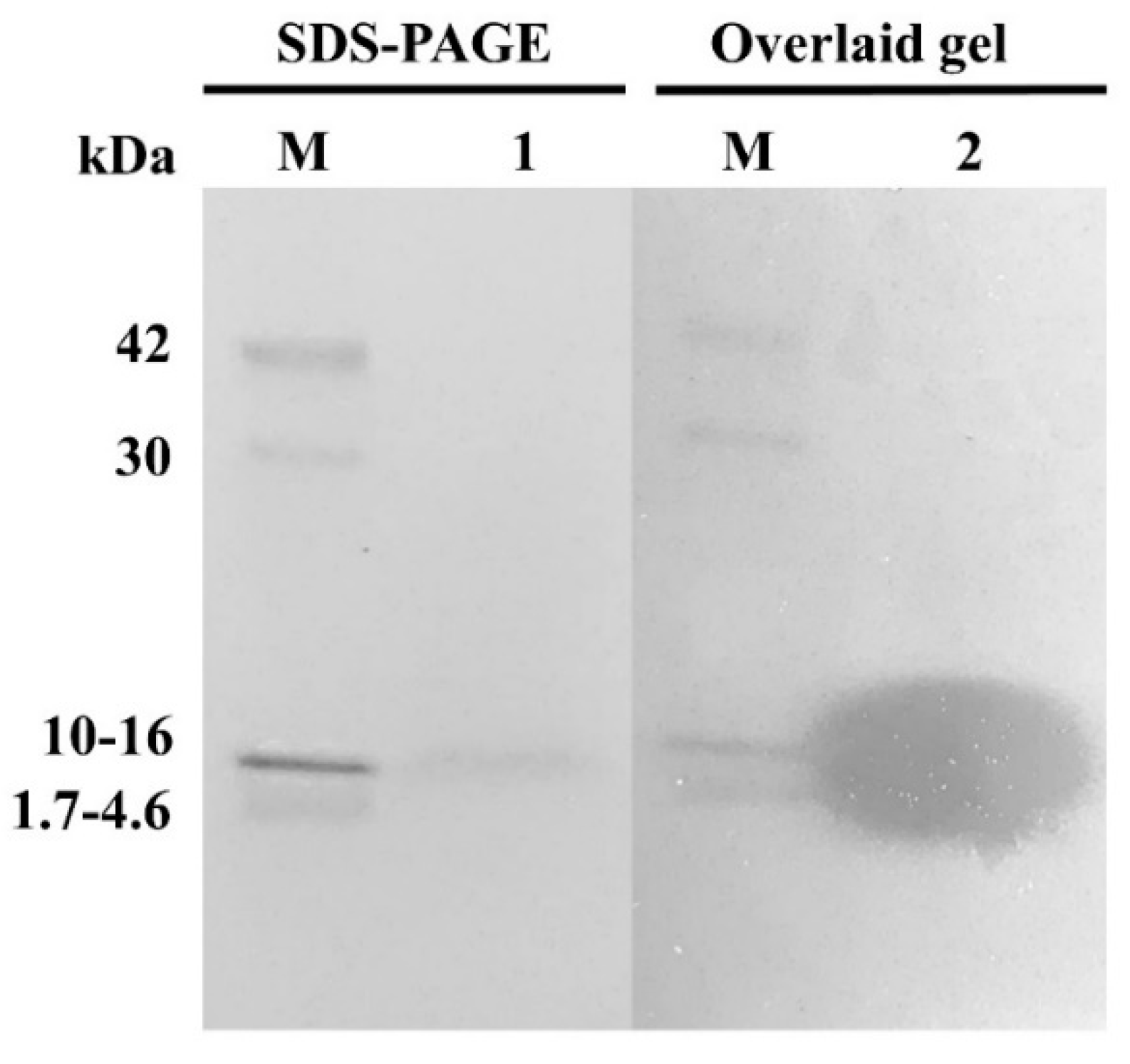
3.6. Partial Purification of AMPs
3.7. Molecular Mass and Amino Acid Sequence Determination of AMPs
3.8. Enzyme Sensitivity, Physicochemical Treatments and Long-Term Storage of AMPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. Toxins in fermented foods: Prevalence and preventions—A mini review. Toxins 2019, 11, 4. [Google Scholar] [CrossRef] [Green Version]
- García, C.; Rendueles, M.; Díaz, M. Liquid-phase food fermentations with microbial consortia involving lactic acid bacteria: A review. Food Res. Int. 2019, 119, 207–220. [Google Scholar] [CrossRef]
- Patel, A.; Prajapati, J.; Holst, O.; Ljungh, A. Determining probiotic potential of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented food products. Food Biosci. 2014, 5, 27–33. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization); WHO (World Health Organization). Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria: Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria; Food and Agriculture Organization; World Health Organization: Córdoba, Argentina, 2001. [Google Scholar]
- Le, B.; Yang, S.H. Efficacy of Lactobacillus plantarum in prevention of inflammatory bowel disease. Toxicol. Rep. 2018, 5, 314–317. [Google Scholar] [CrossRef]
- Ness, I.F.; Diep, D.B.; Ike, Y. Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, 1st ed.; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 466–488. [Google Scholar]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus Enterococcus: Between probiotic potential and safety concerns—An update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Davoodvandi, A.; Marzban, H.; Goleij, P.; Sahebkar, A.; Morshedi, K.; Rezaei, S.; Mahjoubin-Tehran, M.; Tarrahimofrad, H.; Hamblin, M.R.; Mirzaei, H. Effects of therapeutic probiotics on modulation of microRNAs. Cell Commun. Signal. 2021, 19, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Qiu, L.; Xu, X.; Liu, Z.; Zhan, H.; Tao, X.; Shah, N.P.; Wei, H. Beneficial effects of probiotic cholesterol-lowering strain of Enterococcus faecium WEFA23 from infants on diet-induced metabolic syndrome in rats. J. Dairy Sci. 2017, 100, 1618–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadova, A.; Todorov, S.D.; Choiset, Y.; Rabesona, H.; Zadi, T.M.; Kuliyev, A.; de Melo Franco, B.D.G.; Chobert, J.-M.; Haertlé, T. Evaluation of antimicrobial activity, probiotic properties and safety of wild strain Enterococcus faecium AQ71 isolated from Azerbaijani Motal cheese. Food Control 2013, 30, 631–641. [Google Scholar] [CrossRef]
- Santos, J.C.; Sousa, R.C.; Otoni, C.G.; Moraes, A.R.; Souza, V.G.; Medeiros, E.A.; Espitia, P.J.; Pires, A.C.; Coimbra, J.S.; Soares, N.F. Nisin and other antimicrobial peptides: Production, mechanisms of action, and application in active food packaging. Innov. Food Sci. Emerg. Technol. 2018, 48, 179–194. [Google Scholar] [CrossRef]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Ma, Z.; Qu, B.; Yao, L.; Gao, Z.; Zhang, S. Identification and functional characterization of ribosomal protein S23 as a new member of antimicrobial protein. Dev. Comp. Immunol. 2020, 103730. [Google Scholar] [CrossRef]
- Pidutti, P.; Federici, F.; Brandi, J.; Manna, L.; Rizzi, E.; Marini, U.; Cecconi, D. Purification and characterization of ribosomal proteins L27 and L30 having antimicrobial activity produced by the Lactobacillus salivarius SGL 03. J. Appl. Microbiol. 2018, 124, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Choeisoongnern, T.; Sivamaruthi, B.S.; Sirilun, S.; Peerajan, S.; Choiset, Y.; Rabesona, H.; Haertlé, T.; Chaiyasut, C. Screening and identification of bacteriocin-like inhibitory substances producing lactic acid bacteria from fermented products. Food Sci. Technol. 2019, 40, 571–579. [Google Scholar] [CrossRef] [Green Version]
- Hwanhlem, N.; Ivanova, T.; Biscola, V.; Choiset, Y.; Haertlé, T. Bacteriocin producing Enterococcus faecalis isolated from chicken gastrointestinal tract originating from Phitsanulok, Thailand: Isolation, screening, safety evaluation and probiotic properties. Food Control 2017, 78, 187–195. [Google Scholar] [CrossRef]
- Oh, A.; Daliri, E.B.-M.; Oh, D.H. Screening for potential probiotic bacteria from Korean fermented soybean paste: In Vitro and Caenorhabditis elegans model testing. LWT 2018, 88, 132–138. [Google Scholar] [CrossRef]
- Shehata, M.; El Sohaimy, S.; El-Sahn, M.A.; Youssef, M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016, 61, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Nueno-Palop, C.; Narbad, A. Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int. J. Food Microbiol. 2011, 145, 390–394. [Google Scholar] [CrossRef]
- Hwanhlem, N.; Biscola, V.; El-Ghaish, S.; Jaffrès, E.; Dousset, X.; Haertlé, T.; Aran, H.; Chobert, J.-M. Bacteriocin-producing lactic acid bacteria isolated from mangrove forests in Southern Thailand as potential bio-control agents: Purification and characterization of bacteriocin produced by Lactococcus lactis subsp. lactis KT2W2L. Probiotics Antimicrob. Proteins 2013, 5, 264–278. [Google Scholar] [CrossRef]
- Vankerckhoven, V.; Van Autgaerden, T.; Vael, C.; Lammens, C.; Chapelle, S.; Rossi, R.; Jabes, D.; Goossens, H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 2004, 42, 4473–4479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moroni, O.; Kheadr, E.; Boutin, Y.; Lacroix, C.; Fliss, I. Inactivation of adhesion and invasion of food-borne Listeria monocytogenes by bacteriocin-producing Bifidobacterium strains of human origin. Appl. Environ. Microbiol. 2006, 72, 6894–6901. [Google Scholar] [CrossRef] [Green Version]
- Vimont, A.; Fernandez, B.; Hammami, R.; Ababsa, A.; Daba, H.; Fliss, I. Bacteriocin-producing Enterococcus faecium LCW 44: A high potential probiotic candidate from raw camel milk. Front. Microbiol. 2017, 8, 865. [Google Scholar] [CrossRef] [Green Version]
- Bengoa, A.A.; Zavala, L.; Carasi, P.; Trejo, S.A.; Bronsoms, S.; de los Ángeles Serradell, M.; Garrote, G.L.; Abraham, A.G. Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res. Int. 2018, 103, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Koryszewska-Bagińska, A.; Gawor, J.; Nowak, A.; Grynberg, M.; Aleksandrzak-Piekarczyk, T. Comparative genomics and functional analysis of a highly adhesive dairy Lactobacillus paracasei subsp. paracasei IBB3423 strain. Appl. Microbiol. Biotechnol. 2019, 103, 7617–7634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aspri, M.; O′Connor, P.M.; Field, D.; Cotter, P.D.; Ross, P.; Hill, C.; Papademas, P. Application of bacteriocin-producing Enterococcus faecium isolated from donkey milk, in the bio-control of Listeria monocytogenes in fresh whey cheese. Int. Dairy J. 2017, 73, 1–9. [Google Scholar] [CrossRef]
- Xi, Q.; Wang, J.; Du, R.; Zhao, F.; Han, Y.; Zhou, Z. Purification and Characterization of Bacteriocin Produced by a Strain of Enterococcus faecalis TG2. Appl. Biochem. Biotechnol. 2018, 184, 1106–1119. [Google Scholar] [CrossRef]
- Ke, D.; Picard, F.J.; Martineau, F.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 1999, 37, 3497–3503. [Google Scholar] [CrossRef] [Green Version]
- Aran, H.; Biscola, V.; El-Ghaish, S.; Jaffres, E.; Dousset, X.; Pillot, G.; Haertle, T.; Chobert, J.-M.; Hwanhlem, N. Bacteriocin-producing Enterococcus faecalis KT2W2G isolated from mangrove forests in southern Thailand: Purification, characterization and safety evaluation. Food Control 2015, 54, 126–134. [Google Scholar]
- Goh, H.F.; Philip, K. Isolation and mode of action of bacteriocin BacC1 produced by nonpathogenic Enterococcus faecium C1. J. Dairy Sci. 2015, 98, 5080–5090. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bringans, S.; Eriksen, S.; Kendrick, T.; Gopalakrishnakone, P.; Livk, A.; Lock, R.; Lipscombe, R. Proteomic analysis of the venom of Heterometrus longimanus (Asian black scorpion). Proteomics 2008, 8, 1081–1096. [Google Scholar] [CrossRef]
- Wannun, P.; Piwat, S.; Teanpaisan, R. Purification and characterization of bacteriocin produced by oral Lactobacillus paracasei SD1. Anaerobe 2014, 27, 17–21. [Google Scholar] [CrossRef]
- Pinto, A.L.; Fernandes, M.; Pinto, C.; Albano, H.; Castilho, F.; Teixeira, P.; Gibbs, P.A. Characterization of anti-Listeria bacteriocins isolated from shellfish: Potential antimicrobials to control non-fermented seafood. Int. J. Food Microbiol. 2009, 129, 50–58. [Google Scholar] [CrossRef]
- Maia, L.F.; Giazzi, A.; Brandalize, C.; Katsuda, M.S.; Rocha, K.; Real, A.; Terra, M.; Regina, A.; Furlaneto, M.; Cristina, A. Isolation and characterization of potential probiotic enterococci strains from soft cheese flora. Afr. J. Microbiol. Res. 2017, 11, 482–487. [Google Scholar]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Cui, Y.; Qu, X. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch. Microbiol. 2018, 200, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, B.; Stanton, C.; Fitzgerald, G.; Ross, R. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl. Environ. Microbiol. 2005, 71, 3060–3067. [Google Scholar] [CrossRef] [Green Version]
- Dowdell, P.; Chankhamhaengdecha, S.; Panbangred, W.; Janvilisri, T.; Aroonnual, A. Probiotic activity of Enterococcus faecium and Lactococcus lactis isolated from Thai fermented sausages and their protective effect against Clostridium difficile. Probiotics Antimicrob. Proteins 2020, 12, 641–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Jiang, M.; Wan, C.; Chen, X.; Chen, X.; Tao, X.; Shah, N.P.; Wei, H. Screening probiotic strains for safety: Evaluation of virulence and antimicrobial susceptibility of enterococci from healthy Chinese infants. J. Dairy Sci. 2016, 99, 4282–4290. [Google Scholar] [CrossRef] [Green Version]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [Green Version]
- Vandera, E.; Tsirka, G.; Kakouri, A.; Koukkou, A.-I.; Samelis, J. Approaches for enhancing in situ detection of enterocin genes in thermized milk, and selective isolation of enterocin-producing Enterococcus faecium from Baird-Parker agar. Int. J. Food Microbiol. 2018, 281, 23–31. [Google Scholar] [CrossRef]
- İspirli, H.; Demirbaş, F.; Dertli, E. Characterization of functional properties of Enterococcus spp. isolated from Turkish white cheese. LWT 2017, 75, 358–365. [Google Scholar] [CrossRef]
- Gaglio, R.; Couto, N.; Marques, C.; Lopes, M.d.F.S.; Moschetti, G.; Pomba, C.; Settanni, L. Evaluation of antimicrobial resistance and virulence of enterococci from equipment surfaces, raw materials, and traditional cheeses. Int. J. Food Microbiol. 2016, 236, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Golob, M.; Pate, M.; Kušar, D.; Dermota, U.; Avberšek, J.; Papić, B.; Zdovc, I. Antimicrobial Resistance and virulence genes in Enterococcus faecium and Enterococcus faecalis from humans and retail red meat. Biomed. Res. Int. 2019, 2019, 2815279. [Google Scholar] [CrossRef] [Green Version]
- Bagci, U.; Togay, S.O.; Temiz, A.; Ay, M. Probiotic characteristics of bacteriocin-producing Enterococcus faecium strains isolated from human milk and colostrum. Folia Microbiol. 2019, 64, 735–750. [Google Scholar] [CrossRef]
- Basanta, A.; Gómez-Sala, B.; Sánchez, J.; Diep, D.B.; Herranz, C.; Hernández, P.E.; Cintas, L.M. Use of the yeast Pichia pastoris as an expression host for secretion of enterocin L50, a leaderless two-peptide (L50A and L50B) bacteriocin from Enterococcus faecium L50. Appl. Environ. Microbiol. 2010, 76, 3314–3324. [Google Scholar] [CrossRef] [Green Version]
- Toit, M.D.; Franz, C.; Dicks, L.; Holzapfel, W. Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J. Appl. Microbiol. 2000, 88, 482–494. [Google Scholar] [CrossRef]
- Tuncer, B.Ö.; Ay, Z.; Tuncer, Y. Occurrence of enterocin genes, virulence factors, and antibiotic resistance in 3 bacteriocin-producer Enterococcus faecium strains isolated from Turkish tulum cheese. Turk. J. Biol. 2013, 37, 443–449. [Google Scholar] [CrossRef]
- Abouelnaga, M.; Lamas, A.; Quintela-Baluja, M.; Osman, M.; Miranda, J.M.; Cepeda, A.; Franco, C.M. Evaluation of the extent of spreading of virulence factors and antibiotic resistance in Enterococci isolated from fermented and unfermented foods. Ann. Microbiol. 2016, 66, 577–585. [Google Scholar] [CrossRef]
- Mannu, L.; Paba, A.; Daga, E.; Comunian, R.; Zanetti, S.; Duprè, I.; Sechi, L.A. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 2003, 88, 291–304. [Google Scholar] [CrossRef]
- Sharma, C.; Gulati, S.; Thakur, N.; Singh, B.P.; Gupta, S.; Kaur, S.; Mishra, S.K.; Puniya, A.K.; Gill, J.P.S.; Panwar, H. Antibiotic sensitivity pattern of indigenous lactobacilli isolated from curd and human milk samples. 3 Biotech 2017, 7, 53. [Google Scholar] [CrossRef] [Green Version]
- Mathipa, M.G.; Bhunia, A.K.; Thantsha, M.S. Internalin AB-expressing recombinant Lactobacillus casei protects Caco-2 cells from Listeria monocytogenes-induced damages under simulated intestinal conditions. PLoS ONE 2019, 14, e0220321. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Xu, X.; Zhang, F.; Xu, D.; Liu, Z.; Tao, X.; Wei, H. Anti-adhesion of probiotic Enterococcus faecium WEFA23 against 5 pathogens and the beneficial effect of its S-layer proteins against Listeria monocytogenes. Can. J. Microbiol. 2019, 65, 175–184. [Google Scholar] [CrossRef]
- Veljović, K.; Popović, N.; Miljković, M.; Tolinački, M.; Terzić-Vidojević, A.; Kojić, M. Novel aggregation promoting factor AggE contributes to the probiotic properties of Enterococcus faecium BGGO9-28. Front. Microbiol. 2017, 8, 1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, J.; Paula, A.; Casarotti, S.; Penna, A. Lactic acid bacteria antimicrobial compounds: Characteristics and applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Kern, M.; Günzel, D.; Aschenbach, J.R.; Tedin, K.; Bondzio, A.; Lodemann, U. Altered cytokine expression and barrier properties after in vitro infection of porcine epithelial cells with enterotoxigenic Escherichia coli and probiotic Enterococcus faecium. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Yeung, C.Y.; Chiang Chiau, J.S.; Chan, W.T.; Jiang, C.B.; Cheng, M.L.; Liu, H.L.; Lee, H.C. In vitro prevention of Salmonella lipopolysaccharide-induced damages in epithelial barrier function by various Lactobacillus strains. Gastroenterol. Res. Pract. 2013, 2013, 973209. [Google Scholar] [CrossRef] [Green Version]
- Rodes, L.; Khan, A.; Paul, A.; Coussa-Charley, M.; Marinescu, D.; Tomaro-Duchesneau, C.; Shao, W.; Kahouli, I.; Prakash, S. Effect of probiotics Lactobacillus and Bifidobacterium on gut-derived lipopolysaccharides and inflammatory cytokines: An in vitro study using a human colonic microbiota model. J. Microbiol. Biotechnol. 2013, 23, 518–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.C.; Munitic, I.; Mittelstadt, P.R.; Castro, E.; Ashwell, J.D. Suppression of dendritic cell-derived IL-12 by endogenous glucocorticoids is protective in LPS-induced sepsis. PLoS Biol. 2015, 13, e1002269. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Tsen, H.-Y.; Lin, C.-L.; Yu, B.; Chen, C.-S. Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult. Sci. 2012, 91, 2139–2147. [Google Scholar] [CrossRef]
- Li, W.; Huang, Q.; Li, Y.; Rajput, I.R.; Huang, Y.; Hu, C. Induction of probiotic strain Enterococcus faecium EF1 on the production of cytokines, superoxide anion and prostaglandin E2 in a macrophage cell line. Pak. Vet. J. 2012, 32, 530–534. [Google Scholar]
- Tian, Z.; Yang, L.; Li, P.; Xiao, Y.; Peng, J.; Wang, X.; Li, Z.; Liu, M.; Bi, D.; Shi, D. The inflammation regulation effects of Enterococcus faecium HDRsEf1 on human enterocyte-like HT-29 cells. Anim. Cells Syst. 2016, 20, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Rho, M.K.; Kim, Y.E.; Rho, H.I.; Kim, T.R.; Kim, Y.B.; Sung, W.K.; Kim, T.W.; Kim, D.O.; Kang, H. FC-K Derived from Kimchi Is a Probiotic Strain That Shows Anti-Allergic Activity. J. Microbiol. Biotechnol. 2017, 27, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Hussain, N.; Tariq, M.; Saris, P.E.J.; Zaidi, A. Evaluation of the probiotic and postbiotic potential of lactic acid bacteria from artisanal dairy products against pathogens. J. Infect. Dev. Ctries. 2021, 15, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Waheed, S.; Rasool, M.H.; ASLAM, B.; Muzammil, S.; WASEEM, M.; Shahid, M.; Saqib, M.; Hayat, S.; Naeem, M.; Taj, Z. Antagonistic Potential of Dairy Origin Enterococcus faecium Against Multidrug-Resistant Foodborne Pathogens. Rom. Biotechnol. Lett. 2021, 26, 2406–2415. [Google Scholar] [CrossRef]
- De Carvalho, K.G.; Bambirra, F.H.; Kruger, M.F.; Barbosa, M.S.; Oliveira, J.S.; Santos, A.M.; Nicoli, J.R.; Bemquerer, M.P.; de Miranda, A.; Salvucci, E.J. Antimicrobial compounds produced by Lactobacillus sakei subsp. sakei 2a, a bacteriocinogenic strain isolated from a Brazilian meat product. J. Ind. Microbiol. Biotechnol. 2010, 37, 381–390. [Google Scholar] [CrossRef]
- Favaro, L.; Basaglia, M.; Casella, S.; Hue, I.; Dousset, X.; de Melo Franco, B.D.G.; Todorov, S.D. Bacteriocinogenic potential and safety evaluation of non-starter Enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol. 2014, 38, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Vasilchenko, A.; Vasilchenko, A.; Valyshev, A.; Rogozhin, E. A novel high-molecular-mass bacteriocin produced by Enterococcus faecium: Biochemical features and mode of action. Probiotics Antimicrob. Proteins 2018, 10, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Brandão, A.; Almeida, T.; Muñoz-Atienza, E.; Torres, C.; Igrejas, G.; Hernández, P.; Cintas, L.; Poeta, P.; Herranz, C. Antimicrobial activity and occurrence of bacteriocin structural genes in Enterococcus spp. of human and animal origin isolated in Portugal. Arch. Microbiol. 2010, 192, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernandez, P.E.; Nes, I.F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, J.; Criado, R.; Martín, M.; Herranz, C.; Cintas, L.M.; Hernández, P.E. Production of enterocin P, an antilisterial pediocin-like bacteriocin from Enterococcus faecium P13, in Pichia pastoris. Antimicrob. Agents Chemother. 2005, 49, 3004–3008. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Griffiths, M.W.; Wu, P.; Wang, H.; Zhang, X.; Li, P. Enterococcus faecium LM-2, a multi-bacteriocinogenic strain naturally occurring in “Byaslag”, a traditional cheese of Inner Mongolia in China. Food Control 2011, 22, 283–289. [Google Scholar] [CrossRef]
- Carvalho, K.G.; Bambirra, F.H.S.; Nicoli, J.R.; Oliveira, J.S.; Santos, A.M.C.; Bemquerer, M.P.; Miranda, A.; Franco, B.D.G.M. Characterization of multiple antilisterial peptides produced by sakacin P-producing Lactobacillus sakei subsp. sakei 2a. Arch. Microbiol. 2018, 200, 635–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokhtar, N.F.K.; Hashim, A.M.; Hanish, I.; Zulkarnain, A.; Nhari, R.M.H.R.; Sani, A.A.A.; Abbasiliasi, S.; Ariff, A.; Mustafa, S.; Rahim, R.A. The Discovery of New Antilisterial Proteins From Paenibacillus polymyxa Kp10 via Genome Mining and Mass Spectrometry. Front. Microbiol. 2020, 11, 960. [Google Scholar] [CrossRef]
- Herranz, C.; Cintas, L.M.; Hernández, P.E.; Moll, G.N.; Driessen, A.J. Enterocin P causes potassium ion efflux from Enterococcus faecium T136 cells. Antimicrob. Agents Chemother. 2001, 45, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abanoz, H.S.; Kunduhoglu, B. Antimicrobial activity of a bacteriocin produced by Enterococcus faecalis KT11 against some pathogens and antibiotic-resistant bacteria. Korean J. Food Sci. Anim. Resour. 2018, 38, 1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Treatments | Secretion of Cytokines (pg/mL) | ||
|---|---|---|---|
| IL-6 | IL-12 | IL-10 | |
| Untreated | 5.67 ± 0.41 f | 0.34 ± 0.03 e | 8.32 ± 1.33 f |
| LPS | 45.30 ± 2.54 a | 6.04 ± 0.52 a | 99.84 ± 4.05 e |
| E. coli | 39.27 ± 4.00 b | 5.59 ± 1.38 a | 112.20 ± 1.20 d,e |
| S. Typhi | 39.92 ± 3.56 b | 6.38 ± 1.06 a | 97.20 ± 2.80 e |
| S. Typhimurium | 37.00 ± 0.61 b | 4.03 ± 1.00 b | 126.18 ± 15.10 c,d |
| E. faecium OV3-6 | 12.36 ± 2.80 e | 1.80 ± 0.31 d | 247.59 ± 17.43 a |
| LPS + E. faecium OV3-6 | 31.74 ± 2.29 c | 3.57 ± 0.30 b,c | 110.02 ± 2.24 d,e |
| E. coli + E. faecium OV3-6 | 26.58 ± 5.48 c,d | 2.45 ± 0.87 c,d | 130.57 ± 10.30 c |
| S. Typhi+ E. faecium OV3-6 | 25.42 ± 2.86 d | 2.71 ± 0.37 b,c,d | 137.44 ± 2.80 c |
| S. Typhimurium+ E. faecium OV3-6 | 22.80 ± 2.73 d | 1.97 ± 0.42 c,d | 156.11 ± 16.18 b |
| Tested Strains | Inhibition Zone (mm) |
|---|---|
| Gram-positive bacteria | |
| Bacillus cereus ATCC 11778 | 7.00 ± 0.10 d |
| Brochothrix thermosphacta DSMZ 20171T | NI |
| Carnobacterium maltaromaticum NCDO 2760 | 14.70 ± 0.24 b |
| Clostridium perfringens | NI |
| Listeria innocua CIP 80.11T | 12.05 ± 0.65 c |
| Listeria ivanovii SLCC 2379 | 14.60 ± 0.30 b |
| Propionibacterium acnes ATCC 6919 | NI |
| Staphylococcus aureus ATCC 5923 | NI |
| Staphylococcus aureus CIP 76.25 | 15.63 ± 0.03 a |
| Staphylococcus epidermidis ATCC 12228 | NI |
| Streptococcus mutans ATCC 25175 | 6.25 ± 0.25 e |
| Gram-negative bacteria | |
| Cronobacter sakazakii ATCC BAA-894 | NI |
| Escherichia coli ATCC 5922 | NI |
| Escherichia coli CIP 76.24 | NI |
| Pseudomonas aeruginosa ATCC 27853 | NI |
| Salmonella Typhi DMST 22842 | NI |
| Salmonella Typhimurium TISTR 1469 | NI |
| Shigella sonnei | NI |
| Vibrio harveyi ATCC BAA-1119 | NI |
| Yeast | |
| Candida albicans ATCC 90028 | NI |
| Step of Purification | Total Volume (mL) | Activity (AU/mL) | Protein (mg/mL) | Total Activity (AU) | Total Protein (mg) | Specific Activity (AU/mg) | Yield (%) | Increase in Specific Activity (Fold) |
|---|---|---|---|---|---|---|---|---|
| Cell-free supernatant | 1000 | 400 | 1.26 | 400,000 | 1260.00 | 317.46 | 100.00 | 1.00 |
| Crude (NH4)2SO4 precipitation | 100 | 3200 | 1.81 | 320,000 | 181.00 | 1767.96 | 80 | 5.57 |
| Molecular weight cut off | 24 | 12,800 | 2.54 | 307,200 | 58.80 | 5224.49 | 76.80 | 16.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choeisoongnern, T.; Sirilun, S.; Waditee-Sirisattha, R.; Pintha, K.; Peerajan, S.; Chaiyasut, C. Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation. Foods 2021, 10, 2264. https://doi.org/10.3390/foods10102264
Choeisoongnern T, Sirilun S, Waditee-Sirisattha R, Pintha K, Peerajan S, Chaiyasut C. Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation. Foods. 2021; 10(10):2264. https://doi.org/10.3390/foods10102264
Chicago/Turabian StyleChoeisoongnern, Thiwanya, Sasithorn Sirilun, Rungaroon Waditee-Sirisattha, Komsak Pintha, Sartjin Peerajan, and Chaiyavat Chaiyasut. 2021. "Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation" Foods 10, no. 10: 2264. https://doi.org/10.3390/foods10102264
APA StyleChoeisoongnern, T., Sirilun, S., Waditee-Sirisattha, R., Pintha, K., Peerajan, S., & Chaiyasut, C. (2021). Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation. Foods, 10(10), 2264. https://doi.org/10.3390/foods10102264







